Open Access Article
Open Access ArticleZwitterionic poly(sulfobetaine methacrylate) hydrogels incorporated with angiogenic peptides promote differentiation of human adipose-derived stem cells†
Chung-Wei Kaoa,
Po-Hsiu Chenga,
Po-Ting Wua,
Shih-Wen Wanga,
I.-Chun Chena,
Nai-Chen Chengb,
Kai-Chiang Yang c and
Jiashing Yu
c and
Jiashing Yu *a
*a
aDepartment of Chemical Engineering, National Taiwan University, Taipei, 106, Taiwan. E-mail: jiayu@ntu.edu.tw
bNational Taiwan University Hospital, Department of Surgery, Taipei, 100, Taiwan
cSchool of Dental Technology, College of Oral Medicine, Taipei Medical University, Taipei, 106, Taiwan
First published on 3rd November 2017
Abstract
The superhydrophilic and ultralow biofouling properties as well as the resistance to foreign-body reaction make zwitterionic polymer promising in biomedical applications. Incorporation of arginine–glycine–aspartate (RGD) sequence and/or vascular endothelial growth factor mimicking (QK) peptide to zwitterionic poly(sulfobetaine methacrylate) (PSBMA) hydrogels may not only improve cell adhesion but also influence stem cell differentiation. In this study, PSBMA hydrogels were modified with RGD or RGD/QK peptides and characterized. Multipotent human adipose-tissue derived stem cells (hASCs) were seeded to peptides-grafted PSBMA hydrogels, and the cell adhesion, proliferentiation, and differentiation of hASCs on hydrogels were demonstrated. The results showed that grafting of RGD or RGD/QK peptides improved hASCs adhesion, while the mechanical strength of peptides-grafted PSBMA hydrogels was decreased. Incorporation of RGD or RGD/QK peptides did not influence the adsorption adsorption behaviors of PSBMA hydrogels. Relative to RGD-grafted PSBMA hydrogels, the RGD/QK incorporation further improved hASCs differentiated into adipogenic lineage, while RGD or RGD/QK additions had no substantial effects on osteogenesis under osteogenic induction. Moreover, the endothelial differentiation was also enhanced when hASCs cultured on RGD/QK-modified PSBMA hydrogels. In conclusion, the graftings of RGD and RGD/QK did not interfere the nonfouling property of PSBMA hydrogels. The incorporation of RGD/QK peptides can improve hASCs differentiate toward adipogenic/endothelial lineages.
Introduction
Nonfouling materials, which can prevent nonspecific protein absorption and undesirable cell adhesion, are proposed for use in implantable blood-contacting devices and antimicrobial applications.1 With the properties of superhydrophilic and ultralow biofouling, zwitterionic polymers are promising in some biomedical applications. Zwitterionic poly(carboxybetaine methacrylate) (PCBMA) hydrogels are reported to resist foreign-body reactions in mice, which can prevent the formation of a dense fibrous capsule.2 In addition, PCBMA hydrogel is shown to induce less cell adhesions of fibroblasts than that of poly(2-hydroxyethyl methacrylate)(PHEMA), a widely used anti-adhesive polymer.3 Un like the poly(ethylene glycol) or PHEMA, zwitterionic polymers provide abundant functional groups, which are more convenient for the functionalization with biomolecules. Among these zwitterionic materials, poly(sulfobetaine methacrylate) (PSBMA) contains a large number of anionic SO3− groups for the grafting of different peptides, which make zwitterionic polymers not only resist nonspecific protein adsorption but improve selective cell adhesion.In normal tissues, cells bind to the extracellular matrix through the interaction between integrin and adhesive molecules. For artificial microenvironments, cells do not recognize the real surface but the adsorbed proteins on the surface mediate cell adhesions and subsequently cellular functions. Lacking of cell recognition motifs, synthetic polymers may have poor cell attachments and thus hamper biomedical applications.4 Otherwise, surface free energy, electrostatic interactions, steric repulsion, hydration, and topography all influence cell adhesions.5 Therefore, modification of the synthetic polymers is required to improve the cell attachment.
Enrichment of the material surface with extracellular matrix (ECM) components or their functional motifs is a widely used biological approach to promote cell adhesion. Arginine-glycine-aspartate (RGD) sequence, the principal integrin-binding domain in ECM proteins, is the most widely used synthetic binding motif and can enhance cell adhesion efficiently.6 Likewise, the de novo engineered vascular endothelial growth factor (VEGF) mimicking peptide (QK), a region of the VEGF binding interface, is shown to promote angiogenesis.7 Since long-term survival and success of tissue-engineered constructs depend on neovascularization, QK peptides have also been introduced to scaffolds to improve vascular endothelial cell growth.8,9
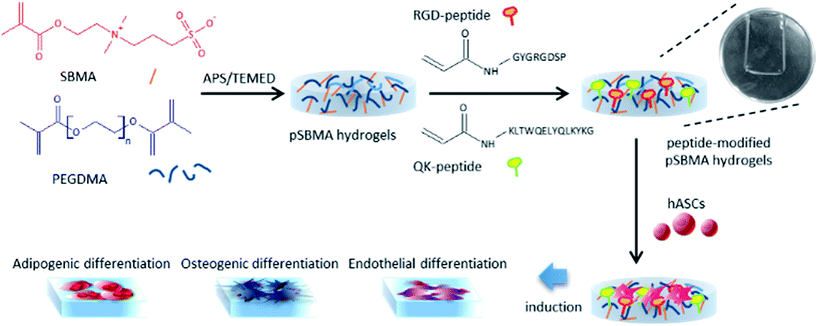
Previously, we showed that incorporation of RGD and RGD/QK peptides into zwitterionic hydrogels improves endothelial cell adhesions and regulates mRNA expressions.10 In addition to having impacts on cell seeding, the matrix stiffness can also direct stem cell differentiation.11 Accordingly, human adipose-tissue derived stem cells (hASCs), a reliable cell source for regenerative medicine, was used to test the zwitterionic PSBMA hydrogels chemically modified with RGD and QK peptides.12 The purpose of this study is to demonstrate the effects of functionalized zwitterionic hydrogels on hASCs adhesion and differentiation to clarify the possible applications in tissue engineering (Fig. 1).
Materials and methods
Preparation of zwitterionic hydrogels
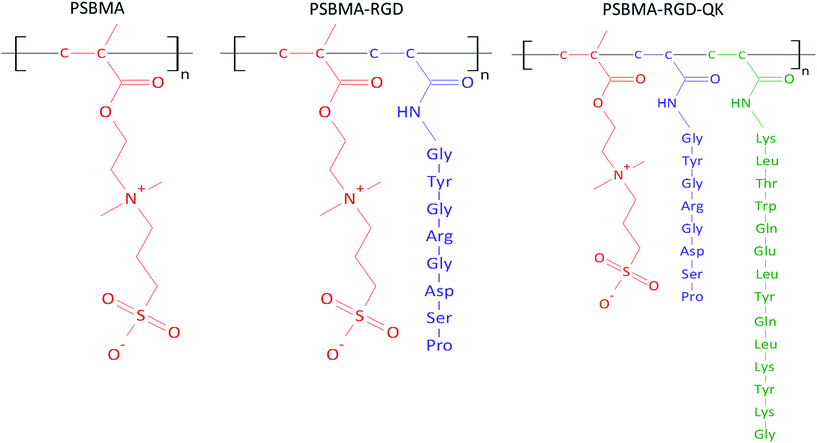
The zwitterionic hydrogel is prepared through free radical polymerization13 and the molecular structures of PSBMA, PSBMA-RGD, and PSBMA-RGD/QK are illustrated in Fig. 2. In brief, sulfobetaine methacrylate monomer (SBMA, 537284, Sigma-Aldrich) was dissolved in phosphate-buffered saline (PBS) at a concentration of 20% (w/v). The crosslinker poly(ethylene glycol) dimethacrylate (PEGDMA, 437468, Sigma-Aldrich) and initiator ammonium persulfate (APS, A3678, Sigma-Aldrich)/N,N,N′,N′-tetramethylethylenediamine (TEMED, T7024, Sigma-Aldrich) were added into the SBMA solution at the concentrations of 1% (w/v) and 5 mM, respectively. The mixed solution was then poured into two glass slides with 1 mm-thick polytetrafluoroethylene spacer and placed at room temperature for 30 minutes. The prepared PSBMA hydrogels were than immersed in PBS for 3 days, and the PBS was changed daily to remove the un-react chemicals.For the peptide-incorporated PSBMA hydrogels, RGD and RGD/QK were added into the solution of SBMA monomer at the final concentrations of 5 mM and 5 mM/2 mM, respectively. The peptide-containing PSBMA solution was then reacted with PEGDMA and APS/TEMED for hydrogel gelation.
Characterization of peptides-incorporated PSBMA hydrogels
The peptides-modified PSBMA hydrogels was characterized by the Fourier-Transform Infrared Spectrometer (FT-IR, Perkin Elmer Spectrum 100, PerkinElmer, USA). The PSBMA hydrogels with/without peptides were stored at −20 °C refrigerator overnight, lyophilized in the freeze-drier, and analyzed in the attenuated total reflection (ATR) mode.The content of PSBMA was determined from the 1H NMR spectrum according to the peak ratio of the methyl protons at 3.2–3.4 ppm, the methylene protons of the polymer main chain at 2.0–2.4 ppm and the methylene protons adjacent to the oxygen at 3.6–4.0 ppm (Fig. S1†). The content of PSBMA-RGD and PSBMA-RGD/QK were also determined from 1H NMR (Fig. S2 and S3†).
The Young's modulus of peptide-incorporated PSBMA hydrogels was measured by using an elastic modulus load cells (LTS-200GA, Ultra Small-capacity Load Cell, Kyowa Electronic Instruments, Japan) with a stepping motor driven stages (SGSP(MS)20-85, Sigma Koki, Japan). The fabricated PSBMA hydrogels with/without peptides were first immersed in PBS overnight. The swelled hydrogels were then cut into cylindrical shapes (5 mm in diameter and 5 mm in height) and the residual liquid was removed from the hydrogels using the filter paper. Finally, the PSBMA hydrogels were set into an unconfined uniaxial compression tester to break 70% strain at a compression velocity 10 μm s−1. The compressive stress–strain ratio was then determined and analyzed by continuous stress recording. Since our preliminary study showed that QK-modified PSBMA hydrogels had no substantial effects on hASCs adhesion, this group was removed in this test.
Protein adsorption to peptides-incorporated PSBMA hydrogels
In order to study the influence of incorporation of peptides on low-fouling property, the protein adsorption behaviors of modified PSBMA was evaluated.14 In brief, the PSBMA and peptides-modified PSBMA hydrogels were loaded into a 96-well tissue culture polystyrene plate (TCPS, 50 μL per well). After incubation in 10% (v/v, fetal bovine serum (FBS, Biological Industries, Israel)) at 37 °C for 1 h, the PSBMA hydrogels were rinsed using PBS and subsequently treated with SDS (0.1% w/v) for 1 h to desorb proteins within the PSBMA hydrogels. The absorbed proteins was quantified using the bicinchoninic acid (BCA) protein assay kit (23225, Pierce, ThermoFisher, USA). The protein adsorptions to un-coating TCPS and polystyrene (PS) plate were also determined.Cell seeding into PSBMA hydrogels
The PSBMA hydrogels were molded into a uniform cylindrical shape (8 mm in diameter and 1 mm in height) by a skin puncher. The cut hydrogels were placed into the 48-well culture plates and then were immersed in culture medium before cell seeding. Subcutaneous adipose tissue from the abdomen was obtained from patients undergoing liposuction or abdominoplasty procedures. The protocols were approved and maintained by Research Ethics Committee at National Taiwan University Hospital under the guidelines of Human Subject Research Acts of Taiwan, R.O.C. Informed consent was obtained from human donors. Isolation, cultivation and identification of hASCs from the stromal vascular fraction were followed our previous study, and proliferative cells (P3–P6) were used in this study.15 The hASCs were detached with trypsin–EDTA, counted, and seeded into the PSBMA hydrogels at a cell density of 5 × 104 cells per cm2. The hASCs/PSBMA hydrogels were cultured in proliferation medium which composed of high glucose Dulbecco's modified Eagle's medium/nutrient mixture F-12 Ham (DMEM/F12, SH30003.02, Thermo Hyclon) supplemented with 10% FBS and 1% antibiotic (Biological Industries, Israel) in an incubator set at 37 °C and 5% CO2. The culture medium was changed every two days.Evaluations of hASCs on PSBMA hydrogels
The adhesion and morphology of hASCs on PSBMA hydrogels were observed using an optical microscope. After culturing on peptide-modified PSBMA hydrogels for 7 and 14 days, the cell survival was also evaluated by using a the LIVE/DEAD® cell imaging kit (R37601, Invitrogen). The hASCs/PSBMA hydrogels were washed twice by PBS and subsequently stained with a solution of 2 μM calcein AM and 4 μM Ethidium homodimer-1 (EthD-1) for 30 minutes. After treatment, the cells per gel samples were observed using a fluorescent microscopy (IX-71, Olympus, Japan).The cell viability of hASCs on PSBMA hydrogels was tested by a colorimetric assay (thiazolyl blue tetrazolium bromide, MTT, L11939, Alfa Aesar). After predetermined time points, the culture medium was removed and cells were washed twice by PBS. The ASCs/PSBMA samples were then cultured in serum-free medium containing 0.5 mg mL−1 MTT reagent for additional 3 h. Finally, the formazan crystals were dissolved by dimethyl sulfoxide (SU0155, Scharlau) and the results were determined using a spectrophotometer at the wavelength of 570 nm.
The cell proliferation of hASC on PSBMA hydrogels was identified by using a lactate dehydrogenase (LDH) assay after 1, 4 and 7 days culture. The cells/hydrogels samples were washed twice using PBS and lysed by incubation with 0.2% Triton-X 100 for 30 minutes. The cell lysates were reacted with the LDH kit and the results were measured using a spectrophotometer at the wavelength of 490 nm. The OD values of LDH assay were normalized to cell number.
Adipogenic, osteogenic and endothelial differentiations to hASCs in PSBMA hydrogels
After cultured in proliferation medium for 3 days, hASCs were induced into adipogenic, osteogenic or endothelial lineage by relevant differentiation medium. The hASCs/PSBMA hydrogels were cultured in adipogenic medium which containing 1 μM dexamethasone (D4902, Sigma-Aldrich), 200 μM indomethacin (I0655, TCI), 500 μM 3-isobutyl-1-methylxanthine (IBMX, I5879, Sigma-Aldrich) and 10 μM insulin (A930615, Humulin) for adipogenic differentiation. The culture medium was changed every two days for 7 days.For osteogenic differentiation, cells on hydrogels were cultured in osteogenic medium that made up of DMEM/H-12, 10 nM dexamethasone, 10 mM β-glycerophosphate (G9422, Sigma-Aldrich), 50 μM ascorbic acid 2-phosphate (A8960, Sigma-Aldrich) and 10 nM 1α,25-dihydroxyvitamin D3 (D1530, Sigma-Aldrich) for 7 days.16,17
Endothelial differentiation of hASCs on PSBMA hydrogels was induced by culturing in endothelial growth medium supplemented with 50 ng mL−1 recombinant human vascular endothelial growth factor (293-VE-010, R&D systems) for 14 days. The medium was changed twice a week.18
RNA extraction and gene expression of the differentiated hASCs
Total RNA of cells on PSBMA hydrogels and peptides-incorporated PSBMA hydrogels was extracted (Total RNA miniprep purification kit, TR01, GeneMark). Since the hASCs were difficult to adhere and grow on PSBMA hydrogels, we seeded hASCs into multiple wells for differentiation, and the extracted RNA were pooled together. The RNA yield was quantified by using a nanodrop spectrophotometer. The cDNA was synthesized using a reverse-transcriptase reagent kit (HiScript I™ First Strand cDNA Synthesis, AM0675-0050, Bionovas) with a PCR system (LifePro Thermal Cycler, Bio-Rad, CA).Reverse transcription-polymerase chain reaction (RT-PCR) was used to analyze adipogenic and osteogenic differentiations. Lipoprotein lipase (LPL) and peroxisome proliferator-activated receptor gamma (PPAR-γ) were selected as target genes for the detections of adipogenesis while collagen type I alpha 1 chain (COL1A1) and alkaline phosphatase (ALP) were used for osteogenesis. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified as an endogenous control. The amplified DNA fragments were separated in a 2.5% agarose gel and stained with 0.5 mg mL−1 ethidium bromide, and the densitometric analysis of the gels was performed with the ImageJ software.
Quantitative real time-polymerase chain reaction (qRT-PCR) was used to detect endothelial differentiation. The expressions of CD31 (platelet endothelial cell adhesion molecule-1, PECAM), CD144 (vascular endothelial cadherin), von Willebrand factor (vWF), and VEGF were analyzed with the housekeeping gene GAPDH. A SYBR system (iQTM SYBR® Green supermix, 170-8882AP, Bio-Rad) was used in qRT-PCR (CFX96, Real-time PCR Thermal Cycler, Bio-Rad, USA). Target gene is normalized to housekeeping gene GAPDH to obtain ΔCT, where CT is the cycle threshold. The ΔCT for cells cultured on different PSBMA hydrogels is further subtracted from the cells cultured on TCPS to obtain ΔΔCT. The level of expression of each target gene was calculated using 2−ΔΔCT. The sequences of the gene-specific primers are shown in Table 1.
| Gene | Primer sequence | Gene | Primer sequence |
|---|---|---|---|
| GAPDH | F: CAAGGCTGAGAACGGGAAGC | CD31 | F: CACAGCAATTCCTCAGGCTA |
| R: AGGGGGCAGAGATGATGACC | R: TTCAGCCTTCAGCATGGTAG | ||
| COL1A1 | F: ACCGCCCTCCTGACGCAC | CD144 | F: ACCCCCACAGGAAAAGAATC |
| R: GCAGACGCAGATCCGGCA G | R: GACTTGGCATCCCATTGTCT | ||
| ALP | F: AACATCAGGGACATTGACGTG | vWF | F: TCTTCCAGGACTGCAACAAG |
| R: GTATCTCGGTTTGAAGCTCTTCC | R: TCCGAGATGTCCTCCACATA | ||
| LPL | F: GTCCGTGGCTACCTGTCATT | VEGF | F: AAGGAGGAGGGCAGAATCAT |
| R: TGGCACCCAACTCTCATACA | R: CACACAGGATGGCTTGAAGA | ||
| PPAR-γ | F: TCAGGTTTGGGCGGATGC | GAPDH | F: CAAGGCTGAGAACGGGAAGC |
| R: TCAGCGGGAAGGACTTTATGTATG | R: AGGGGGCAGAGATGATGACC |
Statistical analysis
All experiments were showed with at least three replicates. Results were reported as average value ± standard deviation. One-way analysis of variance (ANOVA) was used to compare multiple groups of data statically; p values lower than 0.05 were considered statistically significant.Results and discussion
Characterization of PSBMA hydrogels
The grafting of peptides to PSBMA hydrogels were verified by FT-IR (Fig. 3). The RGD and RGD/QK peptide conjugations to PSBMA hydrogels were proved by the presences of peaks at 1750 cm−1 and 1500 cm−1, which represent the amide groups of incorporated peptides (O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N–H).
C–N–H).
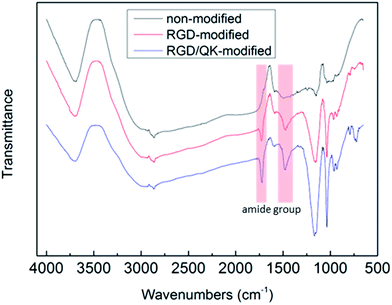 | ||
Fig. 3 FT-IR patterns of peptides-modified PSBMA hydrogels. The peaks at 1750 cm−1 and 1500 cm−1 represent the amide groups of incorporated peptides (O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N–H). C–N–H). | ||
The incorporations of peptides into SBMA decreased the Young's modulus of PSBMA hydrogels significantly. RGD addition decreased Young's modulus of PSBMA hydrogels from 19.9 ± 1.8 kPa to 7.5 ± 1.9 kPa (p < 0.01) while the RGD/QK-modified PSBMA hydrogels has a slightly higher Young's modulus than that of RGD group (10.0 ± 0.6 kPa, significantly lower than that of PSBMA hydrogels but no significant differences to RGD-modified PSBMA group). As the RGD and RGD/QK peptides covalently linked to the SBMA, the available sites for crosslinking were decreased that resulted in a relatively looser hydrogel network. Similar findings were also found in the other studies. The compliance of the hydrogel matrix (Young's modulus) of gelatin methacrylate hydrogel was inversely proportional to the amount of peptide covalently linked to the gelatin matrix.19 A decrease in Young's modulus (ranged from 30-50%) was observed when RGD was grafted onto PSBMA hydrogels.20 Since the stiffness of matrix influences the biological functions of terminally differentiated cells as well as stem cells, this resulting peptides-incorporated PSBMA hydrogels can be used for in the applications of requiring low mechanical strength such as the nucleus pulposus cells21 or tissue engineered muscle.22 Moreover, matrix rigidity is known to effect self-renewal capability of adipose progenitor cells.23 Increase in the SBMA concentration can also improve the compressive strengths of hydrogels, which can extend the applicability of SBMA polymer.10
Protein adsorption to peptides-incorporated PSBMA hydrogels
The adsorbed proteins in PSBMA hydrogels were desorbed by SDS and quantified by BCA kit. The protein adsorption to TCPS was significantly higher than those of PSBMA hydrogels, peptides-incorporated PSBMA hydrogels, and PS (Fig. S4†). However, there was no significant difference in the amounts of protein adsorption among PSBMA hydrogel groups (6.13 ± 0.175 μg cm−2 in un-modified PSBMA, 7.88 ± 0.694 μg cm−2 in RGD-modified PSBMA, and 6.81 ± 1.11 μg cm−2 in RGD/QK-modified PSBMA), which revealed that the incorporation of RGD and RGD/QK peptides did not interfere the nonfouling property of PSBMA hydrogels.Adhesion and survival of hASCs on PSBMA hydrogels
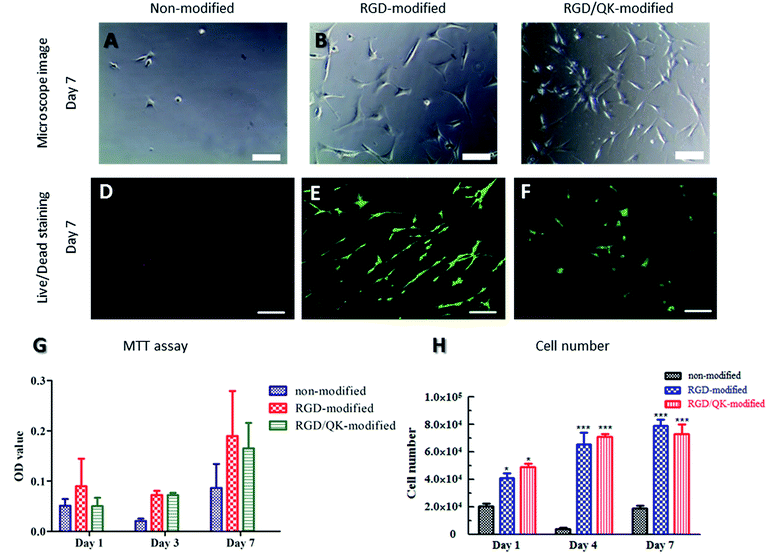
When un-modified PSBMA hydrogels were used in cell culture, the hASCs were difficult to adhere on PSBMA hydrogels. A round shape with un-extending cytoplasm was noticed after 7 days culturing (Fig. 4A). Moreover, the seeded cells were easy to be washed away during medium changing. These findings may be contributed to lack of the adhesion motifs in the matrixes in PSBMA hydrogels and these evidences well represent the non-fouling property of PSBMA polymers. On the contrary, incorporation of RGD into PSBMA hydrogels promoted hASCs adhesion. Cells had a flat shape with extending filopodium on the surface of PSBMA hydrogels (Fig. 4B). Similarly, Santiago et al. reported that poly(caprolactone) modified with RGD improved ASCs adhesion.24 Lin et al. also showed that RGD improved ASCs adhesion on polyethersulfone surfaces.25 Relative to cells on RGD-modified matrixes, the hASCs showed a smaller cell body on RGD/QK-incorporated PSBMA hydrogels (Fig. 4C). The difference of mechanical strength between RGD-modified and RGD/QK-modified PSBMA hydrogels may result in this finding. The stiffness of matrix can modulate integrin-mediated mechanotransduction, regulate focal-adhesion structure and the cytoskeleton, and change cell shape in stem cells.26Live/dead staining was used to evaluate cell survival after seeding for 7 days. Rare cells can be found on non-modified PSBMA hydrogels (Fig. 4D). However, hASCs with extending cytoplasm were observed on modified hydrogels. The hASCs had good survival on both RGD- and RGD/QK-modified PSBMA hydrogels (Fig. 4E and F). Likewise, di Summa et al. showed that RGD can enhance ASCs survival under stress conditions.27 VEGF-transfected ASCs are also shown to improve autologous fat transplantation.28
The viability and proliferation of hASCs on PSBMA hydrogels
The tests of cell metabolic activity proved the benefits of RGD and RGD/QK peptides-grafting to hASCs adhesion and subsequently the viability on PSBMA hydrogels. Relative to consistent low values in none-modified hydrogels, hASCs on peptides-grafted PSBMA hydrogels had a significantly higher MTT value on day 3 and day 7 (all p < 0.05). However, there was no significant differences between RGD and RGD/QK groups (Fig. 4G). This findings showed that hASCs can reduce MTT into formazan which reveal that cells can adhere and metabolize normally. Perviously, Parisi-Amon et al. also reported that RGD improved retention of transplanted ASCs in injectable hydrogels.29 Furthermore, the MTT values increased as cultivation days which implying that hASCs can proliferate on peptides-modified matrixes. This assumption is further supported by the results of cell number (LDH assay, Fig. 4H). The total lysis of cells/hydrogels samples showed that RGD- and RGD/QK-modified PSBMA hydrogels had a significantly higher cell number (both p < 0.001) relative to that of un-modified hydrogels on day 1. In addition, the cell number increased on day 4 for the peptides-modified groups, which reveals that hASCs can proliferate in the modified hydrogels. On the contrary, hASCs in un-modified PSBMA hydrogels remained in a low cell number. This finding is consistent with the results of optical microscope observations as well as MTT assay. For both RGD and RGD/QK groups, there is no significant difference in cell number on day 4 and 7. Likewise, no significant differences were found between RGD and RGD/QK groups in all time points. Besides, VEGF has a critical role in ASCs-mediated regeneration.30 The addition of QK, the functional motif of VEGF, shall improve further applications of PSBMA hydrogels.Adipogenic differentiation
The hASCs cultured on un-modified and peptides-incorporated PSBMA hydrogels were cultured in adipogenic medium for 7 days (Fig. 5). While cells maintained a round shape on un-modified PSBMA hydrogels, a morphology change was noticed when hASCs cultured on RGD- and RGD/QK-modified substrates under adipogenic stimulations (Fig. 5B and C). We further analyzed the mRNA levels of PPAR-γ and LPL. Although no differences were found in LPL levels, the hASCs on RGD-modified PSBMA hydrogels had a significantly higher PPAR-γ expression (p < 0.05) than that of cells on TCPS, and the cells on RGD/QK hydrogels has a highest PPAR-γ level after adipogenic stimulation (p < 0.01 compared to RDG group, and p < 0.001 to TCPS group). Since PPAR-γ modulates downstream genes related to adipogenesis, lipid uptake and lipid metabolism, the peptides-incorporated PSBMA hydrogels may improve adipocyte differentiation to stem cells.31 Previously, Park et al. found that the adipogenic differentiation of ASCs is correlated with adhesion mechanism, which provides a possible explanation to this finding.32 Furthermore, it is known that the activation of receptor tyrosine kinases or inhibition of AMP-activated protein kinase (AMPK) improves adipocyte differentiation.33 PPAR-γ is the downstream targeting of AMPK and thus stimulation of AMPK activity resulted in the down-regulation of PPAR-γ and inhibited the differentiation/growth in adipocytes.34 Therefore, we assume that culture of hASCs on RGD/QK-modified PSBMA hydrogels may down-regulate AMPK to improve PPAR-γ expressions. The expression of LPL, an adipogenic marker in early stages of adipocyte development, is a time-dependent course and also influenced by culture conditions.35 Therefore, a relative longer stimulation period may require for LPL induction.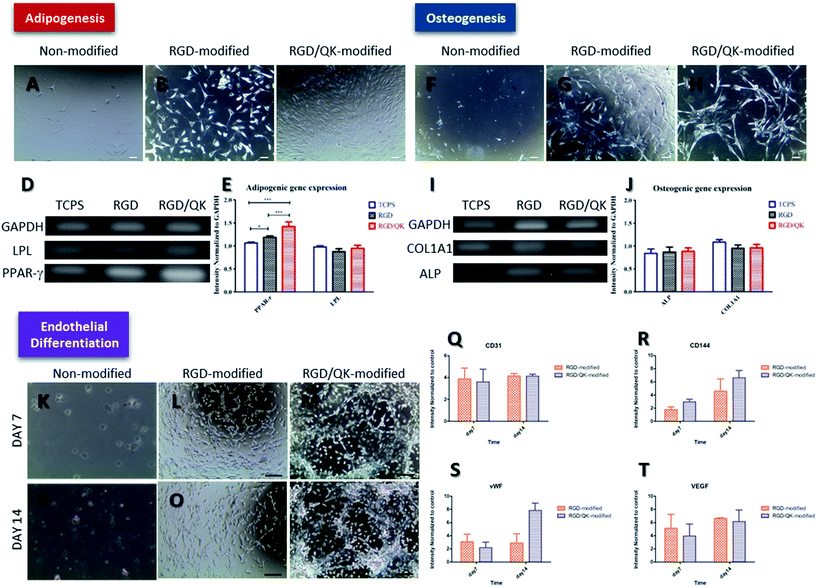 | ||
| Fig. 5 Cell differentiation of hASCs on PSBMA hydrogels. (A–E) adipogenic differentiation, (F–J) osteogenic differentiation and (K–T) endothelial differentiation. Scale bar = 100 μm. | ||
Osteogenic differentiation
Regarding the osteogenesis, hASCs on RGD-modified and RGD/QK-modified PSBMA hydrogels also showed a morphology change after osteogenic inductions (Fig. 5G and H). The mRNA expressions of osteogenic markers ALP and COL1A1 were determined.36 However, there was no any difference in ALP and COL1A1 levels among groups. Although RGD peptide is shown to improve osteogenic differentiation in human bone marrow stromal cells, the addition of QK or 3D environment of hydrogel did not have solid effects in osteogenic differentiation when compared with hASCs on TCPS with osteogenic medium.37 Unlike adipogenic differentiation, Peng et al. reported that cell–cell contacts have a cooperative effect on adipogenic differentiation but a competitive effect on osteogenic differentiation to rat bone marrow mesenchymal stem cells.38 Considering the mechanical strength, the peptides-modified PSBMA hydrogels may more suitable to adipocyte tissue engineering for soft tissue engineering.39Endothelial differentiation
After induction by endothelial growth medium, hASCs on RGD-modified hydrogels formed cord-like structures on day 7. This finding is more pronounced when cells cultured on RGD/QK-modified PSBMA hydrogels for 14 days. Previously, Fischer et al. reported similar results when seeded hASCs into Matrigel, which reveals that PSBMA hydrogels incorporated with RGD/QK may also provide similar microenvironment as Matrigel.40 Likewise, qRT-PCR results showed CD31, an early marker of endothelial cell differentiation, is significantly up-regulated enhanced at day 7 and day 14 when hASCs cultured on peptides-modified PSBMA hydrogels (p < 0.05 to TCPS group).41Moreover, relative to monolayer culture, CD144 (p < 0.05 on day 7 and p < 0.01 on day 14), vWF (p < 0.05 on day 7 and p < 0.01 on day 14) and VEGF (p < 0.05 on day 7 and p < 0.01 on day 14) were all up-regulated significantly. Similarly, Liu et al. showed that 3D RGD peptide hydrogels improved the secretion of angiogenic growth factors to ASCs.42 Zhang also showed 3D microenvironment improved endothelial cell differentiation to human induced pluripotent stem cells.43 Although no difference was found in the expressions of CD31, CD144, and VEGF, the RGD/QK-modified PSBMA hydrogels had a significant higher vWF expression when compared to hASCs cultured on RGD-modified PSBMA hydrogels, which reveals that RGD/QK can further improve endothelial differentiation to hASCs. Up-regulation of phosphoinositide 3-kinase (PI3K) is shown to improve the downstream CD31, vWF and endothelial nitric oxide synthase to promote endothelial differentiation.44 However, RGD/QK-grafted PSBMA hydrogel only improved vWF expression but not CD31, which reveals unknown mechanisms behind the PI3K pathway.
Conclusions
Zwitterionic hydrogels incorporated with RGD or RGD/QK peptides can improve hASCs adhesion. However, the graftings of RGD and RGD/QK did not interfere with the non-fouling property of PSBMA hydrogels. Relative to RGD-grafted PSBMA hydrogels, the RGD/QK incorporation further improved hASCs differentiated into adipogenic lineage. Moreover, the endothelial differentiation was also enhanced when hASCs were cultured on RGD/QK-modified PSBMA hydrogels. In conclusion, the peptides-modified PSBMA hydrogels can improve hASCs differentiate toward adipogenic/endothelial lineages.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the research funding from Ministry of Science and Technology, Taiwan (103-2221-E-002-210).Notes and references
- K. Yu, Y. Mei, N. Hadjesfandiari and J. N. Kizhakkedathu, Colloids Surf., B, 2014, 124, 69–79 CrossRef CAS PubMed.
- L. Zhang, Z. Cao, T. Bai, L. Carr, J. R. Ella-Menye, C. Irvin, B. D. Ratner and S. Jiang, Nat. Biotechnol., 2013, 31, 553–556 CrossRef CAS PubMed.
- L. R. Carr, H. Xue and S. Jiang, Biomaterials, 2011, 32, 961–968 CrossRef CAS PubMed.
- M. Tallawi, E. Rosellini, N. Barbani, M. G. Cascone, R. Rai, G. Saint-Pierre and A. R. Boccaccini, J. R. Soc., Interface, 2015, 12, 20150254 CrossRef PubMed.
- E. Lih, S. H. Oh, Y. K. Joung, J. H. Lee and D. K. Han, Prog. Polym. Sci., 2015, 44, 28–61 CrossRef CAS.
- M. B. Rahmany and M. Van Dyke, Acta Biomater., 2013, 9, 5431–5437 CrossRef CAS PubMed.
- J. E. Leslie-Barbick, J. E. Saik, D. J. Gould, M. E. Dickinson and J. L. West, Biomaterials, 2011, 32, 5782–5789 CrossRef CAS PubMed.
- S. Prakash Parthiban, D. Rana, E. Jabbari, N. Benkirane-Jessel and M. Ramalingam, Acta Biomater., 2017, 51, 330–340 CrossRef CAS PubMed.
- Y. Yang, Q. Yang, F. Zhou, Y. Zhao, X. Jia, X. Yuan and Y. Fan, J. Mater. Sci.: Mater. Med., 2016, 27, 106 CrossRef PubMed.
- C. Y. Lin, Y. R. Wang, C. W. Lin, S. W. Wang, H. W. Chien, N. C. Cheng, W. B. Tsai and J. Yu, BioRes. Open Access, 2014, 3, 297–310 CrossRef CAS PubMed.
- J. H. Wen, L. G. Vincent, A. Fuhrmann, Y. S. Choi, K. C. Hribar, H. Taylor-Weiner, S. Chen and A. J. Engler, Nat. Mater., 2014, 13, 979–987 CrossRef CAS PubMed.
- H. Mizuno, M. Tobita and A. C. Uysal, Stem cells, Dayton, Ohio, 2012, vol. 30, pp. 804–810 Search PubMed.
- H. W. Chien, J. Yu, S. T. Li, H. Y. Chen and W. B. Tsai, Biomater. Sci., 2017, 5, 322–330 RSC.
- H. W. Chien, P. H. Cheng, S. Y. Chen, J. Yu and W. B. Tsai, Biomater. Sci., 2017, 5, 523–531 RSC.
- Y.-H. Lin, K.-W. Huang, S.-Y. Chen, N.-C. Cheng and J. Yu, J. Mater. Chem. B, 2017, 5, 4614–4622 RSC.
- G. C. Reilly and A. J. Engler, J. Biomech., 2010, 43, 55–62 CrossRef PubMed.
- S. Van Vlierberghe, P. Dubruel and E. Schacht, Biomacromolecules, 2011, 12, 1387–1408 CrossRef CAS PubMed.
- F. Colazzo, F. Alrashed, P. Saratchandra, I. Carubelli, A. H. Chester, M. H. Yacoub, P. M. Taylor and P. Somers, Growth Factors, 2014, 32, 139–149 CrossRef CAS PubMed.
- S. P. Parthiban, D. Rana, E. Jabbari, N. Benkirane-Jessel and M. Ramalingam, Acta Biomater., 2017, 51, 330–340 CrossRef PubMed.
- C.-Y. Lin, Y.-R. Wang, C.-W. Lin, S.-W. Wang, H.-W. Chien, N.-C. Cheng, W.-B. Tsai and J. Yu, BioRes. Open Access, 2014, 3, 297–310 CrossRef CAS PubMed.
- S. H. Yang, M. H. Hu, W. Y. Lo, Y. H. Sun, C. C. Wu and K. C. Yang, J. Biomed. Mater. Res., Part A, 2017, 105, 1575–1582 CrossRef CAS PubMed.
- C. Fuoco, E. Sangalli, R. Vono, S. Testa, B. Sacchetti, M. V. Latronico, S. Bernardini, P. Madeddu, G. Cesareni, D. Seliktar, R. Rizzi, C. Bearzi, S. M. Cannata, G. Spinetti and C. Gargioli, Front. Physiol., 2014, 5, 203 Search PubMed.
- E. M. Chandler, C. M. Berglund, J. S. Lee, W. J. Polacheck, J. P. Gleghorn, B. J. Kirby and C. Fischbach, Biotechnol. Bioeng., 2011, 108, 1683–1692 CrossRef CAS PubMed.
- L. Y. Santiago, R. W. Nowak, J. Peter Rubin and K. G. Marra, Biomaterials, 2006, 27, 2962–2969 CrossRef CAS PubMed.
- Y. C. Lin, C. A. Brayfield, J. C. Gerlach, J. P. Rubin and K. G. Marra, Acta Biomater., 2009, 5, 1416–1424 CrossRef CAS PubMed.
- Y. R. Shih, K. F. Tseng, H. Y. Lai, C. H. Lin and O. K. Lee, J. Bone Miner. Res., 2011, 26, 730–738 CrossRef CAS PubMed.
- P. G. di Summa, D. F. Kalbermatten, W. Raffoul, G. Terenghi and P. J. Kingham, Tissue Eng., Part A, 2013, 19, 368–379 CrossRef CAS PubMed.
- F. Lu, J. Li, J. Gao, R. Ogawa, C. Ou, B. Yang and B. Fu, Plast. Reconstr. Surg., 2009, 124, 1437–1446 CrossRef CAS PubMed.
- A. Parisi-Amon, W. Mulyasasmita, C. Chung and S. C. Heilshorn, Adv. Healthcare Mater., 2013, 2, 428–432 CrossRef CAS PubMed.
- S. Y. Song, H. M. Chung and J. H. Sung, Expert Opin. Biol. Ther., 2010, 10, 1529–1537 CrossRef CAS PubMed.
- B. B. Mandal and S. C. Kundu, Biomaterials, 2009, 30, 5019–5030 CrossRef CAS PubMed.
- I. S. Park, M. Han, J. W. Rhie, S. H. Kim, Y. Jung, I. H. Kim and S. H. Kim, Biomaterials, 2009, 30, 6835–6843 CrossRef CAS PubMed.
- M. G. Scioli, A. Bielli, P. Gentile, D. Mazzaglia, V. Cervelli and A. Orlandi, Int. J. Mol. Sci., 2014, 15, 6517–6526 CrossRef PubMed.
- S. Chen, Z. Li, W. Li, Z. Shan and W. Zhu, Can. J. Physiol. Pharmacol., 2011, 89, 793–799 CAS.
- A. A. Ghoniem, Y. Acil, J. Wiltfang and M. Gierloff, Anat. Cell Biol., 2015, 48, 85–94 CrossRef PubMed.
- E. D. Rosen and O. A. MacDougald, Nat. Rev. Mol. Cell Biol., 2006, 7, 885–896 CrossRef CAS PubMed.
- N. M. Moore, N. J. Lin, N. D. Gallant and M. L. Becker, Acta Biomater., 2011, 7, 2091–2100 CrossRef CAS PubMed.
- R. Peng, X. Yao, B. Cao, J. Tang and J. Ding, Biomaterials, 2012, 33, 6008–6019 CrossRef CAS PubMed.
- J. H. Choi, J. M. Gimble, K. Lee, K. G. Marra, J. P. Rubin, J. J. Yoo, G. Vunjak-Novakovic and D. L. Kaplan, Tissue Eng., Part B, 2010, 16, 413–426 CrossRef PubMed.
- L. J. Fischer, S. McIlhenny, T. Tulenko, N. Golesorkhi, P. Zhang, R. Larson, J. Lombardi, I. Shapiro and P. J. DiMuzio, J. Surg. Res., 2009, 152, 157–166 CrossRef CAS PubMed.
- Z. J. Li, Z. Z. Wang, Y. Z. Zheng, B. Xu, R. C. Yang, D. T. Scadden and Z. C. Han, J. Cell. Biochem., 2005, 95, 559–570 CrossRef CAS PubMed.
- X. Liu, X. Wang, X. Wang, H. Ren, J. He, L. Qiao and F. Z. Cui, Acta Biomater., 2013, 9, 6798–6805 CrossRef CAS PubMed.
- S. Zhang, J. R. Dutton, L. Su, J. Zhang and L. Ye, Biomaterials, 2014, 35, 3786–3793 CrossRef CAS PubMed.
- D. Arya, S. Chang, P. DiMuzio, J. Carpenter and T. N. Tulenko, J. Biomed. Sci., 2014, 21, 55 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08919h |
| This journal is © The Royal Society of Chemistry 2017 |