Open Access Article
Open Access ArticlePentasaccharide resin glycosides with multidrug resistance reversal activities from the seeds of Pharbitis nil†
Jun Liab,
Wen-Qiong Wangab,
Shuai Tangab,
Wei-Bin Songab,
Min Huangab and
Li-Jiang Xuan *ab
*ab
aState Key Laboratory of Drug Research, Shanghai Institute of Meteria Medica, Chinese Academy of Sciences, 501 Haike Road, Shanghai 201201, People's Republic of China. E-mail: ljxuan@simm.ac.cn; Fax: +86-21-20231968; Tel: +86-21-20231968
bUniversity of Chinese Academy of Sciences, No. 19A Yuquan Road, Beijing 100049, People's Republic of China
First published on 8th November 2017
Abstract
Resin glycosides are novel P-glycoprotein inhibitors. In order to evaluate their multidrug resistance (MDR) reversal activities, we isolated seven new resin glycosides, pharbitins A–G (1–7) from the seeds of Pharbitis nil. Their chemical structures were determined by extensive application of high resolution 2D NMR techniques, HRESIMS and chemical methods. Compounds 1–4 and 6 were evaluated for their MDR reversal activities in KB/VCR, A549/T and K562/ADR cells. Among them, compound 2 showed moderate MDR reversal activity in KB/VCR cells, and increased the cytotoxicity of vincristine by 2.2-fold when incorporated at 25 μM. A structure–activity relationship study revealed that substituting Rha′′ C-3 with a trans-cinnamoyl group improves the MDR reversal activity. Also, an intracellular Rh123 accumulation assay demonstrated that compound 2 could inhibit the function of P-gp.
Introduction
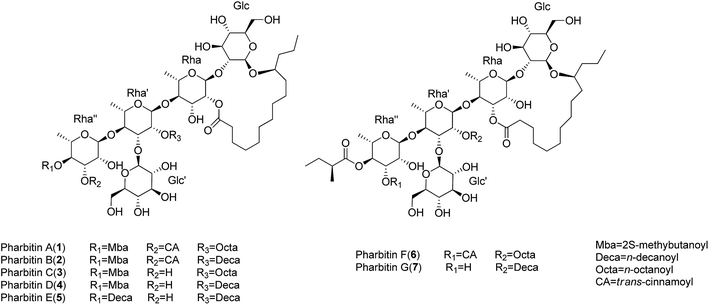
Resin glycosides are unusual amphipathic metabolites with structures including hydrophobic (fatty acid aglycone) and hydrophilic (oligosaccharide) moieties, which are mainly found in the family Convolvulaceae.1 Resin glycosides exhibit various pharmacological activities, such as cytotoxicity, multidrug resistance (MDR) reversal, ionophoresis, and phytogrowth–inhibitory activities.2–4 Because of their intriguing structures and varied activities, resin glycosides have attracted more attention in recent times. Moreover, as novel P-glycoprotein inhibitors, the resin glycosides from Merremia hederacea with MDR reversal activities have been investigated in our previous work.5 However, considering the unclear structure–activity relationship (SAR) and pharmacological mechanism of the resin glycosides, we intend to isolate various analogues from the family Convolvulaceae, investigate their MDR reversal activities, and then proceed to establish their SAR and pharmacological mechanism.Pharbitidis Semen, the seeds of Pharbitis nil, is a purgative crude drug widely grown in China and Japan.6 So far, only four pure resin glycoside acids (pharbitic acids A–D) with an acyclic core have been reported from P. nil.6–8 We report the isolation of seven new resin glycosides pharbitins A–G (1–7) (Fig. 1) with macrolactone rings from the seeds of P. nil. According to their structures, the new compounds can be divided into two types: those possessing 18-membered rings (1–5), and compounds 6–7 with 19-membered rings. Among them, 2 showed moderate activity in MDR reversal against KB/VCR cells, and increased the cytotoxicity of vincristine by 2.2-fold when incorporated at 25 μM. Herein, we described the isolation, structural elucidation and MDR reversal activity evaluation of all isolates from the seeds of P. nil.
Results and discussion
Pharbitin A (1) was obtained as a colorless gum. The molecular formula was established as C66H104O27 based on 13C NMR data (Table 3) and HRESIMS ion at m/z 1351.6647 [M + Na]+ (calcd for C66H104O27Na, 1351.6657). Its IR spectrum exhibited absorptions of hydroxyl (3426 cm−1), carbonyl (1724 cm−1), and aromatic (1635 cm−1) groups. The NMR spectra showed five anomeric signals [δH 4.90 (d, J = 7.5 Hz), δH 5.12 (d, J = 7.6 Hz), δH 5.59 (d, J = 1.8 Hz), δH 5.85 (d, J = 1.8 Hz), δH 6.29 (d, J = 1.8 Hz)] and δC 166.8, 173.7, 173.9, 176.3] and signals of long-chain fatty acids, which indicated that 1 was a resin glycoside.9 The NMR data of 1 could be divided into two parts: resonances in the anomeric region and those representative of the aglycone moieties.For the aglycone part, the 1H NMR data of 1 (Table 1) exhibited two trans-coupled olefinic protons at δH 6.59 (d, J = 16.0 Hz) and δH 7.85 (d, J = 16.0 Hz) due to trans-cinnanoyl group (CA). The signals at δH 0.79 (t, J = 7.0 Hz) and δH 2.31 (m) due to the n-octanoyl group (Octa). Also a methyl triplet signal at δH 0.82, a methyl doublet signal at δH 1.14 and a methine multiplet signal at δH 2.49 due to 2-methybutanoyl moiety (2-Mba). After alkaline hydrolysis, the S absolute configuration of 2-Mba was determined by chiral gas chromatography (GC) analysis. In addition, the 11-hydroxytetradecanoic acid moiety (convolvulinolic acid, Con) was suggested by the diagnostic signals of methyl triplet at δH 0.85, methylene group at δH 2.46 and 2.34, and oxygenated methine at δH 3.88. After alkaline and acid hydrolysis, the resulting 11-hydroxytetradecanoic acid showed a fragment ion at m/z 183 [M − CH3(CH2)2 − H2O]+ by the EIMS data, suggesting the 11-OH group of Con. Its absolute configuration was determined to be S by the Mosher's method.10
| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| a Chemical shifts (ppm) referenced to pyridine-d5 (δH 7.58) at 500 MHz.b Chemical shifts marked with an asterisk (*) indicate overlapped signals. | |||||
| Glc-1 | 4.90 d (7.5) | 4.91 d (7.4) | 4.88 d (7.5) | 4.86 d (7.5) | 4.89 d (7.4) |
| 2 | 3.89 m*, b | 3.90 m* | 3.87 m* | 3.86 m* | 3.89 m* |
| 3 | 4.18 m* | 4.19 m* | 4.14 m* | 4.14 m* | 4.17 m* |
| 4 | 4.12 m* | 4.13 m* | 4.11 m* | 4.10 m* | 4.13 m* |
| 5 | 3.85 m* | 3.86 m* | 3.83 m* | 3.82 m* | 3.84 m* |
| 6 | 4.45 m*, 4.30 dd (11.7, 5.2) | 4.46 m*, 4.31 dd (12.2, 5.2) | 4.45 m*, 4.30 dd (12.5, 4.0) | 4.44m*, 4.28 dd (11.9, 5.5) | 4.46m*, 4.32 dd (10.7, 5.1) |
| Rha-1 | 5.59 d (1.8) | 5.60 d (1.8) | 5.58 d (1.8) | 5.56 d (1.8) | 5.59 d (1.8) |
| 2 | 6.02 dd (3.4, 1.8) | 6.03 dd (3.4, 1.8) | 6.02 dd (3.3, 1.8) | 6.01 dd (3.3, 1.8) | 6.04 dd (3.4, 1.8) |
| 3 | 5.07 m* | 5.07 m* | 5.04 m* | 5.03 m* | 5.03–5.09 m |
| 4 | 4.16 m* | 4.17 m* | 4.18 m* | 4.17 m* | 4.19 m* |
| 5 | 4.44 m* | 4.45 m* | 4.43 m* | 4.42 m* | 4.44 m* |
| 6 | 1.60 d (6.2) | 1.61 d (6.1) | 1.60 d (6.2) | 1.59 d (6.1) | 1.62 d (6.0) |
| Rha′-1 | 5.85 d (1.8) | 5.86 d (1.9) | 5.91 d (1.9) | 5.91 d (1.8) | 5.94 d (1.9) |
| 2 | 6.34 br s | 6.35 dd (3.2, 1.9) | 6.30 dd (3.4, 1.9) | 6.29 br s | 6.33 dd (3.4, 1.9) |
| 3 | 4.81 dd (8.9, 3.3) | 4.80–4.86 m | 4.75–4.81 m | 4.74–4.80 m | 4.77–4.83 m |
| 4 | 4.37 m* | 4.38 m* | 4.34 m* | 4.33 m* | 4.36 m* |
| 5 | 4.36 m* | 4.37 m* | 4.35 m* | 4.34 m* | 4.37 m* |
| 6 | 1.65 d (5.7) | 1.66 d (5.4) | 1.66 d (5.1) | 1.64 d (5.5) | 1.67 d (5.5) |
| Rha′′-1 | 6.29 d (1.8) | 6.30 d (1.8) | 6.24 d (1.9) | 6.23 d (1.9) | 6.28 d (1.9) |
| 2 | 5.27 dd (3.1, 1.8) | 5.28 br s | 4.95 br s | 4.94 br s | 4.99 m* |
| 3 | 5.99 dd (9.8, 3.1) | 6.00 dd (9.8, 3.1) | 4.49–4.57 m | 4.51 dd (9.4, 3.8) | 4.55–4.61 m |
| 4 | 6.09 t (9.8) | 6.10 t (9.8) | 5.76 t (9.4) | 5.75 t (9.4) | 5.82 t (9.4) |
| 5 | 4.50 dd (9.8, 6.3) | 4.51 dd (9.8, 6.3) | 4.36 m* | 4.35 m* | 4.38 m* |
| 6 | 1.43 d (6.3) | 1.44 d (6.3) | 1.40 d (6.2) | 1.39 d (6.2) | 1.45 d (6.3) |
| Glc′-1 | 5.12 d (7.6) | 5.13 d (7.6) | 5.08 d (7.7) | 5.07 d (7.6) | 5.11 d (7.6) |
| 2 | 3.96 m* | 3.97 m* | 3.96 m* | 3.95 m* | 3.98 m* |
| 3 | 4.06 m* | 4.07 m* | 4.03 m* | 4.02 m* | 4.05 m* |
| 4 | 3.93 m* | 3.94 m* | 3.94 m* | 3.93 m* | 3.96 m* |
| 5 | 3.76–3.82 m | 3.80 ddd (9.8, 5.9, 2.5) | 3.74 ddd (8.9, 5.8, 2.5) | 3.73 ddd (8.9, 5.8, 2.5) | 3.76 ddd (8.9, 5.9, 2.5) |
| 6 | 4.40 m*, 4.09 m* | 4.41 m*, 4.10 m* | 4.38 m*, 4.08 m* | 4.37 m*, 4.07 m* | 4.41 m*, 4.10 m* |
| Con-2 | 2.46 m*, 2.34 m* | 2.47 m*, 2.36 m* | 2.46 m*, 2.35 m* | 2.45 m*, 2.34 m* | 2.45 m*, 2.37 m* |
| 11 | 3.88 m* | 3.89 m* | 3.86 m* | 3.85 m* | 3.88 m* |
| 14 | 0.85 t (6.9) | 0.85 t (7.3) | 0.85 t (7.3) | 0.83 t (7.2) | 0.85 t (7.1) |
| 2-Mba-2 | 2.49 m* | 2.50 m* | 2.51 m* | 2.50 m* | |
| 3 | 1.70 m*, 1.40 m* | 1.71 m*, 1.41 m* | 1.78 m*, 1.49 m* | 1.77 m*, 1.47 m* | |
| 4 | 0.82 t (6.9) | 0.82 t (7.5) | 0.93 t (7.4) | 0.92 t (7.4) | |
| 2-Me | 1.14 d (6.9) | 1.15 d (7.0) | 1.20 d (7.0) | 1.19 d (7.0) | |
| CA-2 | 6.59 d (16.0) | 6.59 d (16.0) | |||
| 3 | 7.85 d (16.0) | 7.86 d (16.0) | |||
| 2′/6′ | 7.45 2H m | 7.46 2H m | |||
| 3′/5′ | 7.34 2H m* | 7.34 2H m* | |||
| 4′ | 7.34 m* | 7.34 m* | |||
| Octa-2 | 2.31 m* | 2.31 m* | |||
| 8 | 0.79 t (7.0) | 0.79 t (6.9) | |||
| Deca-2 | 2.33 m* | 2.30 m* | 2.32 m* | ||
| 10 | 0.85 t (7.2) | 0.83 t (7.2) | 0.85 t (7.1) | ||
| Deca′-2 | 2.48 m* | ||||
| 10 | 0.85 t (7.1) | ||||
| Position | 6 | 7 |
|---|---|---|
| a Chemical shifts (ppm) referenced to pyridine-d5 (δH 7.58) at 500 MHz.b Chemical shifts marked with an asterisk (*) indicate overlapped signals. | ||
| Glc-1 | 5.04 d (7.8) | 5.03 d (8.0) |
| 2 | 4.28 m*, b | 4.28 m*, b |
| 3 | 4.37 m* | 4.35 m* |
| 4 | 4.18 m* | 4.17 m* |
| 5 | 3.91 m* | 3.91 m* |
| 6 | 4.56 m*, 4.38 m* | 4.50 m*, 4.38 m* |
| Rha-1 | 6.47 d (1.8) | 6.48 d (1.7) |
| 2 | 5.24 br s | 5.25 br s |
| 3 | 5.70 dd (10.0, 2.8) | 5.68 dd (9.7, 2.7) |
| 4 | 4.71 m* | 4.72 t (9.7) |
| 5 | 5.07 m* | 5.06 m* |
| 6 | 1.72 d (6.2) | 1.70 d (6.1) |
| Rha′-1 | 5.63 d (1.8) | 5.65 d (1.8) |
| 2 | 6.03 dd (3.4, 1.8) | 6.01 dd (3.5, 1.8) |
| 3 | 4.74 dd (8.9, 3.4) | 4.66 dd (8.8, 3.5) |
| 4 | 4.37 m* | 4.36 m* |
| 5 | 4.44 m* | 4.39 m* |
| 6 | 1.66 d (5.8) | 1.65 d (5.9) |
| Rha′′-1 | 6.28 d (2.5) | 6.22 d (2.0) |
| 2 | 5.22 br s | 4.93 br s |
| 3 | 5.93 dd (9.8, 2.5) | 4.47 m* |
| 4 | 6.06 t (9.8) | 5.75 t (9.7) |
| 5 | 4.47 m* | 4.36 m* |
| 6 | 1.44 d (6.2) | 1.41 d (6.2) |
| Glc′-1 | 5.15 d (7.7) | 5.10 d (7.6) |
| 2 | 3.96 m* | 3.97 m* |
| 3 | 4.16 m* | 4.14 m* |
| 4 | 4.18 m* | 4.17 m* |
| 5 | 3.91 m* | 3.91 m* |
| 6 | 4.56 m*, 4.38 m* | 4.50 m*, 4.38 m* |
| Con-2 | 2.74 ddd (14.2, 10.8, 2.7), 2.32 m* | 2.70 ddd (14.2, 10.7, 2.8), 2.28 m* |
| 11 | 3.94 m* | 3.93 m* |
| 14 | 1.02 t (7.0) | 0.95 t (7.1) |
| Jal-2 | ||
| 11 | ||
| 14 | ||
| 2-Mba-2 | 2.46 m* | 2.50 m* |
| 3 | 1.69 m*, 1.40 m* | 1.77 m*, 1.48 m* |
| 4 | 0.81 t (7.4) | 0.93 t (7.4) |
| 2-Me | 1.13 d (7.0) | 1.19 d (7.1) |
| CA-2 | 6.56 d (16.0) | |
| 3 | 7.85 d (16.0) | |
| 2′/6′ | 7.44 2H m | |
| 3′/5′ | 7.33 2H m* | |
| 4′ | 7.33 m* | |
| Octa-2 | 2.42 m* | |
| 8 | 0.81 t (6.9) | |
| Deca-2 | 2.43 m* | |
| 10 | 0.85 t (7.1) | |
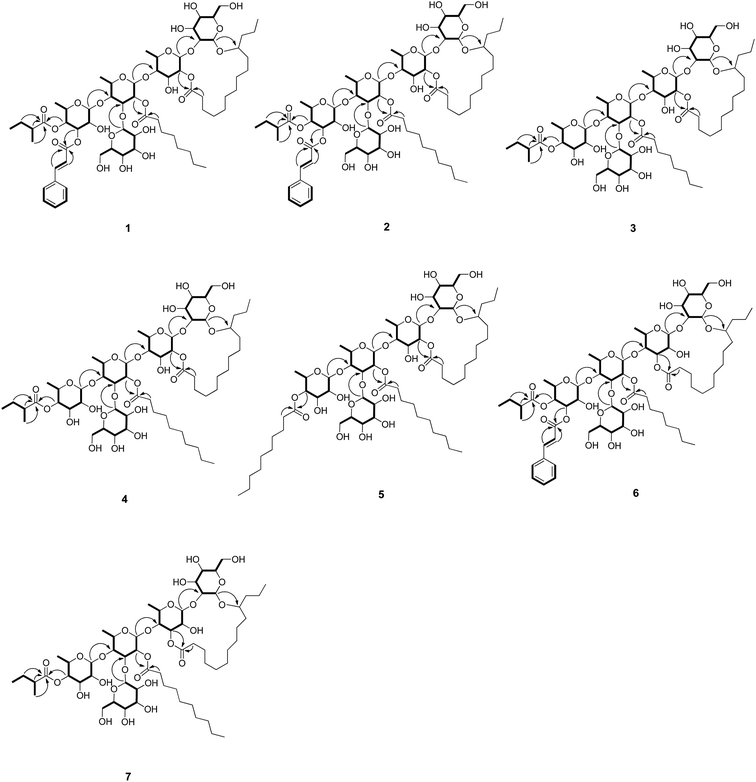
In the anomeric region, the 1H–1H COSY data (Fig. 2) indicated five spin systems, which were attributed to three 6-deoxyhexose and two hexose units. The sugars obtained from the acidic hydrolysates were identified as L-rhamnose and D-glucose through the HPLC analysis and their corresponding optical rotations. The β-configurations of D-glucose was determined by a large coupling constant (J = 7.5 Hz and 7.6 Hz) for the anomeric protons at δH 4.90 and δH 5.12 in the 1H NMR spectrum, while α-configuration of L-rhamnose was revealed by the chemical shift of C-5 of rhamnose (δC 69.3, 68.8, 68.5) in the 13C NMR spectrum.11 The long-range HMBC correlations (Fig. 2) between H-1 of β-Glc (δH 4.90) and C-11 (δC 82.7) of the 11-hydroxytetradecanoyl moiety indicated that β-Glc was the first hexose unit in the sugar moiety. The sequence of the sugar moiety was determined to be glucosyl-(1 → 3)-[rhamnosyl-(1 → 4)]-rhamnosyl-(1 → 4)-rhamnosyl-(1 → 2)-glucosyl by their long-range HMBC correlations: H-1 of α-Rha (δH 5.59) to C-2 of β-Glc (δC 82.2), H-1 of α-Rha′ (δH 5.85) to C-4 of α-Rha (δC 82.2), H-1 of α-Rha′′ (δH 6.29) to C-4 of α-Rha′ (δC 79.3), and δH H-1 of β-Glc′ (δH 5.12) to C-3 of α-Rha′ (δC 80.2). In addition, the positions of esterification, i.e., 2-Mba located at OH-4 of α-Rha′′, CA at OH-3 of α-Rha′′, Octa at OH-2 of α-Rha′ were inferred from the long-range correlations: H-4 of α-Rha′′ (δH 6.09) to C-1 of 2-Mba (δC 176.3), and H-3 of α-Rha′′ (δH 5.99) to C-1 of CA (δC 166.8), H-2 of α-Rha′ (δH 6.34) to C-1 of Octa (δC 173.9) respectively. The C-2 (Rha) site of lactonization was corroborated by the correlation between C-1 of Con (δC 173.7) and H-2 of α-Rha (δH 6.02). Thus, the structure of compound 1 was identified as (11S)-convolvulinolic acid 11-O-β-D-glucopyranosyl-(1 → 3)-O-[3-O-(trans-cinnamoyl)-4-O-(2S-methylbutanoyl)-α-L-rhamnopyranosyl-(1 → 4)]-O-[2-O-n-octanoyl]-α-L-rhamnopyranosyl-(1 → 4)-O-α-L-rhamnopyranosyl-(1 → 2)-O-β-D-glucopyranoside-(1, 2′-lactone).
The molecular formulas of pharbitins B–E (2–5) were determined as C68H108O27, C57H98O26, C59H102O26 and C64H112O26, respectively, based on the 13C NMR (Table 3) and HRESIMS data. The 1H and 13C NMR spectra indicated that 2–5 consisted of the same complex sugar moiety and (11S)-hydroxytetradecanoyl group as 1. They differed at the kinds of acyl residue and the sites of acylation. The identities of the acyl moieties were determined by LC-HRMS after basic hydrolysis. 2-Mba, CA, Deca (n-decanoic acid) in 2, 2-Mba, Octa in 3, 2-Mba, Deca in 4 and Deca in 5 were identified. The positions of acylation were determined by the HMBC correlations as follows: OH-4 of Rha′′ was acylated by 2-Mba in 2–4 and by Deca in 5, OH-2 of Rha′ was acylated by Deca in 2, 4, 5 and by Octa in 3, OH-3 of Rha′′ was acylated by CA only in 2. Moreover, the site of lactonization was corroborated as C-2 of Rha according to the correlation between C-1 of Con and H-2 of Rha. Consequently, the structures of 2–5 were determined as shown.
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| a Chemical shifts (ppm) referenced to pyridine-d5 (δC 135.9) at 125 MHz. | |||||||
| Glc-1 | 104.6 | 104.6 | 104.6 | 104.6 | 104.7 | 101.7 | 101.7 |
| 2 | 82.2 | 82.3 | 82.3 | 82.3 | 82.3 | 75.6 | 75.8 |
| 3 | 76.9 | 76.9 | 76.9 | 76.9 | 77.0 | 79.9 | 79.7 |
| 4 | 72.0 | 72.0 | 72.2 | 72.1 | 72.2 | 72.2 | 72.4 |
| 5 | 78.3 | 78.4 | 78.3 | 78.2 | 78.4 | 78.4 | 78.5 |
| 6 | 63.1 | 63.2 | 63.2 | 63.1 | 63.2 | 63.1 | 63.2 |
| Rha-1 | 98.8 | 98.8 | 98.9 | 98.8 | 98.9 | 100.3 | 100.5 |
| 2 | 73.8 | 73.9 | 73.9 | 73.9 | 73.9 | 70.2 | 70.2 |
| 3 | 69.6 | 69.7 | 69.8 | 69.7 | 69.8 | 78.1 | 78.3 |
| 4 | 82.2 | 82.2 | 81.6 | 81.6 | 81.5 | 77.7 | 77.1 |
| 5 | 69.3 | 69.4 | 69.3 | 69.3 | 69.4 | 68.5 | 68.5 |
| 6 | 19.3 | 19.4 | 19.2 | 19.2 | 19.3 | 19.6 | 19.6 |
| Rha′-1 | 100.5 | 100.6 | 100.3 | 100.2 | 100.3 | 99.8 | 99.7 |
| 2 | 73.6 | 73.6 | 73.5 | 73.4 | 73.5 | 72.6 | 72.5 |
| 3 | 80.2 | 80.3 | 80.5 | 80.4 | 80.6 | 80.5 | 80.7 |
| 4 | 79.3 | 79.3 | 79.0 | 78.9 | 78.9 | 79.4 | 78.9 |
| 5 | 68.8 | 68.8 | 68.8 | 68.8 | 68.9 | 68.5 | 68.6 |
| 6 | 19.4 | 19.4 | 19.5 | 19.4 | 19.5 | 19.2 | 19.1 |
| Rha′′-1 | 103.7 | 103.7 | 103.6 | 103.5 | 103.6 | 103.7 | 103.8 |
| 2 | 70.2 | 70.3 | 72.8 | 72.8 | 72.8 | 70.2 | 72.7 |
| 3 | 73.3 | 73.4 | 70.7 | 70.6 | 70.7 | 73.2 | 70.6 |
| 4 | 72.2 | 72.2 | 75.5 | 75.5 | 75.9 | 72.4 | 75.6 |
| 5 | 68.5 | 68.5 | 68.5 | 68.5 | 68.6 | 68.5 | 68.5 |
| 6 | 18.2 | 18.3 | 18.4 | 18.3 | 18.4 | 18.3 | 18.4 |
| Glc′-1 | 105.8 | 105.9 | 105.8 | 105.7 | 105.8 | 105.1 | 105.2 |
| 2 | 75.5 | 75.6 | 75.6 | 75.5 | 75.6 | 75.9 | 75.6 |
| 3 | 78.8 | 78.8 | 78.8 | 78.7 | 78.9 | 78.8 | 78.6 |
| 4 | 71.8 | 71.9 | 71.8 | 71.7 | 71.8 | 71.2 | 71.1 |
| 5 | 78.4 | 78.5 | 78.5 | 78.4 | 78.6 | 78.5 | 78.8 |
| 6 | 63.3 | 63.3 | 63.3 | 63.2 | 63.3 | 62.9 | 62.9 |
| Con-1 | 173.7 | 173.7 | 173.7 | 173.7 | 173.7 | 175.0 | 175.0 |
| 2 | 34.6 | 34.6 | 34.6 | 34.6 | 34.6 | 34.5 | 34.5 |
| 11 | 82.7 | 82.7 | 82.7 | 82.7 | 82.8 | 79.8 | 80.0 |
| 14 | 14.9 | 14.9 | 14.9 | 14.8 | 14.9 | 15.2 | 15.1 |
| 2-Mba-1 | 176.3 | 176.3 | 176.7 | 176.6 | 176.3 | 176.7 | |
| 2 | 41.9 | 41.9 | 41.9 | 41.9 | 41.9 | 41.9 | |
| 3 | 27.3 | 27.4 | 27.5 | 27.4 | 27.3 | 27.4 | |
| 4 | 12.1 | 12.2 | 12.1 | 12.1 | 12.1 | 12.1 | |
| 5 | 17.3 | 17.3 | 17.4 | 17.3 | 17.2 | 17.4 | |
| CA-1 | 166.8 | 166.8 | 166.5 | ||||
| 2 | 118.9 | 118.9 | 119.0 | ||||
| 3 | 145.7 | 145.7 | 145.6 | ||||
| 1′ | 135.1 | 135.1 | 135.1 | ||||
| 2′/6′ | 128.8 | 128.9 | 128.8 | ||||
| 3′/5′ | 129.6 | 129.6 | 129.6 | ||||
| 4′ | 131.1 | 131.1 | 131.0 | ||||
| Octa-1 | 173.9 | 174.0 | 173.9 | ||||
| 2 | 34.9 | 34.9 | 34.8 | ||||
| 8 | 14.6 | 14.6 | 14.6 | ||||
| Deca-1 | 173.9 | 174.0 | 174.0 | 174.0 | |||
| 2 | 34.9 | 34.9 | 34.9 | 34.9 | |||
| 10 | 14.6 | 14.6 | 14.7 | 14.7 | |||
| Deca′-1 | 173.9 | ||||||
| 2 | 35.1 | ||||||
| 10 | 14.7 | ||||||
Pharbitin F (6) has the same molecular formula, C66H104O27, as 1, according to its HRESIMS data at m/z 1351.6663 [M + Na]+ (calcd 1351.6657). Alkaline hydrolysis of 1 and 6 afforded 2-Mba, CA and Octa in the CHCl3 layer. The sites of acylation of 6 were the same as 1, which was supported by the key HMBC correlation from Rha′′ H-4 (δH 6.06) to 2-Mba C-1 (δC 176.3), Rha′′ H-3 (δH 5.93) to CA C-1 (δC 166.5), Rha′ H-2 (δH 6.03) to Octa C-1 (δC 173.9). Moreover, 1H and 13C NMR data of 1 and 6 indicated that they share the same pentasaccharide skeleton. The only difference between 1 and 6 was the position of the lactonization. In the HMBC spectrum of 6, the H-3 proton of Rha resonated at δH 5.70 and showed an HMBC correlation to the carbonyl group that resonated at δC 175.0 (C-1 of Con), which suggested the lactone bond was linked at C-3 of Rha in 6 rather than at C-2 of Rha as in 1. Therefore, the structure of 6 was determined as shown.
Pharbitin G (7) has the same molecular formula, C59H102O26, as 4, according to its HRESIMS data at m/z 1249.6550 [M + Na]+ (calcd 1249.6552). 1D NMR and the HMBC spectrum, together with the alkaline hydrolysis analysis, suggested 4 and 7 share the same pentasaccharide skeleton, same acyl functions (2-Mba, Deca) and same sites of acylation (OH-4 of Rha′′ was acylated by 2-Mba, OH-2 of Rha′ was acylated by Deca). The only difference between 4 and 7 was the position of the lactonization. The lactonization site was bonded at C-3 of Rha for 7, while at C-2 of Rha for 4, on the basis of corresponding HMBC correlation: δH 5.68 (Rha H-3) to δC 175.0 (Con C-1) in 7, δH 6.01 (Rha H-2) to δC 173.7 (Con C-1) in 4. Thus, the structure of 7 was determined as shown.
Isolates (1–4 and 6) were examined for their multidrug resistance (MDR) reversal activities in KB/VCR and A549/T cells using the SRB method, while in K562/ADR cell using the MTT method (Tables 4 and S1†). However, only the KB/VCR cell line showed favorable results and as such we proceeded to carry out further experiments with this cell line. The MDR reversal activity in KB/VCR is described below.
| Sample | Inhibition ratio % (25 μM) | Vincristine + Sampleb | |
|---|---|---|---|
| IC50 value (μM) | RFc value | ||
| a MDR: multidrug resistance.b Serial dilutions ranging from 0.125 to 1 μM of vincristine in the presence or absence of 25 μM sample.c RF: IC50 of VCR alone/IC50 of VCR in presence of 25 μM sample.d Serial dilutions ranging from 0.125 to 1 μM of vincristine in the presence or absence of 5 μM sample.e RF: IC50 of VCR alone/IC50 of VCR in presence of 5 μM sample. | |||
| 2 | 5.62 | 0.22 | 2.2 |
| 4 | 4.17 | 0.44 | 1.1 |
| Vincristine | 0.48 | ||
The cytotoxicity assay showed that the inhibition ratios of 2 and 4 were less than 50% at 25 μM, indicating that these compounds were noncytotoxic at 25 μM. However, the inhibition ratios of 1, 3, 6 were more than 50% at 25 μM, indicating that these compounds were cytotoxic at 25 μM, thus, these compounds were tested at 5 μM. Compounds 2 and 4 enhanced the cytotoxicity of vincristine by 1.1–2.2-fold when incorporated at 25 μM, while compound 1, 3, 6 increased the cytotoxicity of vincristine by 0.9–1.3-fold when incorporated at 5 μM. Compound 2 with a CA substituent group at Rha′′ C-3 was 2 times more active than compound 4 with no substituent group at the same site. Similar result was observed for compound 1 and 3, which demonstrated that, substituting Rha′′ C-3 with a CA group improves MDR reversal activity.
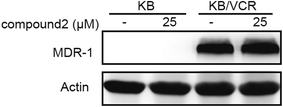
Moreover, compound 2 was tested for its effects on both P-glycoprotein (P-gp) function and expression (Fig. 3 and 4). Intracellular rhodamine 123 (Rh123)-associated mean fluorescence intensity in KB and KB/VCR cells was used to study the effects of compound on the inhibition of P-gp function. The result showed that the Rh123 accumulation of KB/VCR was a lot less than the parental KB cells and the Rh123 accumulation of KB/VCR pretreated with compound 2 was twice higher than untreated. However, the expression of P-gp detected by Western blot showed that P-gp was overexpressed in KB/VCR cell and compound 2 had no effect on P-gp expression.
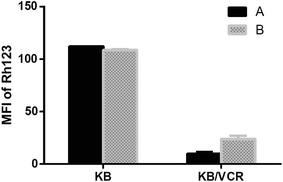 | ||
| Fig. 3 Effects of compound 2 on Rh123 accumulation in drug-sensitive KB and multidrug-resistant KB/VCR cells. Cells were respectively untreated (A) and pretreated with 25 μM of compound 2 (B). | ||
Conclusions
The chemical investigation of the seeds of P. nil led to the isolation of seven new pentasaccharide resin glycosides pharbitins A–G (1–7), and it is the first time resin glycosides with macrolactone rings is being reported from P. nil. The isolated resin glycosides shared the same oligosaccharide core, and differed at the kinds of acyl residue, and the sites of acylation. According to the sites of acylation, compounds 1–5 possess 18-membered rings, whereas compounds 6–7 possess 19-membered rings. Compound 2 showed moderate MDR reversal activity in KB/VCR cell, and increased the cytotoxicity of vincristine by 2.2-fold when incorporated at 25 μM. SAR study showed that, substituting Rha′′ C-3 with a CA group improves MDR reversal activity. This work therefore contributes to the SAR study of resin glycosides with MDR reversal activities. Furthermore, we found that compound 2 inhibited the function of P-gp but had no effect on P-gp expression.Experimental
General experimental procedures
Optical rotations were determined with a Perkin Elmer 341 polarimeter. UV spectra were obtained on a Shimadzu UV-2450 spectrophotometer. IR spectra were measured by a Bruker Tensor 27 spectrometer. NMR experiments were recorded in pyridine-d5 on Bruker AM-400 and Bruker AM-500 spectrometers referenced to solvent peaks (δH 7.58; δC 135.9). EIMS analyses were performed on a Thermo-DFS mass spectrometer. HRESIMS analyses were performed on an Agilent 6224 TOF mass spectrometer with an ESI interface. Semipreparative HPLC was carried out on an Agilent 1100 series HPLC system with a YMC-Pack ODS-A column (250 × 10 mm, 5 μm). Analytical HPLC was performed on an Agilent 1200 series HPLC system with an Agilent ZORBAX NH2 column (150 × 4.6 mm, 5 μm). Silica gel (200–300 mesh, Qingdao Haiyang Chemical Co. Ltd., People's Republic of China), C18 reversed-phase (RP-18) silica gel (20–45 μm; Fuji Silysia Chemical Ltd., Japan) and Sephadex LH-20 gel (Amersham Biosciences, Sweden) were used for column chromatography (CC). Pre-coated silica gel GF254 plates (Qingdao Haiyang Chemical Co. Ltd., People's Republic of China) were used for TLC.Plant material
Seeds of Pharbitis nil were collected in Guangxi, China, in August 2013 and identified by Prof. Heming Yang. A voucher specimen (no. SIMM810) was deposited at the Herbarium of the Shanghai Institute of Materia Medica, Chinese Academy of Sciences and People's Republic of China.Extraction and isolation
Air–dried, powdered seeds of Pharbitis nil (5.0 kg) were refluxed with 95% EtOH three times and concentrated in vacuo to give a crude extract (230 g), which was suspended in 2 L of water and then extracted with EtOAc to give the EtOAc-soluble fraction (168 g). The EtOAc layer was concentrated, and subjected to silica gel CC, eluting with petroleum ether/acetone in a gradient manner (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 to 0
0 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, v/v) to afford nine fractions (A-I). Fraction G was subjected to silica gel CC and eluted with CHCl3/MeOH (20
100, v/v) to afford nine fractions (A-I). Fraction G was subjected to silica gel CC and eluted with CHCl3/MeOH (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to yield subfractions (G1–G7). Fraction G3 was further subjected to a Sephadex LH-20 gel column followed by a RP-C18 column using a gradient of MeOH/H2O (80
1) to yield subfractions (G1–G7). Fraction G3 was further subjected to a Sephadex LH-20 gel column followed by a RP-C18 column using a gradient of MeOH/H2O (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 to 100
20 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, v/v) to give 6 (12 mg). Fraction H was applied to silica gel CC and eluted with CHCl3/MeOH (20
0, v/v) to give 6 (12 mg). Fraction H was applied to silica gel CC and eluted with CHCl3/MeOH (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford subfractions (H1–H6). Fraction H2 was further purified by a Sephadex LH-20 gel column followed by preparative HPLC (MeOH/H2O, 100
1) to afford subfractions (H1–H6). Fraction H2 was further purified by a Sephadex LH-20 gel column followed by preparative HPLC (MeOH/H2O, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, v/v) to obtain 1 (60 mg) and 2 (60 mg). Fraction H3 was chromatographed on a Sephadex LH-20 gel column followed by a RP-C18 column using a gradient of MeOH/H2O (80
0, v/v) to obtain 1 (60 mg) and 2 (60 mg). Fraction H3 was chromatographed on a Sephadex LH-20 gel column followed by a RP-C18 column using a gradient of MeOH/H2O (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 to 100
20 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, v/v) to yield 7 (8 mg). Same method was applied to fraction H4 to yield compound 5 (10 mg). Fraction H5 was subjected to a RP-C18 column (50
0, v/v) to yield 7 (8 mg). Same method was applied to fraction H4 to yield compound 5 (10 mg). Fraction H5 was subjected to a RP-C18 column (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 to 100
50 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, v/v) to give five subfractions (H5A-H5E). Fraction H5D was chromatographed on a silica gel column and eluted with CHCl3/MeOH (10
0, v/v) to give five subfractions (H5A-H5E). Fraction H5D was chromatographed on a silica gel column and eluted with CHCl3/MeOH (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 5
1 to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give fraction H5D1 and H5D2, which were further purified on an open ODS column (MeOH/H2O, 85
1) to give fraction H5D1 and H5D2, which were further purified on an open ODS column (MeOH/H2O, 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 to 100
15 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, v/v) to obtain 3 (60 mg) and 4 (60 mg), respectively.
0, v/v) to obtain 3 (60 mg) and 4 (60 mg), respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (3.89), 217 (3.91), 280 (4.09) nm; IR (KBr) vmax 3426, 2926, 2855, 1724, 1635, 1052 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1351.6647 [M + Na]+ (calcd for C66H104O27Na, 1351.6657).
ε) 203 (3.89), 217 (3.91), 280 (4.09) nm; IR (KBr) vmax 3426, 2926, 2855, 1724, 1635, 1052 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1351.6647 [M + Na]+ (calcd for C66H104O27Na, 1351.6657).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (3.66), 217 (3.64), 279 (3.80) nm; IR (KBr) vmax 3429, 2929, 2858, 1730, 1632, 1049 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1379.6988 [M + Na]+ (calcd for C68H108O27Na, 1379.6970).
ε) 203 (3.66), 217 (3.64), 279 (3.80) nm; IR (KBr) vmax 3429, 2929, 2858, 1730, 1632, 1049 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1379.6988 [M + Na]+ (calcd for C68H108O27Na, 1379.6970).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (3.64), 275 (2.45) nm; IR (KBr) vmax 3405, 2929, 2852, 1727, 1046 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1221.6258 [M + Na]+ (calcd for C57H98O26Na, 1221.6239).
ε) 203 (3.64), 275 (2.45) nm; IR (KBr) vmax 3405, 2929, 2852, 1727, 1046 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1221.6258 [M + Na]+ (calcd for C57H98O26Na, 1221.6239).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (3.59), 275 (2.43) nm; IR (KBr) vmax 3417, 2926, 2855, 1727, 1052 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1249.6561 [M + Na]+ (calcd for C59H102O26Na, 1249.6552).
ε) 203 (3.59), 275 (2.43) nm; IR (KBr) vmax 3417, 2926, 2855, 1727, 1052 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1249.6561 [M + Na]+ (calcd for C59H102O26Na, 1249.6552).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (3.43), 275 (2.38) nm; IR (KBr) vmax 3396, 2926, 2855, 1727, 1046 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1277.6862 [M + Na]+ (calcd for C61H106O26Na, 1277.6865).
ε) 203 (3.43), 275 (2.38) nm; IR (KBr) vmax 3396, 2926, 2855, 1727, 1046 cm−1; 1H and 13C NMR data, see Tables 1 and 3; HRESIMS m/z 1277.6862 [M + Na]+ (calcd for C61H106O26Na, 1277.6865).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (3.83), 217 (3.85), 280 (4.05) nm; IR (KBr) vmax 3417, 2926, 2861, 1730, 1635, 1043 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 1351.6663 [M + Na]+ (calcd for C66H104O27Na, 1351.6657).
ε) 203 (3.83), 217 (3.85), 280 (4.05) nm; IR (KBr) vmax 3417, 2926, 2861, 1730, 1635, 1043 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 1351.6663 [M + Na]+ (calcd for C66H104O27Na, 1351.6657).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 203 (3.57), 275 (2.66) nm; IR (KBr) vmax 3399, 2926, 2855, 1727, 1049 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 1249.6550 [M + Na]+ (calcd for C59H102O26Na, 1249.6552).
ε) 203 (3.57), 275 (2.66) nm; IR (KBr) vmax 3399, 2926, 2855, 1727, 1049 cm−1; 1H and 13C NMR data, see Tables 2 and 3; HRESIMS m/z 1249.6550 [M + Na]+ (calcd for C59H102O26Na, 1249.6552).Aglycon identification
Compounds 1–7 (5 mg, each) were separately dissolved in 5% NaOH (4 mL) and refluxed at 90 °C for 4 h. Each of the reaction mixtures were acidified to PH 4, followed by extraction with CHCl3 (3 × 4 mL) and concentration.12 The CHCl3 layer was then analyzed by HRESIMS to identify organic acids: 2-methylbutanoic acid (m/z 101.0606 [M − H]−, calcd 101.0608) in 1–4 and 6–7; n-octanoic acid (m/z 143.1080 [M − H]−, calcd 143.1078) in 1, 3 and 6; n-decanoic acid (m/z 171.1390 [M − H]−, calcd 171.1391) in 2, 4, 5 and 7; trans-cinnamic acid (m/z 147.0453 [M − H]−, calcd 147.0452) in 1, 2 and 6. The CHCl3 layer was also analyzed by chiral GC to determined the absolute configuration of 2-methylbutanoic acid. In addition, the aqueous phase via CHCl3 extraction was then extracted with n-BuOH (3 × 4 mL) and concentrated to yield a colorless solid. The residue in 1 N H2SO4 (4 mL) was refluxed at 90 °C for 4 h. The reaction mixture was extracted with CHCl3 (3 × 4 mL), which was further concentrated and purified to afford 11-hydroxytetradecanoic acid in 1–7. The 11-hydroxytetradecanoic acid was further separately treated with (S)-MPTA chloride and (R)-MPTA chloride in pyridine-d5, and allowed to stand for 12 h at room temperature. The S configuration of 11-hydroxytetradecanoic acid was determined by the chemical shift difference (Δδ = δS − δR, ΔδH-14 = −0.067).Sugar analysis
Compound 1 (20.0 mg) was hydrolyzed by alkali and acid, as described in the aglycone identification section. The aqueous phase was extracted with n-BuOH (3 × 4 mL) after acid hydrolysis and concentrated to yield a colorless solid. The residue was dissolved in H2O and directly analyzed by HPLC with authentic samples (MeCN–H2O, 90/10): D-glucose eluted at 11.9 min, L-rhamnose at 5.2 min. Each of these eluates was individually collected, concentrated, and dissolved in H2O. The elutes were identified as D-glucose [α]21D +54.7 (c 0.1, H2O), L-rhamnose [α]21D +8.7 (c 0.1, H2O) through comparisons of their specific rotations with those of the corresponding authentic samples.MDR reversal assays
The results were expressed as IC50 values. The reversal fold (RF) as potency of reversal was obtained from fitting the data to RF = IC50 of vincristine or taxel or adriamycin alone/IC50 of vincristine or taxel or adriamycin in the presence of sample.13
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge grants from the State Key Laboratory of Drug Research (SIMM1601ZZ-03), the National Natural Science Foundation of China (No. 81473111), and the China Postdoctoral Science Foundation (No. 2016M600345).References
- R. Pereda-Miranda, R. Villatoro-Vera, M. Bah and A. Lorence, Rev. Latinoam. Quim., 2009, 37, 144–154 CAS.
- L. Chérigo, R. Pereda-Miranda, M. Fragoso-Serrano, N. Jacobo-Herrera, G. Kaatz and S. Gibbons, J. Nat. Prod., 2008, 71, 1037–1045 CrossRef PubMed.
- G. Figueroa-Gonzál, N. Jacobo-Herrera, A. Zentella-Dehesa and R. Pereda-Miranda, J. Nat. Prod., 2012, 75, 93–97 CrossRef PubMed.
- I. Kitagawa, K. Ohashi, H. Kawanishi, H. Shibiya, K. Shinkai and H. Akedo, Chem. Pharm. Bull., 1989, 37, 1679–1681 CrossRef CAS PubMed.
- W. Q. Wang, W. B. Song, X. J. Lan, M. Huang and L. J. Xuan, J. Nat. Prod., 2014, 77, 2234–2240 CrossRef CAS PubMed.
- M. One, N. Noda, T. Kawasaki and K. Miyahara, Chem. Pharm. Bull., 1990, 38, 1892–1897 CrossRef.
- T. Kawasaki, H. Okabe and I. Nakatsuka, Chem. Pharm. Bull., 1971, 19, 1144–1149 CrossRef CAS.
- H. Okabe and T. Kawasaki, Chem. Pharm. Bull., 1972, 20, 514–520 CrossRef CAS.
- Y. Q. Yin, J. S. Wang, J. G. Luo and L. Y. Kong, Carbohydr. Res., 2009, 344, 466–473 CrossRef CAS PubMed.
- B. W. Yu, J. G. Luo, J. S. Wang, D. M. Zhang, S. S. Yu and L. Y. Kong, J. Nat. Prod., 2011, 74, 620–628 CrossRef CAS PubMed.
- S. M. Sang, A. N. Lao, Y. Leng, Z. P. Gu, Z. L. Chen, J. Uzawa and Y. Fji-moto, Tetrahedron Lett., 2000, 41, 9205–9208 CrossRef CAS.
- B. Y. Fan, Y. C. Gu, Y. He, Z. R. Li, J. G. Luo and L. Y. Kong, J. Nat. Prod., 2014, 77, 2264–2272 CrossRef CAS PubMed.
- B. S. Ji, L. He and G. Q. Liu, Life Sci., 2005, 77, 2221–2232 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09026a |
| This journal is © The Royal Society of Chemistry 2017 |