Open Access Article
Open Access ArticleBinding behaviors and structural characteristics of ternary complexes of β-lactoglobulin, curcumin, and fatty acids†
Jingwen Liuab,
Lun Jiangab,
Yunyue Zhangab,
Zhongyao Duab,
Xiaoxia Qiuab,
Lingyan Kongc and
Hao Zhang *ab
*ab
aBeijing Advanced Innovation Center for Food Nutrition and Human Health, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing 100083, P. R. China. E-mail: zhanghaocau@cau.edu.cn; Fax: +86-10-62736344; Tel: +86-10-62736344
bKey Laboratory of Functional Dairy, Co-constructed by Ministry of Education and Beijing Government, Beijing Laboratory of Food Quality and Safety, China Agricultural University, Beijing 100083, P. R. China
cDepartment of Human Nutrition & Hospitality Management, The University of Alabama, Tuscaloosa 35487, AL, USA
First published on 26th September 2017
Abstract
In this study, we evaluated the binding behaviors and structural characteristics of multi-ligand complex of β-lactoglobulin (β-lg) with curcumin and fatty acids. Methods of fluorescence spectroscopy, circular dichroism spectroscopy, dynamic light scattering, Turbiscan stability analysis, HPLC quantification, molecular docking studies, and molecular dynamics simulation were used. The results of fluorescence measurements indicated that β-lg was able to interact with curcumin and fatty acids at the same time, and the presence of fatty acids enhanced the binding constant of curcumin. The formation of ternary complexes with curcumin and fatty acids did not change the secondary structure of β-lg, while this process resulted in a considerable increase in particle size and negative charges compared to the native β-lg (ending up at around 145 nm and −30 mV in the ternary complexes). The loading efficiencies of the fatty acids were 32 ± 2% in β-lg-myristic acid (C14:0)-curcumin and 34 ± 3% in β-lg-palmitic acid (C16:0)-curcumin, and those of curcumin were 58 ± 2% in β-lg-C14:0-curcumin and 57 ± 2% in β-lg-C16:0-curcumin. These findings provide basic information to clarify the binding mechanism for the interactions of β-lg with ligands at multiple sites on the molecular level.
Introduction
In recent years, there has been a growing interest in the food industry to incorporate various bioactive ingredients due to their health benefits. Among them, curcumin (diferuloylmethane), a natural polyphenolic compound derived from the herb turmeric (Curcuma longa), is known to exhibit numerous biological and pharmacological activities, such as anti-tumor, anti-oxidative, and anti-inflammatory properties.1,2 In addition, several clinical trials showed that curcumin is not toxic to humans, even at high doses (up to 8 g per day for 3 months).3 Therefore, curcumin is regarded as a promising nutraceutical to be used in functional foods. However, being a hydrophobic molecule, the poor water solubility and bioaccessibility of curcumin limit its application and biological activities. To enhance the solubility and bioavailability of curcumin, a number of researchers suggested complexation of curcumin with bio-based polymeric ingredients, e.g. albumin, phospholipid, casein micelles, chitosan, and modified starches.4–8Milk protein-based nanoparticles are promising delivery vehicles for nutraceuticals because milk proteins have naturally evolved to deliver nutrients from mother to neonate. Bovine β-lactoglobulin (β-lg) is the major whey protein from bovine milk. Bovine β-lg consists of a 162 amino acid-long mature polypeptide with a molecular weight of approximately 18.3 kDa.9 It has a hydrophobic core, containing eight antiparallel β-strands (A–H), which is called the calyx and is the main binding site of the protein. Furthermore, β-lg is conditional resistant to acidity at low pH of stomach, and to digestion by gastric proteases.10,11 Therefore, it is an attractive candidate to serve as an encapsulant for delivering hydrophobic nutrients and improving their bioavailability. For example, β-lg-based delivery strategies have been developed to carry vitamin D and naringenin to improve their bioavailability.12,13 Moreover, a number of studies demonstrated that the binding of curcumin to β-lg could enhance the solubility and antioxidant activity of curcumin.14,15
However, most of the studies have focused on transporting single ligand or the interferences and competitions of secondary ligand to protein–monoligand complex.16,17 In fact, the trend to incorporate different bioactive compounds into food formulations and offer multiple benefits simultaneously has increased remarkably.18 It is thus required to develop carriers that can bind a plurality of bioactive components at the same time. β-lg has more than one site for ligand binding, with the main site in the calyx and other possible sites at the dimer interface, the outer surface near Trp19-Arg124, and the hydrophobic pocket in a groove between the α-helix and the β-barrel.19–22 Hence, we speculated that β-lg could be used as a carrier for more than one type of hydrophobic molecule at a time. However, the specific binding behaviors of β-lg delivering multiple ligands simultaneously are still not clear. To this end, three fatty acids, i.e. caprylic acid (C8:0), myristic acid (C14:0), and palmitic acid (C16:0), were selected as the second type of hydrophobic molecule to form ternary complexes with β-lg and curcumin. Previous studies noted that β-lg has different affinities to fatty acids of different structures.23 Therefore, the binding of curcumin to β-lg may be affected by different fatty acids, and vice versa.
The main objective of this study was to clarify the binding mechanism and structural characteristics of ternary complexes of β-lg, curcumin, and fatty acids. Complimentary techniques, i.e. fluorescence spectroscopy, circular dichroism (CD) spectroscopy, dynamic light scattering (DLS), Turbiscan stability analysis, loading efficiencies measurements, molecular docking studies, and molecular dynamics (MD) simulation were applied to measure the binding kinetics and probe the structural details of the complexes. This study provides valuable information for developing new β-lg-based delivery systems for multiple ligands with applications in the food industry.
Materials and methods
Materials
Bovine β-lg (purity > 90%) was purchased from Sigma-Aldrich Chemical Co. (St. Louis, Missouri). Curcumin (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 3.6, purity > 98%) was obtained from J&K chemical (Shanghai, China). C8:0 (log
P = 3.6, purity > 98%) was obtained from J&K chemical (Shanghai, China). C8:0 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 2.9, purity > 98%), C14:0 (log
P = 2.9, purity > 98%), C14:0 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 6.1, purity > 99%), and C16:0 (log
P = 6.1, purity > 99%), and C16:0 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 7.2, purity > 97%) were purchased from Tokyo Chemical Industry (Tokyo, Japan). Other reagents were of analytical grade, and double distilled water was used throughout.
P = 7.2, purity > 97%) were purchased from Tokyo Chemical Industry (Tokyo, Japan). Other reagents were of analytical grade, and double distilled water was used throughout.
Preparation of β-lg, curcumin, and fatty acids complexes
To obtain a protein solution, β-lg was dissolved in an aqueous solution of 10 mM Tris–HCl buffer (pH 7.4). The protein concentration was determined using a molar absorption coefficient of ε278 nm = 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1 on a UV-vis spectrophotometer.24 Fresh curcumin solution (4 mM) was prepared by dissolving it in ethanol, and the solutions were protected from light throughout the experiments. The exact concentration of curcumin was measured using a molar absorption coefficient of ε429 nm = 55
600 M−1 cm−1 on a UV-vis spectrophotometer.24 Fresh curcumin solution (4 mM) was prepared by dissolving it in ethanol, and the solutions were protected from light throughout the experiments. The exact concentration of curcumin was measured using a molar absorption coefficient of ε429 nm = 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1.25 Ethanol solutions of C8:0, C14:0, and C16:0 (1 mM) were also prepared in advance.
000 M−1 cm−1.25 Ethanol solutions of C8:0, C14:0, and C16:0 (1 mM) were also prepared in advance.
The above solutions were diluted in Tris–HCl to various concentrations required for further preparing the complex samples. The complex samples were prepared by mixing the diluted solutions in varying proportions. The resulting ethanol concentration never exceeded 10% (v/v), and this concentration had no appreciable effect on protein structure.16
Fluorescence measurements
To reveal the binding behaviors between β-lg and ligands, changes of β-lg tryptophan (Trp) fluorescence and curcumin fluorescence emission intensity were measured. First, curcumin (0–50 μM) was added into the β-lg solution and the β-lg-fatty acid complexes, in which the concentrations of β-lg and fatty acid were 2 μM and 300 μM, respectively. Moreover, in order to clarify the effect of fatty acids on the binding kinetics between curcumin and β-lg, the binding constants, Ka, were calculated according to the fluorescence intensities of β-lg. In this case, the concentrations of curcumin were 0, 1.6, 6, 10, 16, 30, 50, 60, 80, 100, 150, 200 μM. Secondly, to assess the effect of curcumin presence on the binding between fatty acids and β-lg, each fatty acid (0–200 μM) was added into the β-lg solution (2 μM) and the β-lg-curcumin complex containing 2 μM of β-lg and 150 μM of curcumin.Fluorescence measurements were performed on a CARY Eclipse spectrofluorometer (Varian, Palo Alto, California). Binding of these ligands was evaluated using the fluorescence emission quenching of β-lg Trp (λex = 295 nm, λem = 330 nm) and the change in fluorescence emission intensity for curcumin (λex = 425 nm, λem = 500 nm). The fluorescence intensities were shown as the average of three replicates. Because some ligands possess intrinsic fluorescence, the intensities of β-lg and curcumin in the samples were automatically subtracted from the emission intensities of the corresponding solutions. In addition, some ligands are able to absorb energy both at the excitation wavelength and emission wavelength, which is known as the inner-filter effect. Therefore, the measured fluorescence emission intensity was corrected according to eqn (1):26
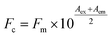 | (1) |
CD measurements
CD spectra were recorded using a CD spectropolarimeter (Pistar π-180, Applied Photophysics Ltd., Surrey, U.K.). The concentration of β-lg was 10 μM for the far UV region (190–260 nm). The molar ratio of β-lg, curcumin, and fatty acids was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the ternary complexes. The path length was 0.1 cm. Ellipticity was recorded at a speed of 100 nm min−1, 0.2 nm resolution, 20 accumulations, and 2.0 nm bandwidth. The collected data were analyzed on Dichroweb (http://dichroweb.cryst.bbk.ac.uk). The proportions of α-helix, β-sheet, turn, and unordered coil were estimated using the SELCON3 program.27 The buffer background was subtracted from the raw spectra, and each spectrum was obtained as the average of three consecutive scans.
1 in the ternary complexes. The path length was 0.1 cm. Ellipticity was recorded at a speed of 100 nm min−1, 0.2 nm resolution, 20 accumulations, and 2.0 nm bandwidth. The collected data were analyzed on Dichroweb (http://dichroweb.cryst.bbk.ac.uk). The proportions of α-helix, β-sheet, turn, and unordered coil were estimated using the SELCON3 program.27 The buffer background was subtracted from the raw spectra, and each spectrum was obtained as the average of three consecutive scans.
DLS measurements and stability analysis
The concentrations of β-lg, curcumin, and fatty acids were 200 μM in the β-lg-curcumin, β-lg-fatty acids, and the ternary complexes. For DLS measurements, the samples (1.5 mL) were analyzed at 25 °C with a Zetasizer Nano-ZS90 (Malvern Instruments, Worcestershire, U.K.). The intensity of light scattered was monitored at 173° for size measurements. The results were recorded as the cumulant mean diameter (nm) and polydispersity index for particle size and size distribution, respectively. All experiments were performed in triplicates. For the zeta potential, the values reported are the average of measurements for three freshly prepared samples, and three readings were recorded for each sample.The stability of the ternary complexes at 4 °C during 14 days storage were monitored by two methods, DLS measurement and Turbiscan analysis. The particle sizes, zeta potentials and Turbiscan curves were recorded at 1, 2, 3, 4, 5, 6, 7, 10, 12, and 14 d. The Turbiscan stability analysis was determined using Turbiscan LAB (Formulaction, Toulouse, France). The Turbiscan scanned the samples from the bottom to the top with near infrared light (λ = 880 nm) and measured the percentage of transmission through the samples as a function of the height of the tube, in order to quantify the rate of destabilisation.
Loading efficiencies of curcumin and fatty acids
To assess the loading efficiencies of curcumin and fatty acids within the complexes, 500 μL β-lg-curcumin, β-lg-fatty acids, and the ternary complexes were prepared. The concentrations of β-lg, curcumin, and fatty acids were 37.5 μM, 75 μM, and 75 μM, respectively. The samples were transferred into Amicon Ultra centrifugal filter devices with a molecular weight cutoff of 10 kDa (Millipore, Boston, MA). The curcumin and fatty acids bound to β-lg were separated from the free ligands by centrifugal ultrafiltration (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 1 h). The retentate above the membrance was removed. After this, the ligands remaining in the membrance were collected by running another washing step based on the method described in previous study.28 The free ligands in the both two permeates were analyzed by HPLC. For reference samples, curcumin and fatty acids dissolved in ethanol were added to Tris–HCl solutions at the same pH and composition as that used for the complexes, but without the protein. Before this, an experiment was performed to confirm that after an additional washing step, the vast majority of the free ligands could be collected. The loading efficiency was calculated using the following equation:28
000 × g, 1 h). The retentate above the membrance was removed. After this, the ligands remaining in the membrance were collected by running another washing step based on the method described in previous study.28 The free ligands in the both two permeates were analyzed by HPLC. For reference samples, curcumin and fatty acids dissolved in ethanol were added to Tris–HCl solutions at the same pH and composition as that used for the complexes, but without the protein. Before this, an experiment was performed to confirm that after an additional washing step, the vast majority of the free ligands could be collected. The loading efficiency was calculated using the following equation:28
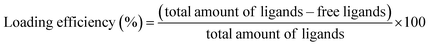 | (2) |
The amounts of curcumin and fatty acids were quantified using a Shimadzu LC-20AT HPLC instrument (Kyoto, Japan). The analysis was performed on a C18 HPLC column, with UV detection at 425 nm for curcumin and 310 nm for the fatty acids after derivatization. Fatty acids (50 μL) were directly derivatized by treatment with 2-nitrophenylhydrazine hydrochloride (100 μL) in the presence of 200 μL of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride, to obtain non-volatile hydrazine derivatives. The derivatized fatty acids were separated from other acid hydrazides and interfering components by a solvent extraction using 1.5 mL of n-hexane. After that, the samples were treated under a stream of nitrogen gas, and the residues were dissolved in methanol. The mobile phase was a mixture of water (A) and acetonitrile (B) at flow rate of 0.8 mL−1 min−1, and acetic acid (0.12% v/v) was added to the mobile phase. For curcumin, the injection volume was 100 μL, and the gradient elution program was as follows: 0–9 min, 44% B to 62% B; 9–30 min, 62% B. For the derivatized fatty acids, the injection volume was 40 μL, and the gradient elution condition was as follows: 0–5 min, 75% B; 5–30 min, 95% B. Four replicates were made to construct the calibration curve with curcumin and fatty acids standard solutions, ranging from 0 to 100 μM.
Molecular docking studies and MD simulation
The three dimension structure of β-lg was downloaded from RCSB Protein Data Bank (http://www.rcsb.org) with the ID 2Q2M. The structure of curcumin, C14:0, and C16:0 were obtained from NCBI with the PubChem compound CIDs of 969516, 11005, and 985, respectively. Binding interactions between ligands and protein were predicted by molecular docking method using AutoDock 4.0 software. All water molecules were removed and polar hydrogens were added at the beginning of the docking experiment. The docking experiment was performed on the whole protein. To recognize binding sites in the protein, the grid center was set at 80 Å, 80 Å, 80 Å. For more accurate docking, the optimized AutoDocking parameters were as follows: the maximum number of energy evaluations were 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per run; the iterations of Solis and Wets local search were 3000; the number of generations was 100, and the number of individuals in population was 300. All other parameters were kept as default values. Result differing by less than 2 Å in a positional root mean square deviation (RMSD) were clustered together. The results of molecular docking were shown by Discovery Studio 4.0 (Accelrys, San Diego, California) to understand the binding interactions and get the lowest energy conformations of the obtained complexes. To investigate the stability of ligand–protein complex, MD simulation was also used. The most favorable ligand–protein complex obtained from docking calculations was regarded as the initial geometry for MD via Materials Studio Version 8.0 (Accelrys, San Diego, California). All MD simulations were performed by using the Amorphous Cell and Forcite module of the Material Studio simulation package with COMPASS. In the MD simulation, first, the amorphous cell module was employed to construct a box, which was inundated with 200 water molecules and the ligand–protein complex. Then, to equilibrate the system, the solute was subjected to the position-restrained dynamics simulation of NPT at 300 K for 20 ns. The RMSD of a selected element can be calculated by least-squares fitting the structure to the reference structure. The results were displayed in Discovery Studio 3.0 (Accelrys, San Diego, California).
000 per run; the iterations of Solis and Wets local search were 3000; the number of generations was 100, and the number of individuals in population was 300. All other parameters were kept as default values. Result differing by less than 2 Å in a positional root mean square deviation (RMSD) were clustered together. The results of molecular docking were shown by Discovery Studio 4.0 (Accelrys, San Diego, California) to understand the binding interactions and get the lowest energy conformations of the obtained complexes. To investigate the stability of ligand–protein complex, MD simulation was also used. The most favorable ligand–protein complex obtained from docking calculations was regarded as the initial geometry for MD via Materials Studio Version 8.0 (Accelrys, San Diego, California). All MD simulations were performed by using the Amorphous Cell and Forcite module of the Material Studio simulation package with COMPASS. In the MD simulation, first, the amorphous cell module was employed to construct a box, which was inundated with 200 water molecules and the ligand–protein complex. Then, to equilibrate the system, the solute was subjected to the position-restrained dynamics simulation of NPT at 300 K for 20 ns. The RMSD of a selected element can be calculated by least-squares fitting the structure to the reference structure. The results were displayed in Discovery Studio 3.0 (Accelrys, San Diego, California).
Statistical analysis
All of the data were analyzed using SPSS 17.0 package (SPSS Inc., Chicago, Illinois) and are expressed as means ± standard deviations. One-way analysis of variance was used to estimate the statistical differences. Significant differences (p < 0.05) between means were determined using Duncan's multiple range tests.Results and discussion
Fluorescence measurements
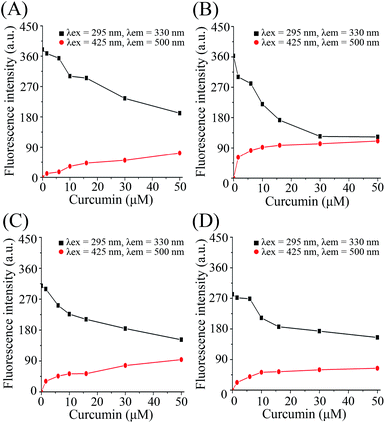
Fluorescence spectroscopy was used to monitor the intermolecular binding behavior of β-lg and curcumin, both of which produce fluorescence emission upon different excitation wavelengths. Binding of β-lg and curcumin will change the polarity of the surrounding environment of the fluorophores, and thus affect their fluorescence intensities. At a fixed curcumin concentration (2 μM), the fluorescence intensity of curcumin increased with increasing β-lg concentration (Fig. S1†). Apparently, with the addition of β-lg, curcumin was transferred from a relatively more hydrophilic aqueous environment to a more hydrophobic environment, i.e. the binding sites on β-lg.When curcumin was gradually added to the β-lg and β-lg-fatty acids complexes, a decrease in β-lg fluorescence intensity at 330 nm and a concurrent increase in curcumin fluorescence emission intensity at 500 nm were observed (Fig. 1). It indicated that curcumin bound to β-lg despite the presence of fatty acids. Furthermore, the binding constants of curcumin to β-lg in the absence and in the presence of fatty acids were calculated. To this end, the fluorescence intensities data were analyzed according to the following equation:26
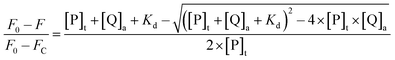 | (3) |
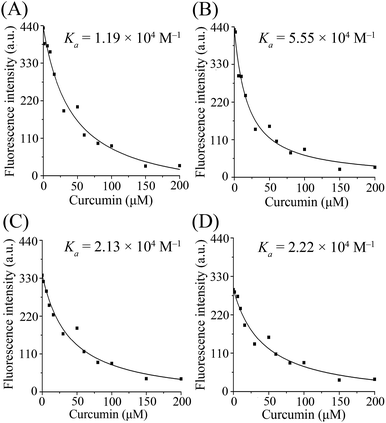
The values of binding constants and the non-linear regressions fit based on eqn (3) are shown in Fig. 2. The binding constant of curcumin, Ka, is 1.19 × 104 M−1 for β-lg-curcumin, and the Ka is higher in the ternary complexes, 5.55 × 104 M−1 in β-lg-C8:0-curcumin, 2.13 × 104 M−1 in β-lg-C14:0-curcumin, and 2.22 × 104 M−1 in β-lg-C16:0-curcumin. Therefore, it was evident that fatty acids facilitate the binding of curcumin to β-lg.
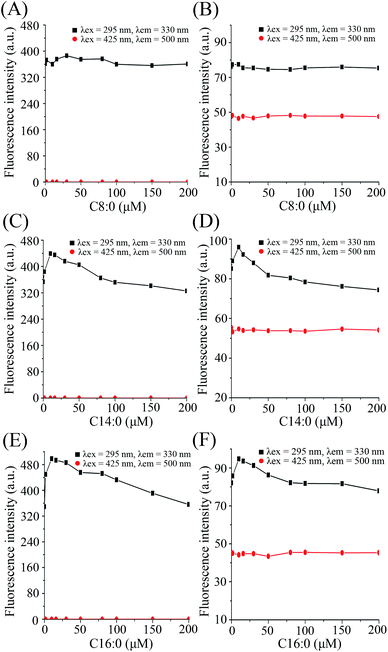
When C14:0 and C16:0 were gradually added into the β-lg solution and β-lg-curcumin complex, the Trp fluorescence intensities approached a maximum at 10 μM fatty acid concentration, and then gradually decreased (Fig. 3). It revealed that when β-lg was saturated with curcumin in advance, C14:0 and C16:0 could still interact with β-lg. While the addition of C8:0 had little influence on the fluorescence intensity of β-lg Trp. Similar observations were seen in previous study which clarified the crystallographic structure of C8:0 binding to β-lg.29 In fact, the results were due to the lower affinity of β-lg-C8:0 compared to those of other fatty acids with longer chains. It was demonstrated that the binding affinity of fatty acids to β-lg increased as the hydrocarbon chain lengthened, and the highest binding affinity was observed in β-lg-C16:0 complex.23,30 The main reasons were that the addition of methylene groups in hydrocarbon chain could increase the hydrophobic interactions between fatty acids and β-lg, and furthermore, β-lg binding pocket for fatty acid accommodated the best with an aliphatic chain constituted by 16 carbon atoms.23 Similar results were reported when fatty acids bound to myoglobin.31 In addition, the fluorescence of curcumin at 500 nm showed no significant modification with the additions of fatty acids, which meant that the binding of fatty acids did not induce release of curcumin from the complexes.
CD measurements
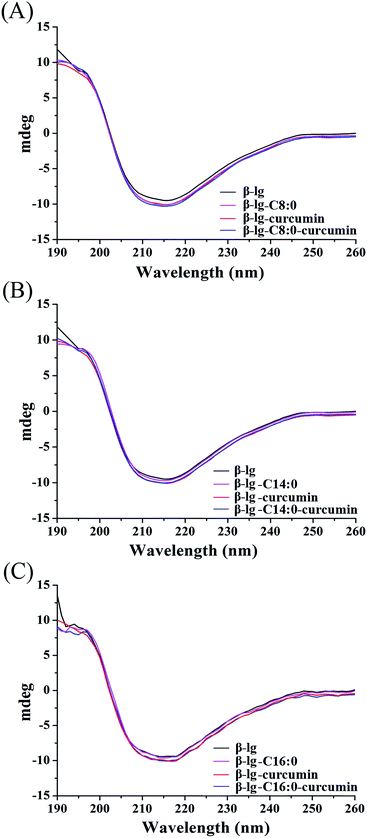
To determine the effect of binding behaviors on the conformation of protein, the conformational transitions of β-lg in β-lg-curcumin, β-lg-fatty acids, and the ternary complexes were monitored by far UV CD spectroscopy (Fig. 4). The far UV CD spectrum showed a broad negative minimum around 217 nm, which was characteristic of β-sheet structure. After the addition of curcumin and fatty acids, the far UV CD spectra remained similar to that of native β-lg, indicating that the binding of ligands to β-lg did not significantly affect the secondary structure of the protein. Furthermore, the proportions of secondary structures also showed little difference among the samples (Table 1). The effect of ligands binding on secondary structure of protein are various in different binding behaviors. It was reported that the interaction of genistein bound to bovine serum albumin led to conversion of the α-helix to β-sheet and turn structures due to a partial protein unfolding.4 While, in our study, the results were in agreement with several previous studies, which had shown that the interaction of ligands with β-lg has no significant effect on protein secondary structure.15,32 It was suggested that the loops of β-lg was relatively flexible, and β-lg did not require extensive changes the structures to allow ligand binding in the calyx.33,34| Samples | α-Helix (±1%) | β-Sheet (±3%) | Turn (±1%) | Unordered (±2%) |
|---|---|---|---|---|
| β-lg | 10.4 | 47.5 | 15.4 | 26.7 |
| β-lg-curcumin | 10.3 | 47.5 | 16.5 | 25.7 |
| β-lg-C8:0 | 10.4 | 47.1 | 15.3 | 26.1 |
| β-lg-C14:0 | 10.1 | 48.3 | 15.5 | 25.9 |
| β-lg-C16:0 | 9.4 | 48.7 | 16.8 | 25.7 |
| β-lg-C8:0-curcumin | 10.3 | 47.7 | 15.7 | 24.7 |
| β-lg-C14:0-curcumin | 9.4 | 49.2 | 15.8 | 25.6 |
| β-lg-C16:0-curcumin | 8.9 | 48.8 | 16.7 | 25.6 |
DLS measurements and stability analysis
The interactions between β-lg and curcumin or the fatty acids were evaluated in terms of particle sizes and zeta potentials (Table 2), and the sample particle size distribution curves are shown in Fig. S2.† The polydispersity index remained within an acceptable range (< 0.5) in all samples (data not shown). The particle sizes and particle size distributions of the β-lg-fatty acids were close to those of β-lg, as the dimeric structures were mostly unaffected by ligand binding. By contrast, the values of mean size increased dramatically with the addition of curcumin (from 6.4 ± 0.3 nm to over 110 nm). The previous study also found that complexation with curcumin was seen to significantly increase the particle size of soy protein isolate nanoparticles.35 It was reported that the addition of curcumin accompanying with ethanol could expose more hydrophobic sites of β-lg, and deprived the surface charge. This process caused the formation of larger particles since hydrophobic association increased and electrostatic repulsion reduced.11 When C8:0 was added to form ternary complex, the particle size of complex showed little difference. While, with the addition of C14:0 and C16:0, further significant increases in particle sizes compared to β-lg-curcumin were observed (p < 0.05), from 113 ± 2 nm to 145 ± 2 nm and 146 ± 4 nm, respectively. The reason of the increased particle size in ternary complexes may be related to the aggregation and cross-linking of β-lg caused by curcumin. The specific mode might be similar to the lactoferrin–EGCG aggregates, in which EGCG acted as linkage among proteins.36| Samples | Size (nm) | Zeta potential (mV) |
|---|---|---|
| a Means with different letters in the same column are significantly different at p < 0.05. | ||
| β-lg | 6.4 ± 0.3 a | −11.6 ± 0.2 a |
| β-lg-curcumin | 113 ± 2 b | −30.4 ± 0.3 e |
| β-lg-C8:0 | 6.5 ± 0.2 a | −10.5 ± 0.5 a |
| β-lg-C14:0 | 6.7 ± 0.4 a | −23.5 ± 0.4 c |
| β-lg-C16:0 | 6.9 ± 0.2 a | −20.1 ± 0.5 b |
| β-lg-C8:0-curcumin | 115 ± 3 b | −25.9 ± 1.3 d |
| β-lg-C14:0-curcumin | 145 ± 2 c | −29.1 ± 0.9 e |
| β-lg-C16:0-curcumin | 146 ± 4 c | −30.8 ± 1.2 e |
The zeta potential results indicated that all samples were negatively charged, and did not exceed absolute values of 40 mV, which is the signal of complex stabilization by electrostatic interactions.37 Thus, the physical stabilities of the samples could not be explained by electrostatic stabilization, and this suggested that other forces, including hydrophobic interactions, Van der Waals interactions or steric overlap interactions, were involved. Except for C8:0, all ligands significantly increased the negative charges of β-lg (p < 0.05), revealing a higher colloidal stability upon their addition. The zeta potentials of the β-lg-fatty acids (C14:0 and C16:0) were around −20.0 mV, and that of β-lg-curcumin was −30.4 ± 0.3 mV. The biggest change was observed between β-lg (−11.6 ± 0.2 mV) and β-lg-C16:0-curcumin (−30.8 ± 1.2 mV).
Moreover, the stability of the ternary complexes were evaluated by DLS measurement and Turbiscan stability analysis. As shown in Fig. S3,† the particle sizes and zeta potentials of the ternary complexes showed little differences during storage time. The Turbiscan curves also observed no destabilization phenomena in the complexes (Fig. S4†). Therefore, the results of both two methods verified that the ternary complexes were stable at 4 °C for at least 14 days.
Loading efficiencies
The loading efficiencies of curcumin and fatty acids in their β-lg complexes are presented in Table 3. Since C8:0 resulted in nearly no changes in fluorescence and dynamic light scattering measurements, the loading efficiencies of fatty acids were evaluated in the complexes including C14:0 and C16:0. The loading efficiency of curcumin in β-lg-curcumin was 49 ± 2%, and this significantly increased in the presence of fatty acids (p < 0.05). The biggest increase was observed when C14:0 was added, with a loading efficiency of 58 ± 2% for β-lg-C14:0-curcumin. In terms of fatty acids, the loading efficiency of C14:0 was 32 ± 2% in β-lg-C14:0-curcumin and that of C16:0 was 34 ± 3% in β-lg-C16:0-curcumin. On the basis of the loading efficiencies of curcumin and fatty acids, it showed that the ternary complexes were able to deliver curcumin and the fatty acids simultaneously, and furthermore, the presence of fatty acids enhanced the loading efficiencies of curcumin in the complexes.| Samples | Curcumin (%) | Fatty acids (%) |
|---|---|---|
| a Means with different letters in the same column are significantly different at p < 0.05. N/A, not applicable. | ||
| β-lg-curcumin | 49 ± 2 a | N/A |
| β-lg-C14:0 | N/A | 46 ± 1 b |
| β-lg-C16:0 | N/A | 47 ± 2 b |
| β-lg-C14:0-curcumin | 58 ± 2 b | 32 ± 2 a |
| β-lg-C16:0-curcumin | 57 ± 2 b | 34 ± 3 a |
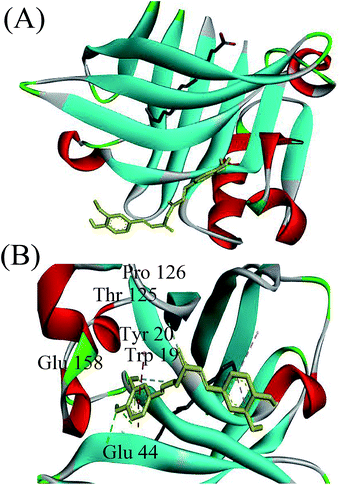
Binding behaviors in the formation of the ternary complexes
To determine the preferred binding sites on β-lg, docking experiments were performed with curcumin. When β-lg interacted with curcumin alone, the results showed that curcumin was surrounded by Phe105, Val41, Lys60, Leu46, Leu54, Ile56, and Val92 with hydrophobic interactions, and Pro38 with hydrogen bonds (Fig. S5†). These amino acids were located near the calyx, with Ile, Leu and Val lining the wall of the calyx, and Phe105 in an orientation that was suitable to establish a π–π interaction with the phenol ring of curcumin.15 Thus, the result indicated that curcumin binding to β-lg in the calyx occurred mainly through hydrophobic interactions. According to a crystallographic study, the primary binding site of C14:0 and C16:0 in β-lg are also the calyx, with end of ligand hydrocarbon chain bound deep in the hydrophobic part of the calyx and carboxyl group located near the calyx entrance.38 With respect to the ternary complexes, when C16:0 occupied the calyx site first, there was another binding site at the surface region for curcumin (Fig. 5). The result for C14:0 was similar to that for C16:0 (data not shown). Curcumin located near Trp19, Tyr20, Thr125, Pro126, Glu158 and Glu44 through both hydrophobic interactions and hydrogen bonds, which was similar to the binding site in β-lg–resveratrol and β-lg–isoxazole interactions.39,40 Moreover, the binding mode for curcumin out of the β-lg calyx was still stable when solvation and flexibility was present in the biological system (Fig. S6†). C14:0 and C16:0 filled available spaces in the calyx with the size and shape corresponding well to geometry of the binding site, and Phe105, a key protein residue in the interaction, could specially provide an additional hydrophobic clamp for C16:0.29,38 Nevertheless, similar phenomenon was not observed with curcumin and thus might result in the preference for fatty acids appearing in the calyx. While, the presence of C8:0 showed little effects on the binding of curcumin since its hydrocarbon tail is shorter compared to the other fatty acids, and thus could not insert deep in the protein calyx.29 Moreover, fatty acid, especially C16:0, is considered to be natural β-lg ligands with relative high affinity compared with other ligands.38 The binding affinities of C14:0 and C16:0 are ranged between 105–107 M−1,30 which is much higher than that obtained for curcumin in this study (1.19 × 104 M−1). Therefore, the possible mechanisms involved in the formation of the ternary complexes are shown in Fig. 6. C14:0 and C16:0 were more likely to be located in the calyx preferentially over curcumin, resulting in the curcumin occupied in a different binding site. Certainly, the simulations provide preliminary information, and further studies need to be carried out to unveil the more detailed mechanism.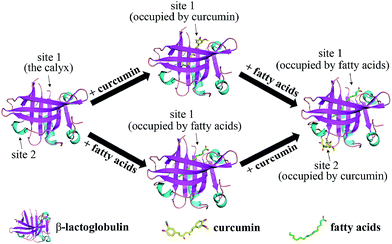 | ||
| Fig. 6 Schematic representation of the possible mechanisms involved in the formation of the ternary complexes of β-lg, curcumin, and fatty acids (C14:0 or C16:0). | ||
Conclusions
Our results showed that β-lg could interact with curcumin and the fatty acids simultaneously. Compared to the interaction of single ligand with β-lg, the binding constant of curcumin enhanced. The particle sizes of the ternary complexes showed significant increases, but the secondary structure of β-lg was unchanged. Furthermore, the loading efficiencies of the fatty acids (C14:0 and C16:0) were more than 30% in the ternary complexes, and those of curcumin were over 50%. Therefore, the results suggest the β-lg-based nanocarrier could be of great potential to deliver more than one nutraceutical simultaneously in the food industry.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 31401492), the Ministry of Science and Technology of China (Grant No. 2016YFD0400804), and Chinese Universities Scientific Fund (Grant No. 2017QC086). The funders had no role in the design, analysis or writing of this article. We also thank Dr Mengchi Sun from the Key Laboratory of Structure-Based Drug Design and Discovery, Shenyang Pharmaceutical University for his kind assistance with computational simulation.References
- P. Anand, A. B. Kunnumakkara, R. A. Newman and B. B. Aggarwal, Mol. Pharmaceutics, 2007, 4, 807–818 CrossRef CAS PubMed.
- A. J. Ruby, G. Kuttan, K. D. Babu, K. N. Rajasekharan and R. Kuttan, Cancer Lett., 1995, 94, 79–83 CrossRef CAS PubMed.
- A. L. Cheng, C. H. Hsu, J. K. Lin, M. M. Hsu, Y. F. Ho, T. S. Shen, J. Y. Ko, J. T. Lin, B. R. Lin, M. S. Wu, H. S. Yu, S. H. Jee, G. S. Chen, T. M. Chen, C. A. Chen, M. K. Lai, Y. S. Pu, M. H. Pan, Y. J. Wang, C. C. Tsai and C. Y. Hsieh, Anticancer Res., 2001, 21, 2895–2900 CAS.
- P. Bourassa, C. D. Kanakis, P. Tarantilis, M. G. Pollissiou and H. A. Tajmir-Riahi, J. Phys. Chem. B, 2010, 114, 3348–3354 CrossRef CAS PubMed.
- L. Li, F. S. Braiteh and R. Kurzrock, Cancer, 2005, 104, 1322–1331 CrossRef CAS PubMed.
- A. Sahu, N. Kasoju and U. Bora, Biomacromolecules, 2008, 9, 2905–2912 CrossRef CAS PubMed.
- A. Anitha, V. G. Deepagan, V. V. D. Rani, D. Menon, S. V. Nair and R. Jayakumar, Carbohydr. Polym., 2011, 84, 1158–1164 CrossRef CAS.
- H. Yu and Q. Huang, Food Chem., 2010, 119, 669–674 CrossRef CAS.
- B. Y. Qin, M. C. Bewley, L. K. Creamer, H. M. Baker, E. N. Baker and G. B. Jameson, Biochemistry, 1998, 37, 14014–14023 CrossRef CAS PubMed.
- N. P. Aditya, H. Yang, S. Kim and S. Ko, Colloids Surf., B, 2015, 127, 114–121 CrossRef CAS PubMed.
- Z. Teng, R. Xu and Q. Wang, RSC Adv., 2015, 5, 35138–35154 RSC.
- M. C. Yang, N. C. Chen, C. J. Chen, C. Y. Wu and S. J. T. Mao, FEBS J., 2009, 276, 2251–2265 CrossRef CAS PubMed.
- A. Shpigelman, Y. Shoham, G. Israeli-Lev and Y. D. Livney, Food Hydrocolloids, 2014, 40, 214–224 CrossRef CAS.
- M. Li, Y. Ma and M. O. Ngadi, Food Chem., 2013, 141, 1504–1511 CrossRef CAS PubMed.
- A. H. Sneharani, J. V. Karakkat, S. A. Singh and A. G. A. Rao, J. Agric. Food Chem., 2010, 58, 11130–11139 CrossRef CAS PubMed.
- C. D. Kanakis, P. A. Tarantilis, M. G. Polissiou and H. A. Tajmir-Riahi, J. Biomol. Struct. Dyn., 2013, 31, 1455–1466 CAS.
- X. Wang, Y. Liu, L. L. He, B. Liu, S. Y. Zhang, X. Ye, J. J. Jing, J. F. Zhang and M. Gao, Food Chem. Toxicol., 2015, 78, 42–51 CrossRef CAS PubMed.
- Y. Wu, H. Cheng, Y. Chen, L. Chen, Z. Fang and L. Liang, J. Agric. Food Chem., 2017, 65, 3019–3030 CrossRef CAS PubMed.
- G. Kontopidis, C. Holt and L. Sawyer, J. Dairy Sci., 2004, 87, 785–796 CrossRef CAS PubMed.
- A. Mensi, Y. Choiset, H. Rabesona, T. Haertle, P. Borel and J. M. Chobert, J. Agric. Food Chem., 2013, 61, 4114–4119 CrossRef CAS PubMed.
- P. Zimet and Y. D. Livney, Food Hydrocolloids, 2009, 23, 1120–1126 CrossRef CAS.
- M. Bello, J. Biomol. Struct. Dyn., 2016, 34, 1884–1902 CAS.
- D. Frapin, E. Dufour and T. Haertle, J. Protein Chem., 1993, 12, 443–449 CrossRef CAS PubMed.
- M. Narayan and L. J. Berliner, Biochemistry, 1997, 36, 1906–1911 CrossRef CAS PubMed.
- F. Mohammadi, A. K. Bordbar, A. Divsalar, K. Mohammadi and A. A. Saboury, Protein J., 2009, 28, 117–123 CrossRef CAS PubMed.
- M. Pantusa, L. Sportelli and R. Bartucci, Eur. Biophys. J., 2012, 41, 969–977 CrossRef CAS PubMed.
- W. Yang, C. Xu, F. Liu, C. Sun, F. Yuan and Y. Gao, J. Agric. Food Chem., 2015, 63, 5046–5054 CrossRef CAS PubMed.
- A. Shpigelman, Y. Cohen and Y. D. Livney, Food Hydrocolloids, 2012, 29, 57–67 CrossRef CAS.
- J. Loch, A. Polit, A. Gorecki, P. Bonarek, K. Kurpiewska, M. Dziedzicka-Wasylewska and K. Lewinski, J. Mol. Recognit., 2011, 24, 341–349 CrossRef CAS PubMed.
- S. Le Maux, S. Bouhallab, L. Giblin, A. Brodkorb and T. Croguennec, Dairy Sci. Technol., 2014, 94, 409–426 CrossRef CAS PubMed.
- S. V. Chintapalli, S. Jayanthi, P. L. Mallipeddi, R. K. Gundampati, S. K. T. Krishnaswamy, D. B. van-Rossum, A. Anishkin and S. H. Adams, J. Biol. Chem., 2016, 291, 251133–251143 CrossRef PubMed.
- L. Liang, H. A. Tajmir-Riahi and M. Subirade, Biomacromolecules, 2008, 9, 50–56 CrossRef CAS PubMed.
- M. Bello and E. Garcia-Hernandez, Biopolymers, 2014, 101, 744–757 CrossRef CAS PubMed.
- L. Dominguez-Ramirez, E. D. Moral-Remirez, P. Cortes-Hernandez, M. Garcia-Garibay and J. Jimenez-Guzman, PLoS One, 2013, 8, e79530 CAS.
- F. P. Chen, B. S. Li and C. H. Tang, J. Agric. Food Chem., 2015, 63, 3559–3569 CrossRef CAS PubMed.
- W. Yang, F. Liu, C. Xu, C. Sun, F. Yuan and Y. Gao, J. Agric. Food Chem., 2015, 63, 5035–5045 CrossRef CAS PubMed.
- O. E. Perez, T. David-Birman, E. Kesselman, S. Levi-Tal and U. Lesmes, Food Hydrocolloids, 2014, 38, 40–47 CrossRef CAS.
- J. I. Loch, A. Polit, P. Bonarek, D. Olszewska, K. Kurpiewska, M. Dziedzicka-Wasylewska and K. Lewinski, Int. J. Biol. Macromol., 2012, 50, 1095–1102 CrossRef CAS PubMed.
- M. Pantusa, R. Bartucci and B. Rizzuti, J. Agric. Food Chem., 2014, 62, 4384–4391 CrossRef CAS PubMed.
- S. Maity, S. Pal, S. Sardar, N. Sepay, H. Parvej, J. Chakraborty and U. C. Halder, RSC Adv., 2016, 6, 112175–112183 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09012a |
| This journal is © The Royal Society of Chemistry 2017 |