Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rapid label-free visual detection of KRAS mutations using peptide nucleic acid and unmodified gold nanoparticles†
Xihong Zhao ab and
Chii-Wann Lin*b
ab and
Chii-Wann Lin*b
aKey Laboratory for Green Chemical Process of Ministry of Education, Key Laboratory for Hubei Novel Reactor & Green Chemical Technology, Research Center for Environmental Ecology and Engineering, School of Chemical Engineering and Pharmacy, Wuhan Institute of Technology, Wuhan 430073, China
bInstitute of Biomedical Engineering, National Taiwan University, No. 1, Sec. 4, Roosevelt Road, Taipei 10617, Taiwan. E-mail: cwlinx@ntu.edu.tw; Fax: +886-2-33665268; Tel: +886-2-33665272
First published on 16th October 2017
Abstract
Colorectal cancer (CRC) is among the most commonly diagnosed cancers worldwide. Point mutations on the Kisten rat sarcoma viral oncogene homologue (KRAS) have been identified as a crucial diagnostic and prognostic biomarker for response to cancer therapy targeting the epidermal growth factor receptor. A rapid label-free colorimetric assay for detection of point mutations in codon 12 and 13 of the KRAS gene using peptide nucleic acid (PNA) and unmodified gold nanoparticles (AuNPs) was presented in this study. Charge neutral PNA induces the AuNPs aggregation in the absence of complementary DNA; when the complementary DNA is present, PNA–DNA hybridization retains the dispersion of AuNPs as the negative charges of the DNA strand adsorbed on the AuNPs surface ensure sufficient charge repulsion. Based on this principle, the developed assay was able to detect the specific sequence in the presence of at least 20 times the amount of interference oligonucleotides, and the limit of detection was low as DNA/PNA ratio of 0.02. The PNA induced colorimetric changes in the presence of the complementary DNA at different ratios of DNA/PNA yielded a standard curve with a linear coefficient of R2 = 0.9371. Furthermore, this assay was validated in order that it could be used to detect the seven known mutant alleles in codon 12 and 13 of the KRAS gene by the naked eye with higher concentration of PNA. This simple colorimetric assay is rapid, sensitive and easily performed at room temperature without additional stringent conditions, which promises the method great potential utility for clinical sample screening and point-of-care testing.
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females. According to WHO statistics, there were estimated over 1.2 million new CRC cases and 608![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 deaths as a result in 2008 worldwide.1 The Kisten rat sarcoma viral oncogene homologue (KRAS) gene is a known downstream signaling molecule in the epidermal growth factor receptor (EFGR) signalling pathway. It has been shown that patients exhibiting KRAS mutations do not benefit from anti-EGFR treatment.2 Mutations in KRAS are found in approximately 30% of adenocarcinoma, principally in codon 12 and 13, making it one of the most commonly mutated genes in cancer.3 As the monoclonal antibody inhibitors of EGFR treatment are toxic and expensive, KRAS mutation status detection in CRC patients has become a crucial diagnostic factor for clinical decisions and designs for future therapy.
700 deaths as a result in 2008 worldwide.1 The Kisten rat sarcoma viral oncogene homologue (KRAS) gene is a known downstream signaling molecule in the epidermal growth factor receptor (EFGR) signalling pathway. It has been shown that patients exhibiting KRAS mutations do not benefit from anti-EGFR treatment.2 Mutations in KRAS are found in approximately 30% of adenocarcinoma, principally in codon 12 and 13, making it one of the most commonly mutated genes in cancer.3 As the monoclonal antibody inhibitors of EGFR treatment are toxic and expensive, KRAS mutation status detection in CRC patients has become a crucial diagnostic factor for clinical decisions and designs for future therapy.
Traditional detection of gene mutations is usually by the direct sequencing method, which is still considered as the gold standard.4 However, the direct sequencing method is too complex, time-consuming, and expensive for routine pretherapeutic screening programs.5,6 Furthermore, the direct sequencing method usually requires a percentage of more than 50% tumour cells and the threshold of detection is around 25% of mutant DNA in the total genetic content.7,8 With the development of mutation-detection techniques over the last decade, many methods have been reported for detection of KRAS mutations. These techniques are based on molecular amplification-related,9–11 sequence-related,12 high resolution melting analysis,13 and other methodologies.14 Some techniques have been proven to be more accurate and sensitive than the previous direct sequencing method, but most of them are still time-consuming, complicated, of high cost and unsuitable for routine clinical screening and resource-limited settings. Motivated by these prospects, we investigated a simple, fast, low-cost and high sensitive method for KRAS mutation detection.
Colorimetric assays based on metallic nanoparticles have obtained significant attention due to the advantages of fast response, room temperature operation, simple performance, cost-effectiveness and high sensitivity. The controlled aggregation and disaggregation of gold nanoparticles (AuNPs) has been a subject of great interest over the past decade due to their simplicity and versatility in diagnostic applications. The AuNP-based colorimetric biosensor, which takes advantage of the colour change that arises from the interparticle plasmon coupling during AuNP aggregation (red to purple or blue) or redispersion of an AuNP aggregate (purple to red), has been developed for detection of a wide range of analytes.15
More recently, the mutation-sensitive hybridisation profile of peptide nucleic acids (PNAs) has been exploited to develop novel detection methods for single nucleotide polymorphisms.16,17 PNA is a synthetic DNA analog in which the phosphodiester backbone is replaced by a peptide-like repeat formed by (2-aminoethyl)-glycine units. Since PNA contains no charged phosphate groups, the binding between PNA/DNA has a higher thermal stability compared to complementary nucleic acids than DNA and RNA. Meanwhile, the PNA/DNA duplex is significantly destabilized in the presence of a single-base mismatch.18 The use of PNA-induced aggregation of gold nanoparticle has been described by Chakrabarti and Klibanov.19 The PNA–AuNPs conjugates were further used to develop a novel colorimetric DNA assay that detected the presence of single base imperfections within minutes due to colloidal stability upon DNA hybridization.20
In this study, we report a novel label-free colorimetric method for rapid detection of KRAS mutations using unmodified AuNPs induced aggregation by PNA.
2. Materials and methods
2.1 Reagents
All the chemicals used in the study were analytical grade without further purification. HAuCl4·3H2O (99.99%) was obtained from Alfa Aesar (MA, U.S.A.). Trisodium citrate dihydrate (99.9%) was obtained from Aldrich and NaBH4 from Fluka. Double distilled water (18.2 MΩ) was used in all solutions with a Milli-Q system (Millipore, Bedford, MA, USA), unless otherwise stated. The PNA oligomers were synthesized by solid phase synthesis from Panagene Corporation (Daejeon, South Korea). Oligonucleotides were synthesized by Purigo Biotech (Taipei, Taiwan) and were further purified by HPLC.The PNA probe (17 mer) and their target DNA of fully complementary sequence (DNAcomp), non-complementary sequence (DNAnc), and seven mutant alleles in codon 12 and 13 of the KRAS gene sequences (from m1 to m7) are shown in Table S1 (ESI†).
2.2 Preparation of AuNPs
The AuNPs were synthesized using a standard citrate method.21 Typically, a 100 mL aqueous solution of HAuCl4 (41 mg, 1.0 mM) was heated with continuous stirring until the solution started boiling, after which 10 mL of trisodium citrate (114 mg, 38.8 mM) solution was quickly added to the boiling solution. This solution was further heated for an additional 10 min and then cooled to room temperature with continuous stirring until a brick-red wine colour appeared. Finally, the resulting solution was filtered and stored at 4 °C until further used. The average diameter of AuNPs was approximately 13 nm, as determined with a particle size analyser; the corresponding absorption band was approximately 520 nm.2.3 Colorimetric assay procedure
PNA was allowed to hybridize to the target oligonucleotide (full complementary, non-complementary or single base mutations) in 10 mM phosphate buffer (pH 7.2) containing 0.1 mM EDTA and 50 mM NaCl. The AuNP solution was added to give a final volume of 100 μL, the colour and absorption spectrum were observed after 5 min at room temperature. To improve the colour discrimination and to detect and distinguish the single base mutations, higher salt concentrations were added to accelerate the aggregation, and the final concentration of 100 mM NaCl was determined to be appropriate. The colour changes of the solutions were recorded using a camera. The absorption spectra of test solutions were recorded using a Varian Cary 50 spectrometer with a wavelength range from 400 to 800 nm.3. Results and discussion
3.1 Detection of the presence of a specific DNA sequence
Typically, free PNA probes induce aggregation in unmodified metallic nanoparticles and lead to colour changes. However, PNA hybridization with the complementary oligonucleotide forms PNA–DNAcomp complex, which retains the stability of colloidal AuNP solutions, thereby preventing a change in colour. This concept is based on the fact that the aggregation of the AuNPs protected by citrate anions occurs immediately in the presence of charge neutral PNA and is retarded when a DNAcomp is present to form a PNA–DNAcomp complex, but not a DNAnc that forms PNA/DNAnc mixture22 (Scheme S1, ESI†). The concentration of PNA was optimized to induce the aggregation with a fixed amount of AuNPs. For observed visual changes, 1 μM PNA was found to be sufficient in a 100 μL AuNPs test solution. The 1 μM PNA concentration was used in later assays.As shown in Fig. S1 (ESI†), when PNA is added, the brick-red wine colour of the untreated AuNP solution (Fig. S1A†) changes to dark purple (Fig. S1A and B†), accompanied with a shift of the adsorption peak to 640 nm. With the distinct backbone properties of PNA (neutrality, high rigidity and peptide composition), PNA base monomers interact more strongly with AuNPs than the corresponding nucleobases.23 There is no charge repulsion between the charge neutral PNA and the citrate anions-protected AuNPs, therefore causing AuNPs aggregation. The degree of aggregation depends on the concentration and length of PNA. A shorter length of PNA aggregates the AuNPs more effectively than a longer one at any certain concentration.24
When the PNA–DNAcomp complex (Fig. S1A and C†) is added, the AuNPs solution retains the brick-red wine colour. This is because the adsorption of the PNA–DNAcomp complex on the AuNPs surface, with which the negative phosphate backbone of the DNA strands ensure sufficient charge repulsion, maintains the AuNPs dispersion.20 In contrast, when the solution contains a mixture of PNA and DNAnc (Fig. S1A and D†), the AuNPs solution changes to purple and increases adsorption at longer wavelengths. It is well known that salt induces aggregation of gold nanoparticles. In this study, 100 mM NaCl can accelerate the aggregation of the AuNPs solutions except with the DNAcomp (Fig. S1B and C′†), in contrast, the AuNPs solution containing DNAnc (Fig. S1B and D′†) changes to purple indicating that the AuNPs aggregation occurred obviously after NaCl was added. Therefore, NaCl is added to accelerate the AuNPs aggregation for improving the selectivity of single-base-mismatch discrimination.
3.2 Selectivity of the AuNPs-based assay
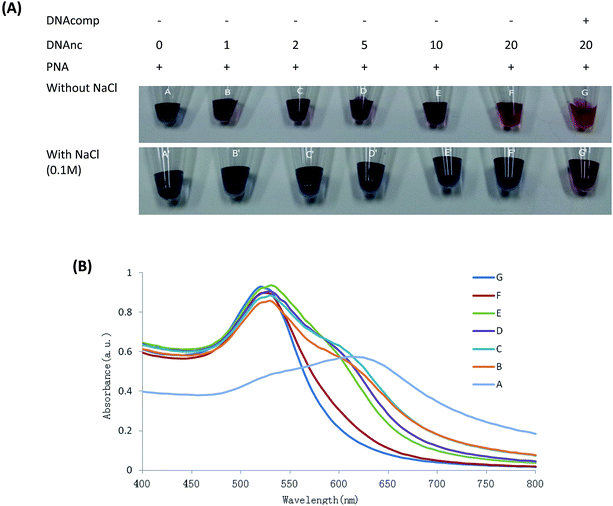
The colorimetric assay was used to detect the different concentrations of DNAnc (0, 1, 2, 5, 10 and 20 times the PNA concentration) in AuNPs solution aggregation by PNA probe with or without the complementary DNA.As shown in Fig. 1, the AuNPs aggregation induced by PNA is gradually reduced with an increase in concentration of DNAnc (Fig. 1(A), A–F), indicating that higher concentration of DNAnc would tend to protect AuNPs aggregation by competing with PNA to encompass the surface of AuNPs. However, the AuNPs aggregation could still be detected by UV spectrum measurement even at 20 times the concentration of DNAnc (Fig. 1(B), F), compared to the stable colloidal as the equivalent DNAcomp present (Fig. 1(B), G). This result indicated that the full match DNA could be detected in the present of at least of 20 times the concentration of interference oligonucleotides. The 0.1 M NaCl acceleration of the AuNPs aggregation with different concentrations of DNAnc (Fig. 1(A), A′–F′) were obviously observed, but not observed when the DNAcomp was also present in the solution (Fig. 1(A), G′). This result indicated that 0.1 M NaCl could enhance the selectivity of DNAcomp detection in high concentrations of interference oligonucleotides by the naked eye. The high selectivity of this colorimetric assay will be of benefit to detection against the complex background of clinical samples, which may include primers, nonspecific amplicons, and oligomer fragments, especially in cancer-related tissue treatment.25
3.3 Sensitivity of the AuNPs-based assay
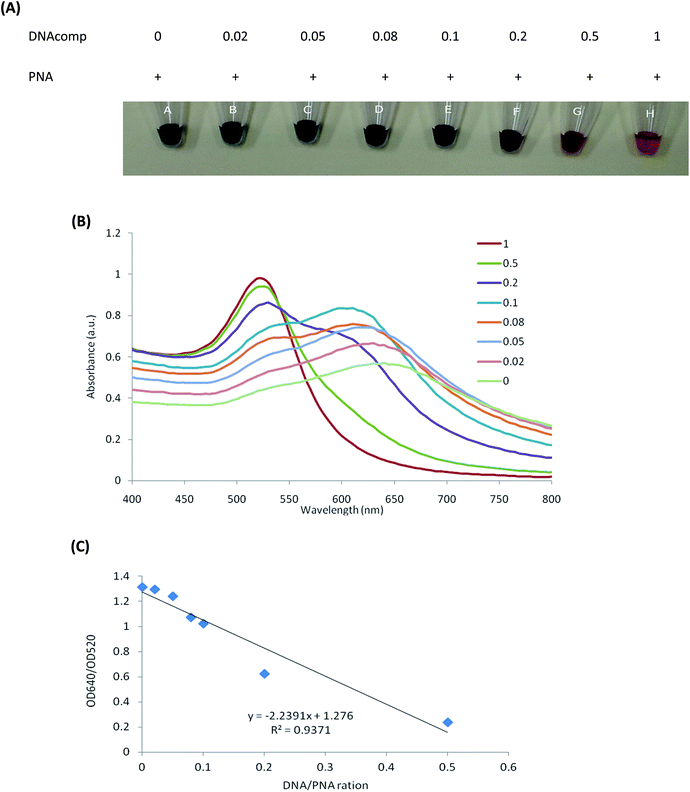
To estimate the sensitivity of this colorimetric assay for quantitative complementary DNA to the PNA probe, the DNAcomp was mixed with the fixed PNA concentration at ratios of 0, 0.02, 0.05, 0.08, 0.1, 0.2, 0.5 and 1.0 for hybridization prior to addition to the AuNPs solution. As shown in Fig. 2(A), the degree of AuNPs aggregation gradually increases with a decrease in the concentration of DNAcomp. The reason is that the less PNA–DNAcomp complex present, the less charge repulsion occurs due to the more charge neutral PNA probes, resulting in an increase in AuNPs aggregation. In the corresponding UV adsorption spectra of Fig. 2(B), a gradual decrease in the absorbance at about 520 nm is seen as the solution containing decreased amounts of PNA–DNAcomp complex indicates a gradual decrease in the popularity of the dispersed AuNPs. From the photographs, there is an obvious difference between untreated AuNPs solution at a DNA/PNA ratio of 0 (deep blue, Fig. 2(A), A) and the PNA–DNAcomp complex at a DNA/PNA ratio of 0.1 (deep purple, Fig. 2(A), E). However, according to the UV adsorption spectra, the absorption of the AuNPs solution at a DNA/PNA ratio of 0.02 (Fig. 2(A), B) is well distinguishable from that with PNA alone (Fig. 2(B)). The absorbance of AuNPs at the DNA/PNA ratio of 0.02 is shown by the second peak at about 640 nm, indicated by the popularity of AuNPs aggregation, which is still distinguishable from the AuNPs solution with PNA alone.In the present study, the limit of detection by the spectrum at a DNA/PNA ratio of 0.02 and by the naked eye at a ratio of 0.1 are lower than those previously published by Rho et al.26 (ratio 0.5), Li et al.27 (ratio 0.2) and Kanjanawarut et al.20 (ratio 0.05), who used unmodified AuNPs for quantification assays of ssDNA and dsDNA. As Kanjanawarut reported,20 single-base-mismatch discrimination was difficult for hybridization with longer probes. In addition to the higher affinity of PNA–DNA hybridization than DNA–DNA, the shorter length of PNA (17 mer) in this study also contributes to the lower complementary DNA detection limit.
In the presence of free PNA, the 520 nm peak of each solution experienced a concomitant decrease and was followed by the inception of a new peak at approximately 640 nm. These difference spectra of each solution were useful for choosing the wavelength peak to use as a ratio of absorbance values at 640–520 nm. This analogy can be applied to design a visual colorimetric assay for the quantification of the target.28 As shown in Fig. 2(C), a plot of these values against the different concentrations of the complementary oligonucleotides yielded high values of linear correlation (R2 = 0.9371), which could be used for quantification of complementary DNA to the PNA probe.
3.4 Colorimetric detection of seven KRAS mutant alleles by AuNPs-based assay using PNA-induced AuNPs aggregation
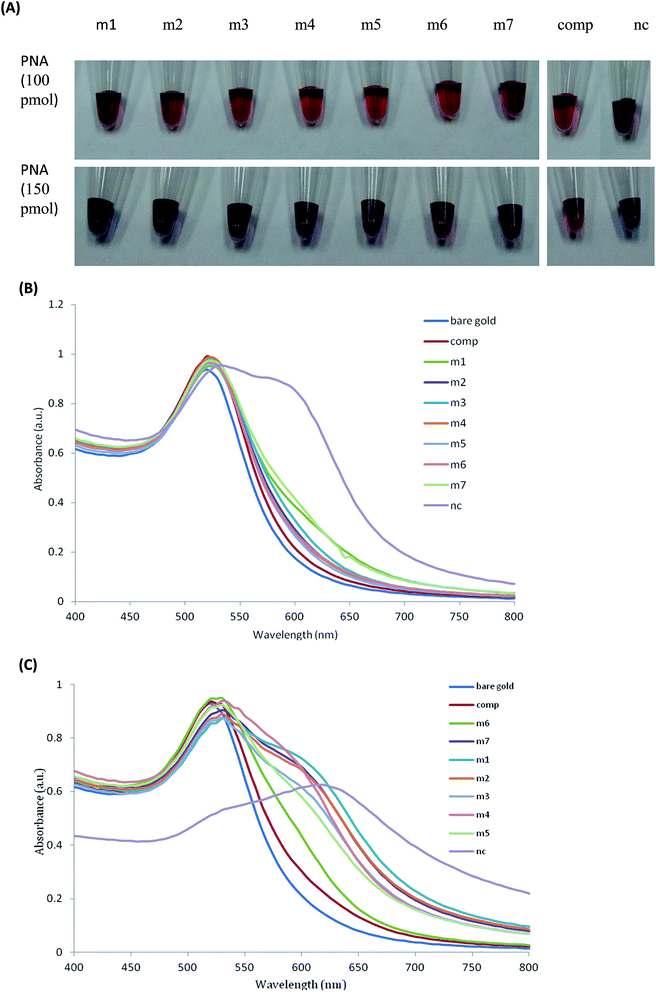
The PNA hybridization with its complementary DNAcomp can abolish PNA-induced AuNPs aggregation. When a single-base-mismatch occurs in the complementary DNA sequence, the hybridization efficiency will reduce and cause free PNA in solution to induce AuNPs aggregation.24This assay was engineered to detect the seven known mutant alleles of KRAS codon 12 and 13. As shown in Fig. 3(B), the absorption of the AuNPs containing PNA annealed with seven KRAS mutants increased at longer wavelengths around 640 nm because free PNA induced AuNPs aggregation increased the absorption at longer wavelengths. The seven KRAS mutants increased absorption ratio, A640, by 129.26% (m1), 37.19% (m2), 54.61% (m3), 35.43% (m4), 24.90% (m5), 40.99% (m6), 122.40% (m7), respectively, relative to those with complementary DNAcomp. The spectra shifts of these seven KRAS mutants were significant compared to the variation of A640 measurement for AuNPs (1.01%). The assay results demonstrated the seven known mutant alleles of KRAS codon 12 and 13 could be detected using UV spectrum by PNA-induced AuNPs aggregation.
However, it was not easy to distinguish the single-base-mismatch by the naked eye in the presence of 100 pmol PNA, as shown in Fig. 3(A). After optimization of the ratio of PNA/DNAcomp, it was found that seven known mutant alleles of KRAS codon 12 and 13 could be visibly detected by the naked eye in the presence of 150 pmol PNA in 100 μL. From Fig. 3(A), the AuNPs containing single-base-mismatch targets changed to a purple-blue colour and DNAcomp changed to brick-red wine colour with light purple. From Fig. 3(C), the absorption of seven KRAS mutants increased obviously at longer absorption wavelengths, which verified the colour changes. All the results were validated by triplicate experiments.
It is more difficult to perform detection of single-base-mismatch in longer sequences because the alternative role of one base mismatch to the affinity of hybridization is reduced.29 In this study, the length of 17 mer sequence was used to detect seven known mutant alleles of KRAS codon 12 and 13. Compared with the surface plasmon resonance29 and electrochemical sensors30 for single-base-mismatch discrimination, which rely on expensive equipment, stringent conditions and complex experimental processes, this assay with the optimization of the ratio of PNA/DNA can be performed easily by the naked eye at room temperature in resource-limit areas.
Gold nanoparticles with controlled geometrical, optical, and surface chemical properties are suitable for several biological and biomedical applications.31 With the development of synthetic chemistry, gold nanoparticles can be made with precise control over physicochemical and optical properties that are desired for specific biological and clinical applications. For example, when the size of the AuNPs was reduced to the Au nanoclusters (AuNCs, typically less than 2 nm) range through size control, AuNCs exhibited an enhanced and wide-spectrum antimicrobial activity, which would allow them to better interact with bacteria.32 AuNPs have been successfully employed in genomics and biosensorics, immunoassays and clinical chemistry, cancer radiotherapy, targeted delivery of drugs and antigens, and optical bioimaging of cells and tissues with state-of-the-art nanophotonic detection systems.33,34
4. Conclusion
In the present study, the unmodified AuNPs aggregation behaviour induced by a PNA probe was manipulated by its complementation with target oligonucleotides for detection of single-base-mismatch. This colorimetric assay was validated in order that it could be used to detect the seven known mutant alleles in codon 12 and 13 of the KRAS gene, which could provide clinical benefits for CRC patients to help predict the efficacy of anti-EGFR treatment. This simple colorimetric assay is robust and offered advantages in terms of rapidity, simplicity, direct visualization of result by the naked eye without additional stringent conditions and advanced instrumentation, and with great potential utility for clinical sample screening and point-of-care testing. However, it could be expected that this colorimetric assay could be further improved in terms of the sensitivity by introducing an enhancement step and clinical sample preparation, which might benefit the direct colorimetric assay of clinical genomic DNA samples without an amplification step, as also demonstrated by other authors on array hybridization.Conflicts of interest
The authors indicated no potential conflicts of interests.Acknowledgements
This work was supported by the Taiwan National Science Council (102-2218-E-002-014-MY3), National Natural Science Foundation of China (Grant No. 31501582) and the Center for Emerging Material and Advanced Devices of National Taiwan University.References
- A. Jemal, F. Bray, M. M. Center, J. Ferlay, E. Ward and D. Forman, Ca-Cancer J. Clin., 2011, 61, 69–90 CrossRef PubMed.
- C. Tan and X. Du, World J. Gastroenterol., 2012, 18, 5171–5180 CrossRef CAS PubMed.
- A. A. Adjei, J. Natl. Cancer Inst., 2001, 93, 1062–1074 CrossRef CAS PubMed.
- E. Massarelli, M. Varella-Garcia, X. Tang, A. C. Xavier, N. C. Ozburn, D. D. Liu, B. N. Bekele, R. S. Herbst and I. I. Wistuba, Clin. Cancer Res., 2007, 13, 2890–2896 CrossRef CAS PubMed.
- X. Zhao, J. Zhong, C. Wei, C. W. Lin and T. Ding, Front. Microbiol., 2017, 8, 580 Search PubMed.
- X. Zhao, C. W. Lin, J. Wang and D. H. Oh, J. Microbiol. Biotechnol., 2014, 24, 297–312 CrossRef CAS PubMed.
- W. Pao and M. Ladanyi, Clin. Cancer Res., 2007, 13, 4954–4955 CrossRef CAS PubMed.
- D. A. Eberhard, G. Giaccone and B. E. Johnson, Non-Small-Cell Lung Cancer Working, J. Clin. Oncol., 2008, 26, 983–994 CrossRef PubMed.
- K. Hoshi, H. Takakura, Y. Mitani, K. Tatsumi, N. Momiyama, Y. Ichikawa, S. Togo, T. Miyagi, Y. Kawai, Y. Kogo, T. Kikuchi, C. Kato, T. Arakawa, S. Uno, P. E. Cizdziel, A. Lezhava, N. Ogawa, Y. Hayashizaki and H. Shimada, Clin. Cancer Res., 2007, 13, 4974–4983 CrossRef CAS PubMed.
- S. Miyano, K. Hanazawa, T. Kitabatake, M. Fujisawa and K. Kojima, Exp. Ther. Med., 2012, 4, 790–794 CrossRef CAS PubMed.
- X. Zhao, C. C. Chang, T. L. Chuang and C. W. Lin, Biotechnol. Biotechnol. Equip., 2016, 30, 1155–1162 Search PubMed.
- S. Ogino, T. Kawasaki, M. Brahmandam, L. Yan, M. Cantor, C. Namgyal, M. Mino-Kenudson, G. Y. Lauwers, M. Loda and C. S. Fuchs, J. Mol. Diagn., 2005, 7, 413–421 CrossRef CAS PubMed.
- M. Krypuy, G. M. Newnham, D. M. Thomas, M. Conron and A. Dobrovic, BMC Cancer, 2006, 6, 295 CrossRef PubMed.
- M. Guha, E. Castellanos-Rizaldos, P. F. Liu, H. Mamon and G. M. Makrigiorgos, Nucleic Acids Res., 2013, 41, e50 CrossRef CAS PubMed.
- W. Zhao, M. A. Brook and Y. F. Li, ChemBioChem, 2008, 9, 2363–2371 CrossRef CAS PubMed.
- H. Lee, A. Kim, I. S. Ahn, S. W. Joo, S. Y. Lee, K. A. Yoon and K. Lee, Chem. Commun., 2011, 47, 11477–11479 RSC.
- J. Dabritz, J. Hanfler, R. Preston, J. Stieler and H. Oettle, Br. J. Cancer, 2005, 92, 405–412 CrossRef CAS PubMed.
- T. Ratilainen, A. Holmen, E. Tuite, G. Haaima, L. Christensen, P. E. Nielsen and B. Norden, Biochemistry, 1998, 37, 12331–12342 CrossRef CAS PubMed.
- R. Chakrabarti and A. M. Klibanov, J. Am. Chem. Soc., 2003, 125, 12531–12540 CrossRef CAS PubMed.
- R. Kanjanawarut and X. D. Su, Anal. Chem., 2009, 81, 6122–6129 CrossRef CAS PubMed.
- C. C. Chang, S. C. Wei, T. H. Wu, C. H. Lee and C. W. Lin, Biosens. Bioelectron., 2013, 42, 119–123 CrossRef CAS PubMed.
- Y. Y. Qi, L. Li and B. X. Li, Spectrochim. Acta, Part A, 2009, 74, 127–131 CrossRef PubMed.
- A. Gourishankar, S. Shukla, K. N. Ganesh and M. Sastry, J. Am. Chem. Soc., 2004, 126, 13186–13187 CrossRef CAS PubMed.
- X. D. Su and R. Kanjanawarut, ACS Nano, 2009, 3, 2751–2759 CrossRef CAS PubMed.
- K. Sato, K. Hosokawa and M. Maeda, Nucleic Acids Res., 2005, 33, e4 CrossRef PubMed.
- S. Rho, S. J. Kim, S. C. Lee, J. H. Chang, H. G. Kang and J. Choi, Curr. Appl. Phys., 2009, 9, 534–537 CrossRef.
- H. X. Li and L. J. Rothberg, J. Am. Chem. Soc., 2004, 126, 10958–10961 CrossRef CAS PubMed.
- V. G. Joshi, K. Chindera, A. K. Singh, A. P. Sahoo, V. D. Dighe, D. Thakuria, A. K. Tiwari and S. Kumar, Anal. Chim. Acta, 2013, 795, 1–7 CrossRef CAS PubMed.
- A. I. K. Lao, X. D. Su and K. M. M. Aung, Biosens. Bioelectron., 2009, 24, 1717–1722 CrossRef CAS PubMed.
- Q. Wang, L. J. Yang, X. H. Yang, K. M. Wang, L. L. He and J. Q. Zhu, Anal. Chim. Acta, 2011, 688, 163–167 CrossRef CAS PubMed.
- J. S. Martinez and J. Xie, APL Mater., 2017, 5, 053001 CrossRef.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, ACS Nano, 2017, 11, 6904–6910 CrossRef CAS PubMed.
- N. Goswami, Z. Luo, X. Yuan, D. T. Leong and J. Xie, Mater. Horiz., 2017, 4, 817–831 RSC.
- L. Dykman and N. Khlebtsov, Chem. Soc. Rev., 2012, 41, 2256–2282 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09088a |
| This journal is © The Royal Society of Chemistry 2017 |