Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Isolation and characterization of a bacterial strain Hydrogenophaga sp. PYR1 for anaerobic pyrene and benzo[a]pyrene biodegradation†
Zaisheng Yan *a,
Yu Zhangab,
Huifang Wu*b,
Mingzhong Yangab,
Haichen Zhangab,
Zheng Haoac and
Helong Jianga
*a,
Yu Zhangab,
Huifang Wu*b,
Mingzhong Yangab,
Haichen Zhangab,
Zheng Haoac and
Helong Jianga
aState Key Laboratory of Lake Science and Environment, Nanjing Institute of Geography and Limnology, Chinese Academy of Sciences, Nanjing, 210008, China. E-mail: zshyan@niglas.ac.cn; Fax: +86-25-8688 2210; Tel: +86-25-8688 2210
bCollege of Environment, Nanjing University of Technology, Nanjing, 211816, China. E-mail: whfkhl@sina.com
cUniversity of Chinese Academy of Sciences, Beijing, China
First published on 3rd October 2017
Abstract
A pyrene-degrading strain Hydrogenophaga sp. PYR1 was isolated from PAH-contaminated river sediments and found to be able to degrade high molecular weight-polycyclic aromatic hydrocarbons under both aerobic and anaerobic conditions. The strain was Gram-negative, rod-shaped, pale yellow and motile flagellum with rounded ends. Under aerobic conditions, 94% pyrene could be degraded by the strain PYR1 within 15 d at 28 °C, while the degradation of benzo[a]pyrene was not obvious. However, this strain could significantly degrade pyrene and even benzo[a]pyrene under iron-reducing anaerobic conditions. Through Fourier transform infrared spectroscopy and amino acids analysis, it was found that ferric citrate as the sole electron acceptor stimulated lipopeptide biosurfactant production by the strain during PAH degradation. The presence of lipopeptide biosurfactant facilitated benzo[a]pyrene degradation possibly through improving the bioavailability of benzo[a]pyrene. Furthermore, six intermediates formed under iron-reducing conditions were identified, and then a pathway for anaerobic benzo[a]pyrene degradation by this strain was proposed. It seemed that this bacterial isolate has great potential in the bioremediation of PAH-contaminated sediments.
1. Introduction
Polycyclic aromatic hydrocarbon (PAH) pollution levels in river sediments have become increasingly serious,1,2 and have negative impacts on the aquatic ecosystems and pose a threat to human health due to their toxicity, mutagenicity, and carcinogenicity.3,4 In aquatic environments, high molecular weight (HMW) PAHs, such as pyrene or benzo[a]pyrene (BaP), are considered to be recalcitrant and persist in the environment due to their strong hydrophobicity.5,6 Microbial degradation and transformation is a major process for HMW-PAH removal in sediments.3,7,8River sediments can be oxic under aerated water flow, but PAH-contaminated sediments usually become anoxic.9 Oxic and anoxic conditions can be a major driver of ecological niche differentiation of bacteria involved in PAH biodegradation in sediments.9,10 Microorganisms can degrade PAHs in sediments with O2, nitrate, sulfate or ferric iron as electron acceptors, even methanogenic cultures.11 And while biodegradation mechanisms of PAHs are well known under aerobic conditions, little is known about the anaerobic bacterial metabolism of pyrene or benzo[a]pyrene. To date, no responsible microorganism has been isolated in pure culture under anaerobic condition. Additionally, the rate and extent of pyrene and benzo[a]pyrene degradation is limited due to the low bioavailability.12,13 Thus, towards understanding the mechanisms by which pyrene or benzo[a]pyrene-degrading microorganisms facing the problems resulted from these poorly available substrates are very necessary.
Biosurfactants produced by bacteria is deemed to be a valid microbial strategy that enhances the bioavailability of HMW-PAHs by solubilizing PAHs, lowing the surface or using as co-substrate.14–18 Many types of biosurfactants have been produced from bacteria belonging to a wide variety of genera, such as Pseudomonas, Bacillus, Acinetobacter, Mycobacterium, Pseudozyma, Paenibacillus, Streptomyces, and Achromobacter.14,16,19,20 Biosurfactants production is related to different environmental conditions, it has been shown that addition of soluble forms of iron can improve the yield of biosurfactant by bacteria.21–23 Until now, it was not clear whether biosurfactants were produced during anaerobic PAH biodegradation, and if so, how biosurfactants were involved in anaerobic PAH degradation.
The objectives of this study are to investigate the process of anaerobic pyrene and benzo[a]pyrene biodegradation through isolating the HMW-PAHs degradation bacterial strains, and to propose one pathway for anaerobic degradation of HMW-PAHs. For this purpose, the PAH-contaminated sediment was collected from estuarine of one freshwater lake, and one Hydrogenophaga sp. PYR1 strain with capabilities of anaerobic pyrene and benzo[a]pyrene degradation as well as biosurfactant production was successfully isolated. While Hydrogenophaga species are known to possess ability to degrade organic pollutants such as methyl tert-butyl ether24 and benzene,25 little is known about the potential of Hydrogenophaga for biosurfactant production and benzo[a]pyrene biodegradation. Therefore, the structural characterization of biosurfactant produced by the strain and involvement of HMW-PAHs degradation under anaerobic condition were evaluated. This study would help develop practical strategies for in situ bioremediation of contaminated sediments.
2. Material and methods
2.1. Sample source
Sediment samples were collected from the PAH-contaminated river sediments in the east area (31°04′ N, 120°24′ E) of Taihu Lake (a large shallow lake in China). The principal properties and concentration of PAHs in the sediments were summarized in Table 1. A high concentration of PAHs especially the HMW-PAHs were detected in the sediments, indicating a heavy pollution in the estuarine zone according to sediment quality guidelines for freshwater ecosystems.26| Moisture content (%) | 40.2 ± 1.9 |
| pH | 8.6 ± 0.7 |
| Total-PAHs (mg kg−1) | 33.16 ± 2.22 |
| 2-3ring-PAHs (mg kg−1) | 9.23 ± 0.47 |
| 4-6ring-PAHs (mg kg−1) | 23.94 ± 1.75 |
| Total nitrogen (g kg−1) | 1.88 ± 0.10 |
| Total phosphorous (g kg−1) | 1.12 ± 0.09 |
| Total organic carbon (g kg−1) | 17.7 ± 0.42 |
| Phosphate (mg kg−1) | 0.58 ± 0.02 |
| Ferric iron (g kg−1) | 1.02 ± 0.14 |
| Sulphate (g kg−1) | 0.87 ± 0.01 |
| Nitrate (mg kg−1) | 0.08 ± 0.11 |
2.2. Chemicals and media
Pyrene and benzo[a]pyrene (98% purity, Alfa Acsar Co., UK) were purchased. Stock solutions of pyrene or benzo[a]pyrene were 1000 mg L−1 in acetonitrile. All other solvents and chemicals used were of reagent grade.Mineral salt medium (MSM) with the following composition was used as a growth medium:5 18 mM (NH4)2SO4; 1 μM FeSO4·7H2O; 100 μM CaCl2·2H2O; 1 mM MgSO4·7H2O; 1 mM FeCl3·6H2O; 0.85 mM NaCl; 7 μM MnCl2·4H2O and 5 μM Na2MoO4·2H2O in 10 mM Na2HPO4–KH2PO4 buffer, pH 7.0. For preparation of MSM agar plates, 1.5% agar was added.
2.3. Enrichment and isolation of pyrene-degrading bacteria
For enrichment of pyrene-degrading bacteria from the river sediment samples, about 1 g wet weight of sediment was added to 100 mL of PYR-MSM (pyrene, 100 mg L−1), and the mixture was incubated at 28 °C on a rotary shaker at 120 rpm for 15 d. After confirmation of bacterial growth, 1 mL of culture was transferred to fresh PYR-MSM (100 mg L−1) and incubated for 15 d. After a third transfer, enrichment cultures were serially diluted. Then, the MSM agar plates were sprayed with acetonitrile containing pyrene (100 mg L−1). The acetonitrile was evaporated immediately and a white spot was left on the plate. An aliquot of 10 μL diluted culture was dropped onto the spot and incubated at 28 °C. The spot was marked as positive if the colony was surrounded by a clear zone.5,6 Then, colonies were picked with an inoculating needle. After repeated restreaking, pure cultures were obtained for further study. The morphology of the strain Hydrogenophaga sp. PYR1 growing on pyrene was observed by a transmission electron microscope (TEM) (JEM 1400-PLUS, Japan) according to the manufacturer' instructions. The isolated bacterial strain was selected for further experimentation and its ability to degrade PAHs under both aerobic and anaerobic conditions.2.4. 16S rRNA gene sequencing and phylogenetic analysis
DNA of the isolate was extracted, and 16S rDNA was amplified by PCR using the forward primer Eubac27F and reverse primer Universal 1492R10 and sequenced by BGI Biological Technology Co., Ltd. (Shanghai, China). Sequence similarity was analyzed in the GenBank nucleotide sequence databases by the BLAST program. A neighbor-joining (NJ) tree was constructed using the MEGA program (version 5.0).7,10The 16S rRNA gene nucleotide sequence obtained in this study has been deposited in GenBank under accession number KY283827.
2.5. Cell growth properties
Effects of different pH (4.0, 6.0, 7.0, 8.0, 9.0 and 11.0) and temperatures (4 °C, 15 °C, 28 °C and 35 °C) on the growth of strain Hydrogenophaga PYR1 were investigated under aerobic and anaerobic conditions. 1 mL pyrene stock solution (1000 mg L−1) in acetonitrile was transferred into the sterilized flasks. After the acetonitrile evaporated completely, 50 mL MSM with adjusted pH (4.0–11.0) was added into each sterilized flask. Then each flask was inoculated with 1 mL of aliquot taken from pyrene-induced cell enrichment culture of Hydrogenophaga PYR1. The flasks were incubated in darkness at 120 rpm at 28 °C. To investigate the optimum cell growth temperature (4 °C, 15 °C, 28 °C and 35 °C), the experiments were conducted at pH 7.0 in darkness at 120 rpm. Cell growth was determined by measuring optical density (OD600) by spectrophotometer (UV-752, Shanghai, China) at the end of experiments. After 7 day aerobic incubation or 35 day anaerobic incubation, pyrene concentrations were also determined in each vial. The anaerobic incubation was used ferric citrate (50 mM) as the electron acceptor.The substrate utilization ability of strain PYR1 was investigated by batch experiments using the medium, with various combinations of electron donors and acceptors. The strain PYR1 was preincubated in the PYR-MSM under oxic conditions for 5 d with shaking, harvested by centrifugation (9600×g, 4 °C, 10 min) and washed twice with phosphate-buffered saline. The obtained biomass was inoculated in the flasks, and incubated at 28 °C for 5 d aerobic incubation. OD600 was measured by a spectrophotometer (UV-752, Shanghai, China). For anaerobic incubation, the composition of medium was prepared according to a previous study:7 0.27 g L−1 K2HPO4; 0.35 g L−1 KH2PO4; 2.7 g L−1 NH4Cl; 0.1 g L−1 MgCl2·6H2O; 0.1 g L−1 CaCl2·2H2O; and 0.02 g L−1 FeCl2·4H2O. The obtained biomass was inoculated in 100 mL serum bottles containing 50 mL of medium in an anaerobic chamber filled with ultra-high purity nitrogen, and incubated at 28 °C for 35 day anaerobic incubation. In addition, sodium nitrate (20 mM), sodium sulfate (20 mM), ferric citrate (50 mM), sodium hydrogen carbonate (20 mM) or oxygen was used as an electron acceptor. Pyrene (20 mg L−1), benzo[a]pyrene (5 mg L−1), yeast extract (100 mg L−1), glucose (10 mM) and acetate (10 mM) was used as an electron donor, respectively.
2.6. Pyrene and benzo[a]pyrene degradation tests
For pyrene and benzo[a]pyrene degradation tests, the strain PYR1 with a cell density OD600 of 0.02 was inoculated into 30 mL vials containing 10 mL of PYR-MSM (20 mg L−1). The cells were also inoculated at an OD600 of 0.02 in 30 mL vials containing 10 mL of BaP-MSM (benzo[a]pyrene, 5 mg L−1). Culture was carried out at 28 °C and pH 9 under 120 rpm shaking. During experiments, the cell density (OD600) was monitored in each vial at 5, 10, 25, 50, 75, 100 and 125 h. Pyrene and benzo[a]pyrene concentrations were also monitored in each vial at hours 48, 120, 168, 288 and 360. Pyrene and benzo[a]pyrene degradation by the strain PYR1 under anaerobic conditions was also performed with different electron acceptors according to the previous study.7 Four different anaerobic redox conditions (nitrate-reducing, iron-reducing, sulfate-reducing, and methanogenic) were maintained through adding four different electron acceptors (20 mM sodium nitrate, 50 mM ferric citrate, 20 mM sodium sulfate, and 20 mM sodium hydrogen carbonate) to serum bottles in an anaerobic chamber filled with ultra-high purity nitrogen respectively. After capped with butyl rubber stoppers, serum bottles were incubated in the dark without shaking for anaerobic treatments. After 35 day anaerobic incubation, pyrene and benzo[a]pyrene concentrations in vials were analyzed.Also, an anaerobic iron-reducing culture was enriched with pyrene and benzo[a]pyrene as the sole carbon and energy source and ferric citrate as the electron acceptor. After 90 day anaerobic incubation, pyrene and benzo[a]pyrene concentrations and ferrous iron concentrations were determined in each vial.
2.7. Biosurfactant detection
Biosurfactant production by the isolated strain PYR1 under anaerobic conditions with different electron acceptors was detected by emulsification index (E24). E24 was measured according to the previous method reported with a slight modification.27 Briefly, a 5 mL anaerobic incubation broth was mixed with an equal volume of kerosene, and vortexed for 5 min followed by anaerobic incubation at 28 °C for 24 h. The E24 is given as percentage of height of emulsified layer (mm) divided by total height of the liquid column.The chemical structure and components of the biosurfactant sample from the iron-reducing treatment were determined using Fourier transform infrared (FTIR) spectroscopy (Model Nicolet380, USA). The infrared scan was performed over 400–4000 cm−1 wave number ranges.
The amino acid in the biosurfactant sample from the iron-reducing treatment was analyzed by o-phthalaldehyde (OPA) pre-column derivatization and HPLC (Agilent 1100, USA) analysis according to a previous study.6 The biosurfactant sample was concentrated by freeze drying and re-dissolved in 0.5 mL deionized water for analysis.
2.8. Analysis of benzo[a]pyrene metabolites and PAH concentrations
Bacterial culture (10 mL) from the iron-reducing enrichment culture on day 90 was sampled and separated from the medium by centrifugation at 9600×g at 4 °C for 10 min. The metabolites in the supernatant were extracted and analysed according to previous studies.5,28 Supernatant was extracted three times through liquid–liquid extraction using 1 mL of dichloromethane, and then organic phases were dehydrated through Na2SO4 anhydride, and followed by evaporation under N2 stream and dissolved with 1 mL acetonitrile for metabolites analysis.5 The metabolites in the supernatant were analysed by gas chromatograph (GC)-mass spectrometer (MS) according to a previous study with a slight modification.28 The gas chromatograph (Agilent, model 7890) equipped with mass spectrometer (model 5975) and a DB-5MS fused silica capillary column was used to analyze metabolites. The column temperature of gas chromatograph was as follows: 60 °C for 1 min, increased to 310 °C at 6 °C min−1, and then maintained at 310 °C for 5 min. The spectral of metabolites matching scores obtained using the Agilent library was cross-validated by the NIST 08 MS spectral library. Metabolites were also confirmed with standard compounds through high-performance liquid chromatography (Agilent 1200, USA).PAHs in sediment and water samples were extracted and analysed according to the procedure described elsewhere.5–7 In brief, 5 g of each sediment sample combined with equal volume quartz sand and 5 g of copper powder were extracted with accelerated solvent extraction (ASE, Dionex ASE 300) with dichloromethane solvent. The extracts were completely condensed to approximately 2 mL, followed by Florisil solid-phase extraction column for clean-up, and concentrated to 1 mL for analysis. To analyze PAH in water samples, 1 mL of water samples were extracted with 1 mL of methanol for 90 min. The extraction liquid was filtrated through 0.22 μm filter membranes, and then analyzed. PAHs were determined by a high-performance liquid chromatograph (HPLC) (Agilent 1200, USA) equipped with a diode array detector (DAD) and a fluorescence detector (FLD). The detailed conditions of the instrument can be found in previous studies.5–8 Recoveries rates of PAH (75.28–103.63%) were determined by adding the known concentration of standards solutions containing 16 priority PAH compounds.
3. Results and discussion
3.1. Isolation and identification of pyrene-degrading bacteria
After repeating domestication, the microbial community capable of degrading the pyrene was continuously enriched. After enrichment, one bacterial strain was isolated from sediment using plate screening techniques. The strain designated as PYR1, was Gram-negative, rod-shaped, pale yellow and motile flagellum with rounded ends (Fig. 1A). The phylogenetic analysis showed that the strain belonged to the genus Hydrogenophaga (Fig. 1B). Similar calculations after neighbour-joining analysis indicated that PYR1 was on the same phylogenetic branch. The closest relatives of PYR1 were H. atypica strain M10 with a 16S rRNA sequence similarity of 99%.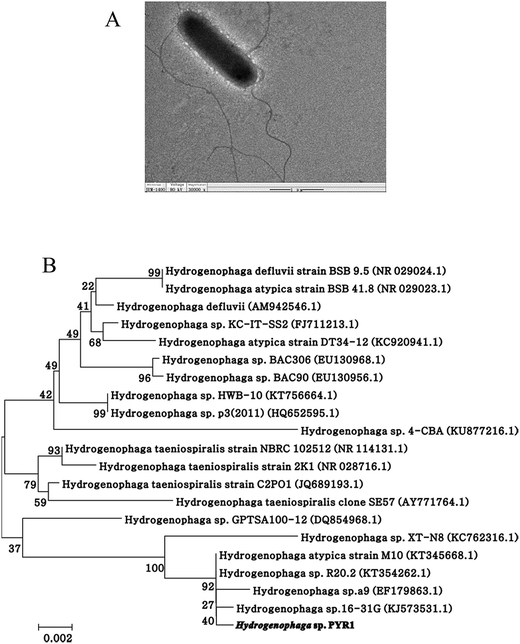 | ||
| Fig. 1 Photographs of cells and colonies of the isolate Hydrogenophaga sp. PYR1 (A) and neighbor-joining phylogenetic tree (B) based on 16S rDNA sequences of the strain Hydrogenophaga sp. PYR1. | ||
3.2. Growth characterization
The strain Hydrogenophaga sp. PYR1 was isolated from anaerobic sedimentary environment, and was found to be able to grow in aerobic liquid medium. Effects of pH and temperature on the growth of the strain under both aerobic and anaerobic conditions were shown in Fig. 2. With using pyrene as substrate, the optimum condition was about 28–35 °C at pH 8.0–11.0 for aerobic growth (Fig. 2A and B), and from 28 to 35 °C at pH 9.0–11.0 for anaerobic growth (Fig. 2C and D). As a result, temperature and initial pH value of medium for both aerobic and anaerobic growths were set as 28 °C and 9.0, respectively. This indigenous alkaliphilic degrader seemed to be suitable for bioremediation of petroleum hydrocarbons contaminated alkaline sediment as observed the genus Hydrogenophaga Rs71 for benzene biodegradation at pH 8.0 in a previous study.25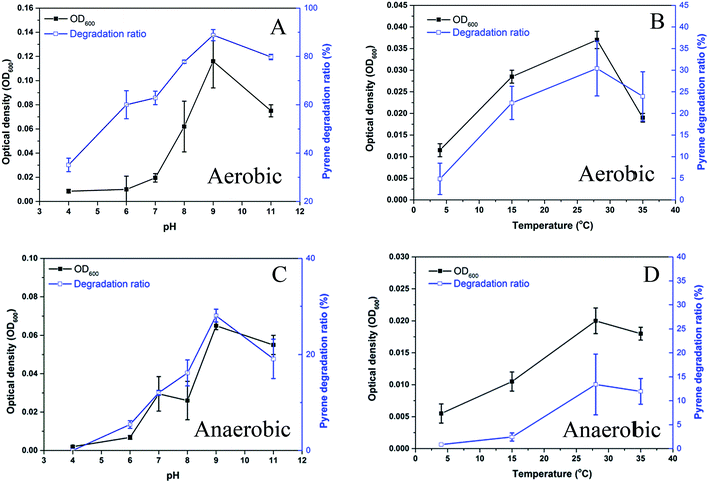 | ||
| Fig. 2 Effect of pH and temperature on growth of the strain Hydrogenophaga sp. PYR1 under aerobic (A, B) and anaerobic (C, D) conditions. | ||
The substrate utilization ability of the strain PYR1 was shown in Table 2. Under aerobic condition, this strain could use pyrene, benzo[a]pyrene, glucose and yeast extract as the electron donors except acetate based on the cell numbers (OD600). Different from aerobic condition, pyrene, benzo[a]pyrene and yeast extract as the electron donor could be utilized by the strain under iron-reducing conditions, only yeast extract as the sole electron donor for nitrate reduction. However, no growth of this strain was observed under sulfate reduction and methanogenic conditions with any electron donors and this strain could not use acetate as the electron donor with any electron acceptors tested.
| Electron acceptors | Electron donors | ||||
|---|---|---|---|---|---|
| Pyrene | Benzo[a]pyrene | Glucose | Acetate | Yeast extract | |
| a Abbreviation: OD, optical density. Growth was determined as OD600 after 5 d aerobic incubation or 35 day anaerobic incubation, −, OD600 < 0.01; +, OD600 > 0.01; ++, OD600 > 0.10; +++, OD600 > 0.30. | |||||
| Oxygen | ++ | ++ | + | − | +++ |
| Ferric citrate | + | + | − | − | + |
| Sodium nitrate | − | − | − | − | ++ |
| Sodium sulfate | − | − | − | − | − |
| Sodium hydrogen carbonate | − | − | − | − | − |
Members of the genus Hydrogenophaga are known to be facultative chemo-organotrophic or chemolithoautotrophic bacteria that can oxidize hydrogen but not acetate.29,30 However, it was reported that H. intermedia and H. atypica are unable to grow chemolithoautotrophically using hydrogen gas, and only a few organic compounds can be used as sole carbon sources.31 To date, the various strains of Hydrogenophaga species have been identified,31,32 and have the ability to degrade various organic pollutants, i.e. methyl tert-butyl ether,24 benzene,25 biphenyl,33 and co-metabolize polychlorinated biphenyls,33 but this genus has not been reported to be able to degrade HMW-PAHs previously.
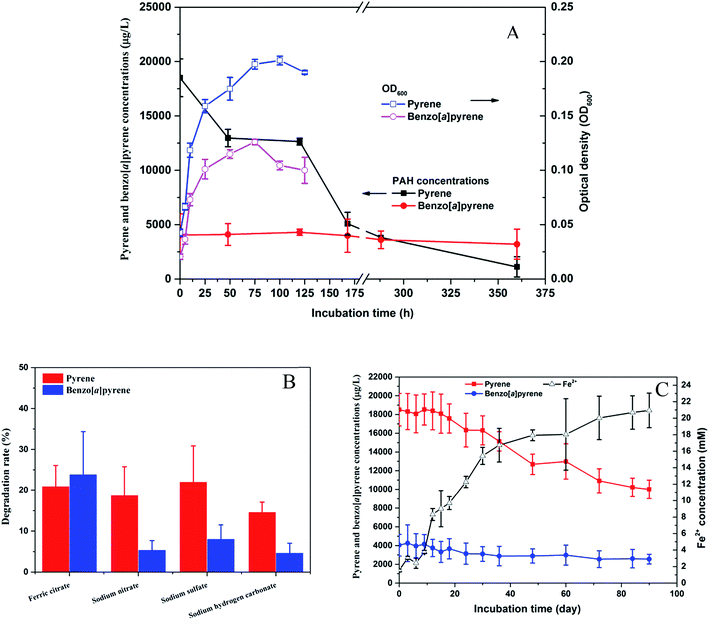
3.3. Degradation of pyrene and benzo[a]pyrene
The strain PYR1 was selected for further degradation studies because it exhibited the pyrene and benzo[a]pyrene degrading abilities under aerobic and anaerobic conditions. The isolated strain could degrade 94% pyrene within 15 d under aerobic condition with concomitant cell growth, while benzo[a]pyrene degradation was not obvious under aerobic condition (Fig. 3A). However, significant (P < 0.05) degradation of benzo[a]pyrene were observed under iron-reducing condition compared to the nitrate reduction, sulfate reduction and methanogenic conditions after 35 day anaerobic incubation (Fig. 3B). No differences in pyrene and benzo[a]pyrene removal were observed when the results for nitrate reduction, sulfate reduction and methanogenic conditions. To examine further the potential role of the strain in the biodegradation of pyrene and benzo[a]pyrene coupled to microbial iron reduction, the oxidation of pyrene and benzo[a]pyrene in the ferric iron-reducing enrichment culture was investigated. The accumulation of Fe2+ concentration (Fig. 3C) during the 90 day anaerobic oxidation of pyrene and benzo[a]pyrene further suggested that ferric iron was a potential electron acceptor.These results imply that the strain PYR1 was able to use oxygen or ferric citrate as the terminal electron acceptor for HMW-PAH degradation. The pyrene-degrading Hydrogenophaga PYR1 strain was able to degrade benzo[a]pyrene under the iron-reducing condition, indicating iron might play an important role in benzo[a]pyrene degradation. As shown in Table 1, ferric iron is an important and highly abundant electron acceptor in anoxic sediments. It seems likely that microbial oxidation of pyrene and benzo[a]pyrene by the strain, coupled to iron reduction, and may be capable of removing HMW-PAHs. The addition of ferric iron not only enhanced the biodegradation of phenanthrene and pyrene in freshwater sediments,7 but microbially mediated the anaerobic benzene degradation during the reduction of ferric iron.34,35 Moreover, iron as a component of the mono- or dioxygenase enzymes, PAHs degradation occurs by successive oxidations catalyzed by these enzymes that require iron as cofactor.22
3.4. Characterization of biosurfactant produced by the strain PYR1
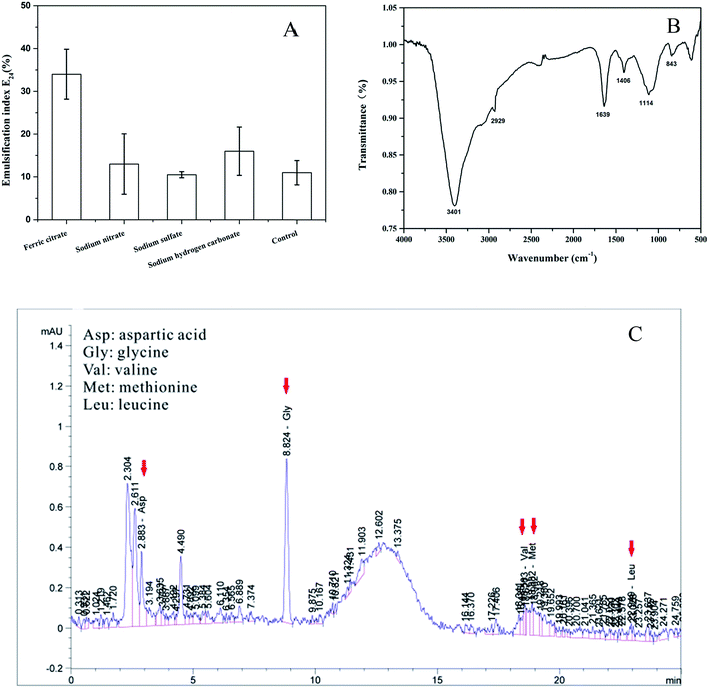
The iron can also stimulate the biosurfactant production by PAHs-degrading bacteria.21,22 In this study, biologically produced biosurfactant- and bioemulsifier-like compounds were detected under anaerobic conditions. Significantly (P < 0.05) highest emulsification index E24 was observed in the ferric citrate treatment which had 34% activity (Fig. 4A). However, the E24 did not show significant (P > 0.05) increase among the nitrate reduction, sulfate reduction and methanogenic conditions compared to the control treatment (Fig. 4A). These results indicated that ferric iron greatly stimulated the biosurfactant production by the strain PYR1 under anaerobic conditions.To further study the structure properties of the biosurfactant in the ferric citrate treatment, the FTIR scans and amino acid composition were analyzed as presented in Fig. 4B and C. The transmittance peaks was around 3401 cm−1, corresponding to O–H stretching and associated hydrogen bonds.7 The peaks at 2929 cm−1 and 1406 cm−1 were ascribed to aliphatic C–H stretching and aliphatic C–H deformation vibrations, and the absorption band at 1639 cm−1 for amide C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond stretching.19,36,37 In addition, peaks range 1114–843 cm−1 indicated the presence of amine groups.27 These transmittance peaks of the biosurfactant were similar to that of lipopeptide biosurfactants in the previous studies by the FTIR scans analysis.19,36,37
O bond stretching.19,36,37 In addition, peaks range 1114–843 cm−1 indicated the presence of amine groups.27 These transmittance peaks of the biosurfactant were similar to that of lipopeptide biosurfactants in the previous studies by the FTIR scans analysis.19,36,37
The amino acid composition of biosurfactants as shown in the Fig. 4C indicated that there were aspartic acid (Asp), glycine (Gly), valine (Val), methionine (Met) and leucine (Leu) in the biosurfactant. It was previously reported that leucine (Leu) and aspartic acid (Asp) were very important in lipopeptides.36
Although most biosurfactant-producing organisms are aerobic, a few examples of anaerobic producers exist as observed in the previous studies. Bacillus licheniformis JF-2 anaerobically produced a biosurfactant for in situ soil decontamination.38 Rhamnolipid production by Pseudomonas aeruginosa was also observed under denitrification condition.39 Pseudomonas strains can produce many types of biosurfactant including lipopeptide under anaerobic conditions, which have great potential for PAHs removal and decrease benzo[a]pyrene toxicity to bacteria.12,27,36,37 However, few literatures have reported that Hydrogenophaga strain could produce biosurfactant under iron-reducing condition albeit unknown soluble microbial product could be excreted by Hydrogenophaga strain AR20 with electrode as the electron acceptor.29
3.5. Degradation mechanisms of benzo[a]pyrene by strain PYR1 and the role of biosurfactant under iron-reducing condition
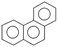
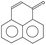
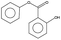
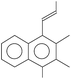
Aerobic degradation of HMW-PAHs has been extensively studied and is well understood, whereas anaerobic degradation using alternative electron acceptors is much less reported.11 Here, the degradation pathway of benzo[a]pyrene by strain PYR1 under ferric iron reduction conditions was investigated. Table 3 and Fig. S1† showed the identified mass/charge ratios with the inferred chemical formula. Six probable metabolites were identified from the GC-MS data. According to the detected mass/charge ratios and chemical standards, a proposed biodegradation pathway of benzo[a]pyrene is shown in Fig. 5. Benzo[a]pyrene became more bioavailable to the strain PYR1 due to lipopeptide biosurfactant solubilization40 under iron-reducing condition. The benzene ring of benzo[a]pyrene was broken first and degraded by carboxylation or methylation as a possible initial activation step,34 and 5-ethylchrysene was generated. The 5-ethylchrysene could be further transformed into pyrene, which could be further transformed into 1H-phenalen-1-one and phenanthrene. Because phenanthrene is relatively easily biodegraded, phenanthrene could be transformed into benzoic acid, 2-hydroxy-phenyl ester and naphthalene,1,2,3-trimethyl-4-propenyl. In addition, intermediate metabolites such as pyrene and phenanthrene were also confirmed with standard compounds.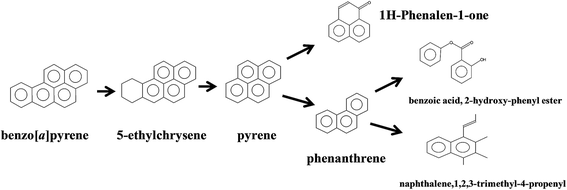 | ||
| Fig. 5 Proposed pathways for anaerobic degradation of benzo[a]pyrene by the strain Hydrogenophaga sp. PYR1 under iron-reducing condition. | ||
For aromatic compounds aerobic metabolism, the degradation pathways were through oxygenases introducing one or two oxygen atoms from O2 leading to a catechol-like structure.35 Under aerobic conditions, the strain PYR1 firstly used oxygen (O2) as the electron acceptor for PAH degradation. So, the ferric iron under aerobic condition might be as cofactor for microbial PAH degradation with slight emulsification index. Also, almost no obvious degradation of benzo[a]pyrene was observed under aerobic condition. However, for aromatic compounds anaerobic metabolism, the degradation pathways were through the benzoyl-CoA pathway.41
In this study, the detection of benzoic acid, 2-hydroxy-phenyl ester and naphthalene,1,2,3-trimethyl-4-propenyl as intermediate metabolites suggested that either carboxylation or methylation could be initial activation steps.34 In fact, benzoate was identified as an intermediate of benzene degradation under different anaerobic conditions.11,34,35,42,43 In addition, the presence of lipopeptides biosurfactant under iron-reducing condition might increase the apparent solubility and desorption rate of the benzo[a]pyrene to the aqueous phase,12,15,22,40,44 thus facilitating direct transfer of the biosurfactant-associated benzo[a]pyrene to the strain PYR1.
4. Conclusions
The strain Hydrogenophaga sp. PYR1 was isolated from river sediments and was able to degrade pyrene and benzo[a]pyrene anaerobically. The lipopeptide biosurfactant produced by the strain PYR1 under iron-reducing condition facilitated anaerobic degradation of pyrene and benzo[a]pyrene. The degradation pathway of benzo[a]pyrene by the strain PYR1 under ferric iron reduction conditions was proposed. Thus, this strain and its biosurfactant produced by ferric iron stimulation could have promising application in the bioremediation of PAH-contaminated sediments.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (41371456, 41671496, 51679228), Major Program of Natural Science Foundation of Jiangsu Province (BE2016703) and State Key Laboratory of Pollution Control and Resource Reuse Foundation (PCRRF13018).References
- Z. H. Zhao, L. Zhang and J. L. Wu, Limnol. Oceanogr., 2016, 61, 47–60 CrossRef CAS.
- Y. Liu, B. Beckingham, H. Ruegner, Z. Li, L. M. Ma, M. Schwientek, H. Xie, J. F. Zhao and P. Grathwohl, Environ. Sci. Technol., 2013, 47, 701–709 CrossRef CAS PubMed.
- A. L. Juhasz and R. Naidu, Int. Biodeterior. Biodegrad., 2000, 45, 57–88 CrossRef CAS.
- A. K. Haritash and C. P. Kaushik, J. Hazard. Mater., 2009, 169, 1–15 CrossRef CAS PubMed.
- Z. S. Yan, H. L. Jiang, X. H. Li and Y. Shi, J. Hazard. Mater., 2014, 272, 66–74 CrossRef CAS PubMed.
- Z. S. Yan, H. L. Jiang, H. Y. Cai, Y. L. Zhou and L. R. Krumholz, Sci. Rep., 2015, 5, 10709 CrossRef PubMed.
- Z. S. Yan, N. Song, H. Y. Cai, J. H. Tay and H. L. Jiang, J. Hazard. Mater., 2012, 199, 217–225 CrossRef PubMed.
- Z. S. Yan, Y. H. He, H. Y. Cai, J. D. Van Nostrand, Z. L. He, J. Z. Zhou, L. R. Krumholz and H. L. Jiang, Environ. Sci. Technol., 2017, 51, 8519–8529 CrossRef CAS PubMed.
- C. Quantin, E. J. Joner, J. M. Portal and J. Berthelin, Environ. Pollut., 2005, 134, 315–322 CrossRef CAS PubMed.
- N. Song, H. Y. Cai, Z. S. Yan and H. L. Jiang, Bioresour. Technol., 2013, 131, 281–287 CrossRef CAS PubMed.
- R. U. Meckenstock and H. Mouttaki, Curr. Opin. Biotechnol., 2011, 22, 406–414 CrossRef CAS PubMed.
- W. J. Xia, Z. F. Du, Q. F. Cui, H. Dong, F. Y. Wang, P. Q. He and Y. C. Tang, J. Hazard. Mater., 2014, 276, 489–498 CrossRef CAS PubMed.
- I. Ghosh and S. Mukherji, Int. Biodeterior. Biodegrad., 2016, 108, 67–75 CrossRef CAS.
- K. V. Sajna, R. K. Sukumaran, L. D. Gottumukkala and A. Pandey, Bioresour. Technol., 2015, 191, 133–139 CrossRef CAS PubMed.
- R. S. Makkar and K. J. Rockne, Environ. Toxicol. Chem., 2003, 22, 2280–2292 CrossRef CAS PubMed.
- F. A. Bezza and E. M. N. Chirwa, J. Hazard. Mater., 2017, 321, 218–227 CrossRef CAS PubMed.
- N. Fuchedzhieva, D. Karakashev and I. Angelidaki, J. Hazard. Mater., 2008, 153, 123–127 CrossRef CAS PubMed.
- W. J. Lin, S. S. Liu, L. Tong, Y. M. Zhang, J. Yang, W. T. Liu, C. L. Guo, Y. Y. Xie, G. N. Lu and Z. Dang, RSC Adv., 2017, 7, 24321–24330 RSC.
- M. C. Deng, J. Li, Y. H. Hong, X. M. Xu, W. X. Chen, J. P. Yuan, J. Peng, M. Yi and J. H. Wang, J. Appl. Microbiol., 2016, 120, 889–899 CrossRef CAS PubMed.
- C. Y. Li, T. T. Jia, M. Fu, N. Hou, H. M. Cao, Q. R. Wang and D. P. Li, RSC Adv., 2017, 7, 4339–4347 RSC.
- Y. H. Wei and I. M. Chu, Enzyme Microb. Technol., 1998, 22, 724–728 CrossRef CAS.
- E. C. Santos, R. J. S. Jacques, F. M. Bento, M. D. R. Peralba, P. A. Selbach, E. L. S. Sa and F. A. O. Camargo, Bioresour. Technol., 2008, 99, 2644–2649 CrossRef CAS PubMed.
- Y. H. Wei, L. F. Wang and J. S. Chang, Biotechnol. Prog., 2004, 20, 979–983 CrossRef CAS PubMed.
- S. H. Streger, S. Vainberg, H. L. Dong and P. B. Hatzinger, Appl. Environ. Microbiol., 2002, 68, 5571–5579 CrossRef CAS PubMed.
- A. Fahy, A. S. Ball, G. Lethbridge, K. N. Timmis and T. J. McGenity, Lett. Appl. Microbiol., 2008, 47, 60–66 CrossRef CAS PubMed.
- D. D. MacDonald, C. G. Ingersoll and T. A. Berger, Arch. Environ. Contam. Toxicol., 2000, 39, 20–31 CrossRef CAS PubMed.
- F. A. Bezza and E. M. N. Chirwa, Chemosphere, 2016, 144, 635–644 CrossRef CAS PubMed.
- Y. Zhong, T. G. Luan, H. W. Zhou, C. Y. Lan and N. F. Y. Tam, Environ. Toxicol. Chem., 2006, 25, 2853–2859 CrossRef CAS PubMed.
- Z. Kimura and S. Okabe, ISME J., 2013, 7, 1472–1482 CrossRef CAS PubMed.
- B. S. Chung, S. H. Ryu, M. Park, Y. Jeon, Y. R. Chung and C. O. Jeon, Int. J. Syst. Evol. Microbiol., 2007, 57, 1126–1130 CrossRef CAS PubMed.
- P. Kämpfer, R. Schulze, U. Jackel, K. A. Malik, R. Amann and S. Spring, Int. J. Syst. Evol. Microbiol., 2005, 55, 341–344 CrossRef PubMed.
- A. Willems, J. Busse, M. Goor, B. Pot, E. Falsen, E. Jantzen, B. Hoste, M. Gillis, K. Kersters, G. Auling and J. Deley, Int. J. Syst. Evol. Microbiol., 1989, 39, 319–333 CAS.
- A. J. Lambo and T. R. Patel, Curr. Microbiol., 2006, 53, 48–52 CrossRef CAS PubMed.
- U. Kunapuli, C. Griebler, H. R. Beller and R. U. Meckenstock, Environ. Microbiol., 2008, 10, 1703–1712 CrossRef CAS PubMed.
- N. Abu Laban, D. Selesi, T. Rattei, P. Tischler and R. U. Meckenstock, Environ. Microbiol., 2010, 12, 2783–2796 CAS.
- M. L. Ibrahim, U. J. J. Ijah, S. B. Manga, L. S. Bilbis and S. Umar, Int. Biodeterior. Biodegrad., 2013, 81, 28–34 CrossRef CAS.
- W. Ismail, I. S. Al-Rowaihi, A. A. Al-Humam, R. Y. Hamza, A. M. El Nayal and M. Bououdina, Int. Biodeterior. Biodegrad., 2013, 84, 168–178 CrossRef CAS.
- M. Javaheri, G. E. Jenneman, M. J. Mcinerney and R. M. Knapp, Appl. Environ. Microbiol., 1985, 50, 698–700 CAS.
- C. Chayabutra, J. Wu and L. K. Ju, Biotechnol. Bioeng., 2001, 72, 25–33 CrossRef CAS PubMed.
- R. A. Kanaly, S. Harayama and K. Watanabe, Appl. Environ. Microbiol., 2002, 68, 5826–5833 CrossRef CAS PubMed.
- M. Boll, G. Fuchs and J. Heider, Curr. Opin. Chem. Biol., 2002, 6, 604–611 CrossRef CAS PubMed.
- R. Chakraborty and J. D. Coates, Appl. Environ. Microbiol., 2005, 71, 5427–5432 CrossRef CAS PubMed.
- F. Luo, R. Gitiafroz, C. E. Devine, Y. C. Gong, L. A. Hug, L. Raskin and E. A. Edwards, Appl. Environ. Microbiol., 2014, 80, 4095–4107 CrossRef CAS PubMed.
- S. Boonchan, M. L. Britz and G. A. Stanley, Biotechnol. Bioeng., 1998, 59, 482–494 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09274a |
| This journal is © The Royal Society of Chemistry 2017 |