Open Access Article
Open Access ArticleStudies on effect of Ginkgo biloba L. leaves in acute gout with hyperuricemia model rats by using UPLC-ESI-Q-TOF/MS metabolomic approach†
WeiJia Chen a,
Yuanyuan Liu
a,
Yuanyuan Liu a,
Mengying Wei
a,
Mengying Wei a,
Liqiang Shi
a,
Liqiang Shi a,
Yi Wu
a,
Yi Wu *a,
ZhongYing Liu
*a,
ZhongYing Liu a,
Shu Liu
a,
Shu Liu b,
FengRui Song
b,
FengRui Song b and
ZhiQiang Liu
b and
ZhiQiang Liu bc
bc
aSchool of Pharmaceutical Sciences, Jilin University, Changchun 130021, China
bNational Center of Mass Spectrometry in Changchun, Jilin Province Key Laboratory of Chinese Medicine Chemistry and Mass Spectrometry, Chemical Biology Laboratory, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, 130022, China
cState Key Laboratory of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun 130022, China
First published on 5th September 2017
Abstract
Gout is a result of sodium urate deposition in and around the joints that is caused by long-standing hyperuricemia. In the present study, Ginkgo biloba L. leaves have been used for the treatment of acute gout and hyperuricemia, but the pathogenesis of acute gout with hyperuricemia and the mechanism of action of Ginkgo biloba L. leaves in gout remain unclear. This study aimed to investigate the pathogenesis of acute gout with hyperuricemia in rats and investigate the therapeutic effects of extract of Ginkgo biloba L. leaves (GBE) by using the metabolomic method. In this study, the rat model of acute gout with hyperuricemia was established by intraperitoneal injection of xanthine and oxonic acid potassium salt and intra-articular injection of monosodium urate (MSU). Serum level of interleukin-1 beta (IL-1β) was evaluated to compare the model group with the group with GBE by enzyme-linked immunosorbent assay (ELISA). Joint swelling was used for testing the effects of MSU and the pathological changes of joint and kidney were assessed by hematoxylin–eosin (H&E) staining. Potential biomarkers were identified from urinary samples by ultra-performance liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC-ESI-Q-TOF/MS) combined with principal component analysis (PCA) which was used to observe the metabonomic alterations and the separations of the scatter points of different groups. Results show that joint swelling and serum level of IL-1β significantly decreased, and pathological abnormalities of the joint and kidney were ameliorated after GBE treatment. 27 potential biomarkers were identified and the primary metabolism pathways involved tryptophan metabolism, pyrimidine metabolism, pentose phosphate pathway, TCA cycle, tyrosine metabolism, lysine degradation and purine metabolism. The disturbed pathways were restored after treatment with GBE. This study indicated that GBE possessed evident therapeutic effects on acute gout with hyperuricemia rats.
Introduction
Gout, a common inflammatory arthritis, is caused by MSU deposition in and around the joints resulting from hyperuricemia. With dietary and lifestyle changes, the incidence of the disease has increased annually. Gout attack can cause a number of important complications such as common renal disease, hypertension, diabetes, metabolic syndromes, and cardiovascular disease.1–3 At present, non-steroidal anti-inflammatory drugs (NSAIDs), corticosteroids, IL-1 inhibitors and urate-lowering drugs (allopurinol and benzbromarone) are often used as the clinical therapies for gout. However, these agents may have serious adverse effects on the gastrointestinal and renal systems.4–6 Thus, in recent years, the development of new anti-gout drugs with high efficacy and less toxicity are eagerly needed, for which, natural resources are important.Traditional Chinese Medicine (TCM) has been used as a natural clinical drug for treating disease and received satisfactory results. TCM usually includes a great diversity of compounds and manifold biological activities in the various herbs. It can hit multiple targets with its complex components, improve therapeutic efficacy and reduce drug-related adverse effects.7,8 Therefore, TCM have caused widely attentions.
Ginkgo biloba L. leaves, also known as “yinxingye” (Chinese), belong to the family of Ginkgoaceae and is used as a TCM to treat different disease. It is recorded in the Pharmacopoeia of the People's Republic of China that can improve blood circulation and eliminate blood stasis, relieve pain, resolve turbidity and reduce lipid. In addition, GBE can inhibit the production of pro-inflammatory cytokines IL-1β and tumour necrosis factor α (TNF-α).9 Serum IL-1β as a crucial mediator for gout is being increasingly concerned. However, the mechanism of action of Ginkgo biloba L. leaves for the treatment of acute gout with hyperuricemia is unclear.
Metabonomics is a rapidly advancing field that is used in detecting novel biomarkers for diagnosis, prognosis and personalized medicine.10,11 It could reveal the essence of life activities of the body through the changes of endogenous metabolites which obtained by using some analytical methods. Liquid chromatography-mass spectrometry (LC-MS) is the most widely used analytical technology in the field of global metabolite profiling,12,13 due to its ability to separate and detect a wide range of molecules.14,15 Urine is commonly used for metabolite analysis for animals because it contains many metabolites and metabolic end products and is free from interfering proteins or lipids and chemically complex. Therefore, the pathogenesis of acute gout with hyperuricemia in rats and the therapeutic effects of GBE may be clarified by using the urinary metabolomic method.
In this study, a urinary metabonomic method based on UPLC-ESI-Q-TOF/MS combined with PCA was developed to find biomarkers of a rat model of acute gout with hyperuricemia and figure out metabonomic alterations to elucidate relevant path mechanisms. Meanwhile, the therapeutic effect of GBE for acute gout with hyperuricemia model rats was evaluated by detecting the degree of metabolic disorder recovery. The findings of this study will be important for future diagnosis and therapy of gout.
Experimental
Reagents and materials
Xanthine and sodium urate were purchased from Sigma (St. Louis, MO, USA), oxonic acid potassium salt was purchased from Xiya Reagent (Chengdu, China), acetonitrile of HPLC grade was obtained from Fisher Scientific (Loughborough, UK), formic acid of HPLC grade was purchased from ROE Scientific Inc. (Newark, USA), and rat IL-1β ELLSA kits were obtained from the Nanjing Jiancheng Bioengineering Institute (Nanjing, China).GBE was brought from Hongjiu Biotech Co., Ltd (batch number: HJ150103; extract solvent: ethanol/water). The total ginkgo flavone glycoside (≥24%) and total terpene lactones (≥6%) were in conformity with the Pharmacopoeia of the People's Republic of China.
Animal experiment
Adult male albino Sprague-Dawley rats (weighing 200–220 g) were obtained from Liaoning Changsheng Technology and Biology Institute and used for the experiments. The rats were housed in a temperature- and humidity-controlled room (22 ± 2 °C) under a 12 h light–dark cycle. The animals were acclimatized for one week before the experiment with freely accessing food and water. After that, the rats were randomly divided into three groups: control group (CG), model group (MG), and GBE-treated group (GbG). The CG was intraperitoneally injected with identical volume of normal saline. The MG was injected with the model drug (60 mg mL−1 of xanthine and oxonic acid potassium salt) at 0.5 mL/100 g of body weight twice daily16 for 7 days. The GbG group was injected with the model drug at 0.5 mL/100 g of body weight twice daily for 7 days and was treated with GBE at 20 mg/100 g of body weight once daily for 7 days simultaneously. On the sixth day, 100 μL sodium urate solution (30 mg mL−1) was injected into the right articular cavity for MG and GbG groups. Joint swelling on the right ankle was respectively measured by a soft rule at 3, 6, 12, 24, and 48 h after sodium urate solution was injected. After 48 h, all the rats were sacrificed.All animal experiments were performed in accordance with the Regulation on the Administration of Laboratory Animals (The state council of the People's Republic of China) for the care and use of laboratory animals and were approved by the Ethics Committee for the Use of Experimental Animals of Jilin University.
Sample collection
After 7 days, the rats were fasted for 12 h, and then the serum and urine samples were collected. In which, the serum was treated by centrifuging blood clot at 4000 rpm for 10 min and was frozen at −80 °C for ELISA. The collected 12 h urine samples were centrifuged at 4000 rpm for 10 min to remove the particle contaminants, and the supernatant was frozen at −80 °C until analysis. The kidney and the right joint of rats were fixed with 10% formalin and embedded in paraffin. Then, the tissues were stained with H&E for general evaluation.Joint swelling
Joint swelling was determined from the circumference of the right ankle by soft rule. Joint swelling ratio was calculated using the following formula: joint swelling ratio (%) = (A − B)/B × 100% where A is the circumference of each rat at different times and B is the circumference of each rat at 0 h.Preparation of metabolomic samples
All the stored frozen urine samples were thawed and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C, and then filtered through a 0.22 μm membrane. Subsequently, the samples were diluted in deionized water (Milli-Q water system) with a ratio of 1
000 rpm for 10 min at 4 °C, and then filtered through a 0.22 μm membrane. Subsequently, the samples were diluted in deionized water (Milli-Q water system) with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 (v/v) before UPLC-ESI-Q-TOF/MS analysis.
8 (v/v) before UPLC-ESI-Q-TOF/MS analysis.
UPLC-ESI-Q-TOF/MS conditions
UPLC analysis was performed on a Waters ACQUITY UPLC system coupled with a Q-TOF SYNAPT G2 High Definition Mass Spectrometer (HDMS, resolution = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) (Waters Corporation, Manchester, UK) that is controlled by Masslynx V4.1 (Waters Corporation, Milford, USA). Chromatographic separation was executed on an UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm, ACQUITY) with a temperature of 35 °C. Elution was carried out at a flow rate of 0.3 mL min−1 by using the mobile phases of solvent A (acetonitrile) and solvent B (0.1% formic acid in water). The following gradient program was used: 0–2 min, 5–10% A; 2–5 min, 10–20% A; 5–6 min, 20–50% A; 6–9 min, 50–70% A; 9–10 min, 70–100% A. The autosampler was maintained at 4 °C during the entire analysis. The processed sample injection volume was 5 μL. The ESI source in both positive and negative ion modes was used in MS analysis.
000) (Waters Corporation, Manchester, UK) that is controlled by Masslynx V4.1 (Waters Corporation, Milford, USA). Chromatographic separation was executed on an UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm, ACQUITY) with a temperature of 35 °C. Elution was carried out at a flow rate of 0.3 mL min−1 by using the mobile phases of solvent A (acetonitrile) and solvent B (0.1% formic acid in water). The following gradient program was used: 0–2 min, 5–10% A; 2–5 min, 10–20% A; 5–6 min, 20–50% A; 6–9 min, 50–70% A; 9–10 min, 70–100% A. The autosampler was maintained at 4 °C during the entire analysis. The processed sample injection volume was 5 μL. The ESI source in both positive and negative ion modes was used in MS analysis.
The source parameters were set as follows: the source temperature was 120 °C, desolvation gas temperature was 350 °C, cone gas flow was 50 L h−1, desolvation gas flow was 700 L h−1, capillary voltage was 3 kV for positive ion mode and 2.5 kV for negative ion mode, sample cone voltage was 40 V, extraction cone voltage was 5.0 V, full scan range was 100–1000 m/z, and scan time was 0.2 s. For ESI mode, leucine–enkephalin was used as the lock mass to ensure mass accuracy and reproducibility, generating reference ions for positive ion mode ([M + H]+ = 556.2771) and negative ion mode ([M − H]− = 554.2615) via the Lock-Spray. Moreover, sodium formate was used to set up mass spectrometer calibration.
Statistical analysis
The UPLC-MS raw data files were processed by using MassLynx V4.1 (Waters Corporation, Milford, USA). The filter method was set as follows: retention time ranging from 0 min to 10 min, mass ranging from 50 amu to 1000 amu, marker intensity threshold was 500, and noise elimination level was 60. MassLynx V4.1 automatically processed ion pair extraction, peak alignment, peak matching, and peak intensity correction for the base peak intensity (BPI) chromatograms. The resulting data matrix contains the retention time and the m/z information. Then the PCA was performed by Umetrics EZinfo 2.0 (Waters Corporation, Milford, USA). Scaling was set to “Pareto,” and transformation was set to “None.” Statistical analysis was performed using one-way analysis of SPSS 18.0, and significance t-test (p < 0.01 or p < 0.05) was carried out. Biochemical databases were used to identify potential biomarkers, such as HMDB (http://www.hmdb.ca/), METLIN (http://metlin.scripps.edu/) and KEGG (http://www.kegg.com/).Results and discussion
Evaluation of joint swelling and inflammatory cytokines
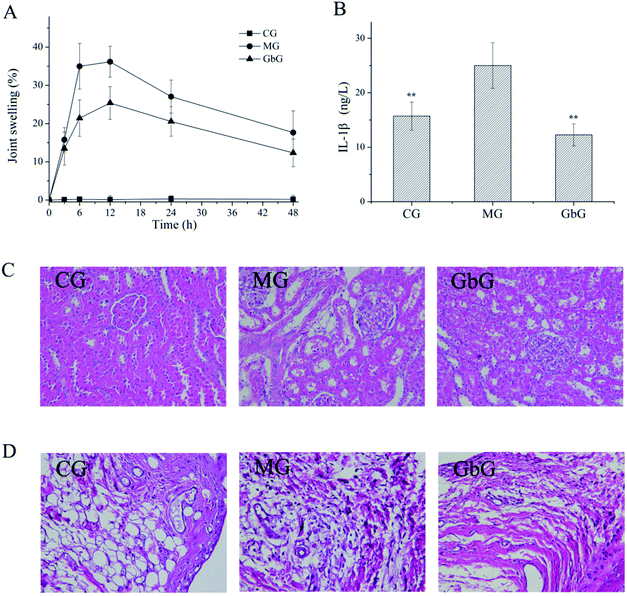
Rat model of acute gout with hyperuricemia was established by intraperitoneal injection of xanthine and oxonic acid potassium salt and intra-articular injection of MSU. The development and progression of acute gouty arthritis caused by MSU were diagnosed mainly based on joint swelling. The result (Fig. 1A) shows that joint swelling reached a peak at 6–12 h and stably decreased after 12 h in MG. Compared with the MG, joint swelling significantly reduced in GbG within 48 h.IL-1β is a crucial mediator for MSU crystal-induced inflammation and is overexpressed in the patients with acute gout.17 The expression of serum IL-1β was detected via ELISA kits, and the results are shown in Fig. 1B. The expression of IL-1β was up-regulated in the MG compared with CG. After the treatment with GBE, the expression of serum IL-1β was down-regulated significantly. The results reveal that GBE had a potential effect on inhibiting expression of inflammatory factor.
Histopathological analysis of kidney and synovial membrane
Histological changes in the kidney and joint were observed by H&E staining. Renal injury in the MG was more serious compared with CG (Fig. 1C). The morphological changes of the kidney were observed, including dilation of tubular lumen, tubular epithelial vacuolization, glomerular capillary congestion and precipitated uric acid crystals. The pathological abnormalities in kidney were ameliorated after the administration of GBE. As shown in Fig. 1D, the morphological changes of the joint were observed, including inflammatory cell infiltration, necrosis, and edema of tissue in MG. In the GbG, the pathological abnormalities were ameliorated. The results indicate that not only the inflammatory factor could be inhibited obviously, but the kidney and joint could be protected markedly by GBE.Urine metabolite profiles
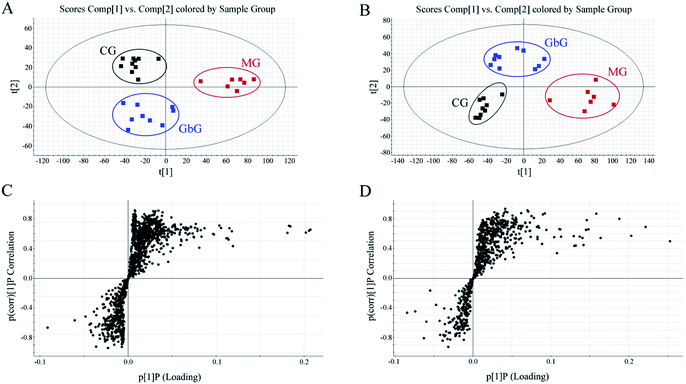
The separation conditions of urine on BEH C18 column were optimized in terms of peak shape and reproducibility, and the mass spectrum parameters were optimized based on the ionization properties of compounds. Typical UPLC-TOF/MS base peak intensity (BPI) chromatograms of urine samples obtained in positive and negative ion modes are shown in Fig. 2, respectively. Fig. 3 shows the score plots in both positive (A) and negative (B) ion modes. It obviously clustered and separated of metabolic data from the groups (CG, MG and GbG) in the scatter diagram of PCA, thereby showing the different metabolic perturbation trends in rats. It shows that the CG and MG groups had significant distinctions after injection of the model drug for 7 days, and the GbG group was close to the CG group. It indicates that GBE has a certain therapeutic effect for diseases.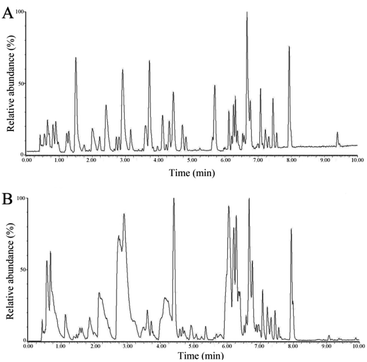 | ||
| Fig. 2 The typical urinary BPI chromatograms of the rat urine obtained in positive mode (A) and negative mode (B) based on UPLC-ESI-Q-TOF/MS. | ||
The S-plot (Fig. 3C and D) was derived from the OPLS and used to select biomarkers. The farthest from the origin variables were the most likely potential endogenous biomarkers due to their high contributions and correlations. The S-plot of each spot represents a substance. To select candidate biomarkers among the thousands of variables, only those with a VIP > 1 and p < 0.05 of an independent t-test between two groups were considered as the potential biomarkers among the groups. The structures of the potential biomarkers were confirmed by using databases such as HMDB, METLIN and KEGG. Moreover, the compound should be endogenous compound, and the source of the compound is urine. Following the criterion above, endogenous metabolites were selected and identified for further study between MG and CG.
Identification of potential biomarkers
The possible molecular formulae of the potential biomarkers were calculated using high-accuracy quasi-molecular ion with mass error of 10 ppm, and the structures were identified according to fragment ions, which were obtained from MS/MS analysis by comparing with standards and database. The selected and identified potential biomarkers were structurally confirmed by comparing the retention times and mass spectra. The biomarkers of cytosine m/z 112.0523 in positive ion and uric acid m/z 167.0221 in negative ion mode were taken as examples to illustrate the identification process of biomarkers. Two major fragment ions of cytosine were found at m/z 95.0264 and m/z 69.0480, which represented the fragments of [M + H − NH2]+ and [M + H − NH2 − C2H2]+, respectively (Fig. 4A). Two major fragment ions of uric acid were found at m/z 124.0164 and m/z 96.0205, which represented the fragments of [M − H − CHNO]− and [M − H− CHNO − CO]−, respectively (Fig. 4B). In the current study, 27 metabolites were confirmed as potential biomarkers. A list of the identified biomarkers is given in Table 1.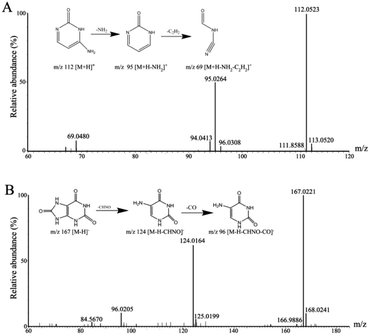 | ||
| Fig. 4 (A) Mass fragment information of cytosine in positive ion mode. (B) Mass fragment information of uric acid in negative ion mode. | ||
| Mode | No. | Rt (min) | Measured (m/z) | VIP | Compounds | MG vs. CG | GbG vs. MG | Metabolic pathway | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Trend | p Value | Trend | p Value | |||||||
| a “↑” represents the potential biomarker up-regulated; “↓” represents the potential biomarker down-regulated. | ||||||||||
| ES+ | 1 | 0.67 | 112.0509 | 2.03 | Cytosine | ↓ | 2.12 × 10−3 | ↑ | 1.37 × 10−3 | Pyrimidine metabolism |
| 2 | 5.36 | 118.0655 | 1.21 | Indole | ↑ | 7.20 × 10−5 | ↓ | 1.44 × 10−4 | Tryptophan metabolism | |
| 3 | 7.08 | 161.1289 | 1.73 | N(6)-Methyllysine | ↑ | 2.40 × 10−4 | ↓ | 4.43 × 10−4 | Lysine degradation | |
| 4 | 4.83 | 162.0551 | 1.58 | Indole-3-carboxylic acid | ↑ | 1.06 × 10−2 | ↓ | 3.26 × 10−2 | Tryptophan metabolism | |
| 5 | 0.9 | 180.087 | 1.25 | 7-Aminomethyl-7-carbaguanine | ↑ | 8.79 × 10−3 | ↓ | 1.25 × 10−2 | Folate biosynthesis | |
| 6 | 0.75 | 182.0812 | 1.02 | Tyrosine | ↑ | 5.41 × 10−4 | ↓ | 9.53 × 10−4 | Tyrosine metabolism | |
| 7 | 7.11 | 189.1595 | 1.03 | N6,N6,N6-Trimethyl-L-lysine | ↑ | 4.33 × 10−4 | ↓ | 7.66 × 10−4 | Lysine degradation | |
| 8 | 2.75 | 208.0607 | 1.28 | 4-(2-Aminophenyl)-2,4-dioxobutanoic acid | ↑ | 1.00 × 10−2 | ↓ | 1.42 × 10−2 | Tryptophan metabolism | |
| 9 | 0.67 | 228.0984 | 1.38 | Deoxycytidine | ↓ | 2.03 × 10−4 | ↑ | 1.27 × 10−3 | Pyrimidine metabolism | |
| 10 | 0.66 | 282.1194 | 1.27 | 1-Methyladenosine | ↓ | 7.45 × 10−5 | ↑ | 5.23 × 10−7 | Purine metabolism | |
| 11 | 7.11 | 357.279 | 8.92 | Tetracosahexaenoic acid | ↑ | 1.13 × 10−3 | ↓ | 1.06 × 10−2 | Fatty acid metabolism | |
| ES− | 12 | 4.26 | 107.0504 | 3.16 | p-Cresol | ↑ | 2.32 × 10−4 | ↓ | 5.74 × 10−3 | Phenylalanine metabolism |
| 13 | 2.04 | 152.0349 | 1.36 | 3-Hydroxyanthranilic acid | ↑ | 8.46 × 10−6 | ↑ | 4.96 × 10−2 | Tryptophan metabolism | |
| 14 | 0.55 | 155.0098 | 2.06 | Orotic acid | ↑ | 3.18 × 10−3 | ↓ | 4.88 × 10−2 | Pyrimidine metabolism | |
| 15 | 0.52 | 157.0369 | 1.45 | Allantoin | ↑ | 3.84 × 10−4 | ↓ | 6.39 × 10−6 | Purine metabolism | |
| 16 | 2.87 | 162.056 | 1.32 | 3-Methyldioxyindole | ↑ | 4.01 × 10−3 | ↓ | 2.13 × 10−2 | Tryptophan metabolism | |
| 17 | 0.65 | 167.0211 | 2.28 | Uric acid | ↑ | 9.62 × 10−4 | ↓ | 1.46 × 10−2 | Purine metabolism | |
| 18 | 4.74 | 173.082 | 1.55 | Suberic acid | ↑ | 3.88 × 10−2 | ↓ | 1.96 × 10−2 | Fatty acid metabolism | |
| 19 | 0.68 | 191.0201 | 6.32 | Citric acid | ↑ | 5.48 × 10−3 | ↓ | 1.24 × 10−2 | TCA cycle | |
| 20 | 0.51 | 195.051 | 1.36 | Gluconic acid | ↓ | 5.29 × 10−3 | ↑ | 9.06 × 10−3 | Pentose phosphate pathway | |
| 21 | 6.34 | 201.1135 | 2.01 | Sebacic acid | ↑ | 2.03 × 10−2 | ↓ | 4.78 × 10−2 | Fatty acid metabolism | |
| 22 | 1.62 | 212.0019 | 1.28 | Indoxyl sulfate | ↑ | 1.14 × 10−3 | ↓ | 1.14 × 10−3 | Tryptophan metabolism | |
| 23 | 0.56 | 287.0517 | 2.68 | Orotidine | ↑ | 6.00 × 10−4 | ↓ | 3.96 × 10−2 | Pyrimidine metabolism | |
| 24 | 5.99 | 297.0981 | 1.85 | 2-Phenylethanol glucuronide | ↓ | 6.52 × 10−3 | ↑ | 5.75 × 10−5 | Pentose and glucuronate interconversions | |
| 25 | 6.27 | 297.11 | 1.61 | 7-Methylguanosine | ↑ | 2.92 × 10−6 | ↓ | 2.71 × 10−3 | Purine metabolism | |
| 26 | 7.32 | 313.2377 | 1.12 | Octadecanedioic acid | ↑ | 3.19 × 10−2 | ↓ | 3.19 × 10−2 | Fatty acid metabolism | |
| 27 | 6.74 | 567.3159 | 1.13 | Deoxycholic acid 3-glucuronide | ↑ | 6.11 × 10−4 | ↓ | 6.11 × 10−4 | Pentose and glucuronate interconversions | |
The pathway of identified biomarkers and their functions
In this study, 27 metabolites were confirmed between MG and CG. The network of the main potential biomarkers is shown in Fig. 5. The primary metabolism pathways involved tryptophan metabolism, pyrimidine metabolism, purine metabolism, pentose phosphate pathway, TCA cycle, tyrosine metabolism, lysine degradation and fatty metabolism. In addition, the network and pathway analysis were also performed with the Cytoscape software. Metscape is a plug-in for Cytoscape which provide the compounds, reactions, enzymes, genes, pathways and the relationships between them. We upload experimental data by Metscape and obtained the compounds–reactions–enzymes–genes network. ESI Fig. S1† shows that 10 metabolites were recognized by the software and matched to the database to build a metabolic network. Metabolites are interrelated with each other to form a huge network. Lots of enzymes and genes participate in the reactions and transformation process of metabolites upstream and downstream. The main pathways, metabolites, enzymes and genes associated with gout are analyzed as following: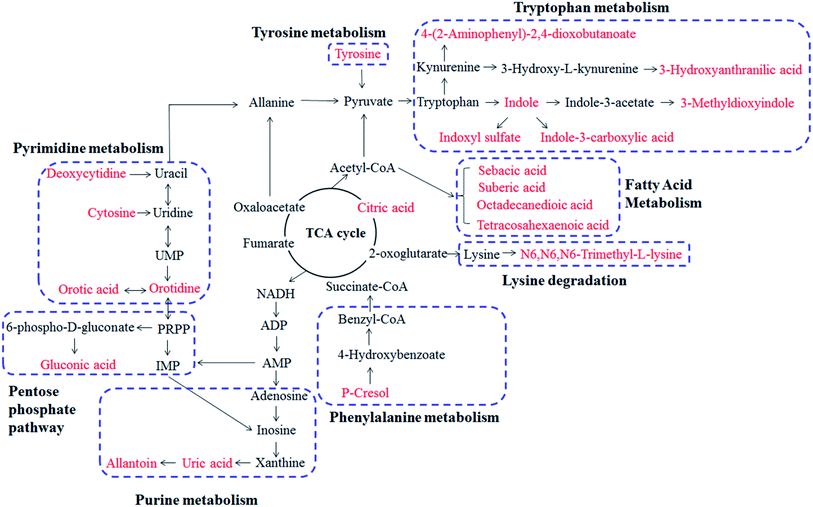 | ||
| Fig. 5 Correlation networks of main potential biomarkers in response to acute gout with hyperuricemia in rats. The red marked metabolites denote that the potential metabolites. | ||
Among these potential metabolites, indole, indole-3-carboxylic acid, 4-(2-aminophenyl)-2,4-dioxobutanoic acid, 3-hydroxyanthranilic acid, 3-methyldioxyindole and indoxyl sulfate were side products of tryptophan metabolism. They were all up-regulated and caused the abnormality of tryptophan metabolism in MG compared with CG. Tryptophan undergoes degradation via two distinct pathways: serotonin pathway and kynurenine pathway. 4-(2-Aminophenyl)-2,4-dioxobutanoic acid and 3-hydroxyanthranilic acid belong to kynurenine pathway and were up-regulated in the process of tryptophan catabolism. Kynureninase (KMO gene encoding protein) and kynurenine 3-momooxygenase (KYNU gene encoding protein) are the key enzymes that catalyze the upstream product into 3-hydroxyanthranilic acids (Fig. S1†). 3-Hydroxyanthranilic acid was demonstrated beneficial effects on anti-inflammation, antioxidant and inhibited the expression of pro-inflammatory cytokine TNF-α and chemokine expression.18,19 It was up-regulated in MG, but there were no significant changes after administrating GBE. This may be due to the increased activities of kynureninase and kynurenine 3-momooxygenase and the body protecting itself from injury after MSU-induced. This indicates that 3-hydroxyanthranilic acid plays an important role in inflammatory diseases, such as gout. Indoxyl sulfate could up-regulate the expressions of intercellular adhesion molecule-1 (ICAM-1), interleukin-6 (IL-6) and activate nuclear transcription factor-κB (NF-κB) pathway.20–22 Indole-3-carboxylic acid and 3-methyldioxyindole were reported to be closely related to chronic kidney disease.23 Therefore, tryptophan metabolism disorder is a key pathway in the development of gout and the importance also has been reported.24 The tendencies of these metabolites were decreased after the administration of GBE except 3-hydroxyanthranilic acid (Table 1), indicating that GBE has a beneficial effect for treating gout.
Regarding pyrimidine metabolism, cytosine, deoxycytidine, orotic acid and orotidine were observed in disturbance metabolites. From Fig. 5 and S1,† we can see that UMP (uridine 5′-monophosphate) is the intermediate material of deoxycytidine, orotic acid and orotidine. 5′-Nucleotidase participates in the synthesis of deoxycytidine. Cytosine and deoxycytidine are the principal nucleosides of DNA. And orotic acid and orotidine are intermediates in the pathway for the synthesis of pyrimidine nucleotides. The perturbations in pyrimidine metabolism were due to increased activity of repair endonucleases or restriction endonucleases.25 In the present study, orotic acid and orotidine were increased in MG, and cytosine and deoxycytidine were decreased compared with the CG (Table 1). The perturbations in pyrimidine metabolism were reversed to normal level after GBE treatment.
The increased urinary tyrosine was observed in MG. MSU could stimulate tyrosine phosphorylation in human neutrophils which is important for the initiation of acute gouty and it may be specific to the microcrystals.26–28 In that process, MSU crystals induced the phosphorylation and activation of non-receptor protein tyrosine kinase Tec. Moreover, activation of Tec leads to the rapid release of IL-1β, IL-8 and IL-1 in human neutrophils upon stimulation.28 Taken together, the neutrophils and tyrosine kinase are activated that correlating with the increased tyrosine by MSU-stimulated. Therefore, tyrosine is a characteristic biomarker of MSU-induced gout and could provide the basis for clinical diagnosis. The level of tyrosine was markedly increased in MG and then decreased after administration of GBE (Table 1). This indicates a potential effect of GBE in the treatment of gout.
As we all know, gout is a disease of purine metabolic disorders and resulted from MSU deposition in and around the joints. Xanthine and oxonic acid potassium salt were used to establish the early hyperuricemia model in rats. Uric acid is an important indicator to measure the model of hyperuricemia to be successful or not. Allantoin is considered as a stable biomarker of urate oxidation. The two types of the compounds are products of purine metabolism and present in urine. Uric acid is filtered from glomeruli, and approximately 90% is reabsorbed by the proximal tubule, thereby resulting in kidney injury or depositions of urate crystals in the joint and kidney.29 Hypoxanthine and xanthine are the precursor of uric acid. The process is catalyzed by xanthine oxidase (XO) and xanthine dehydrogenase (XD). They are encoded by the XDH gene (Fig. S1†). XO is a rate-limiting enzyme promoting purine metabolism.30 Oxonic acid potassium salt inhibits the oxidation of uric acid converting to allantoin through the catalysis of urate oxidase and oxidoreductase (Fig. 5). In this study, the levels of uric acid and allantoin were significantly up-regulated in the MG compared with the CG. Uric acid and allantoin are important biomarkers for gout and hyperuricemia. After treatment with GBE, the levels of uric acid and allantoin were recovered (Table 1). This shows that purine metabolism was regulated to normal level after GBE treatment.
In the conversions of alloxan and uric acid, peroxidase is an important enzyme which is associated with EPX, MPO, LPO and PRDX6 genes (Fig. S1†). MPO (myeloperoxidase) constitutes the major component of neutrophil and is proposed to play a central role in host defense.31 MPO could use hydrogen peroxide to oxidize chloride to hypochlorous acid, but under physiological conditions will also convert urate to the urate radical.32 MPO promotes oxidation of urate may contribute to the sequelae of inflammatory diseases, particularly gout.33 In addition, LPO (lactoperoxidase) and EPX (eosinophil peroxidase) belong to the peroxidase family and associate with oxidation process. Therefore, MPO, LPO and EPX are the key enzymes and genes for the occurrence and development of gout. Above information is provided by the metabolic network. It provides a new direction for studying the pathogenesis of gout.
Citric acid is a key intermediate of the TCA cycle in mitochondria. It is associated with energy metabolism. Among that, aconitate hydratase (ACO1 and ACO2 gene encoding protein) is a key enzyme (Fig. S1†). The function of ACO1 proteins is to catalyze the conversion of citrate to isocitrate. The increased citric acid may be due to the inhibited enzyme activity, which cause it cannot be converted to isocitric acid. ACO2 protein is encoded in the nucleus and functions in the mitochondrion. The increased citric acid suggests that the impaired action of energy metabolism in rats of MG, and GBE could reverse the citric acid level.
p-Cresol is a precursor of uremic retention compound and is generated by intestinal flora. Indoxyl sulfate and p-cresol sulfate have similar features on kidney injury and stimulate significant cellular inflammation on cultured proximal renal tubular cells.34 It was reported that these compounds played significant roles in the pathology associated with chronic kidney injury.22,35 In the study of histopathological analysis of kidney, the pathological abnormalities in MG was more serious compared with CG. This may be related to the up-regulation of indoxyl sulfate and p-cresol (Table 1). In the study, the administration of GBE could reduce the level of p-cresol.
Regarding fatty acid metabolic, the up-regulation of tetracosahexaenoic acid in positive ion mode, and suberic acid, sebacic acid and octadecanedioic acid in negative ion mode were observed in MG compared with CG. These biomarkers connect with fatty acid oxidation (FAO). The process of FAO could produce a large amount of free radicals and peroxides, which led to endothelial cell injury and activate NF-κB signaling pathway, thereby increasing TNF-α, ICAM-1, vascular cell adhesion molecule-1 (VCAM-1) and other inflammatory cytokines.36 The down-regulation of fatty metabolites after the administration of GBE indicated that GBE might change perturbations toward normal level and inhibit oxidative stress.
Conclusions
In this study, the pathogenesis of acute gout with hyperuricemia and the therapeutic effects of GBE were investigated by metabonomic approach. It is benefited for the early detection and diagnosis but also for prediction of gout. It demonstrating the untargeted metabonomic method is a reliable method to investigate the pathogenesis of disease. A total of 27 potential biomarkers were identified to be involved in tryptophan metabolism, pyrimidine metabolism, pentose phosphate pathway, TCA cycle, tyrosine metabolism, lysine degradation and purine metabolism. Lots of enzymes and genes participate in the reactions and transformation process of metabolites upstream and downstream. Moreover, the study provided an overall view of the progression of gout and theoretical basis for treating gout.Conflicts of interest
The authors declare that they have no conflicts of interest regarding the publication of this work.Acknowledgements
The authors wish to give thanks to everyone who participated in this study. They are also grateful to the staff of the National Center of Mass Spectrometry in Changchun & Chemical Biology Laboratory. This work was supported by the National Natural Science Foundation of China (No. 81303280, 81573574, 81530094, 81473537, 31670356).References
- N. A. Kunitskaia and M. A. Andrianova, Adv. Gerontol. Res., 2012, 25, 285–289 CAS.
- K. G. Saag and H. Choi, Arthritis Res. Ther., 2006, 1(suppl. 8), S2 Search PubMed.
- T. Shimizu, H. Kitada, M. Umeyama, H. Hori and N. Takasaki, J. Urol., 2013, 189, 1747–1752 CrossRef PubMed.
- P. J. Fortun and C. J. Hawkey, Curr. Opin. Gastroenterol., 2007, 23, 134–141 CAS.
- C. Hesse, V. Razmovski-Naumovski, C. C. Duke, N. M. Davies and B. D. Roufogalis, Phytother. Res., 2007, 21, 523–530 CrossRef CAS PubMed.
- K. Higuchi, E. Umegaki, T. Watanabe, Y. Yoda, E. Morita, M. Murano, S. Tokioka and T. Arakawa, J. Gastroenterol., 2009, 44, 879–888 CrossRef PubMed.
- D. Normile, Science, 2003, 299, 188–190 CrossRef CAS PubMed.
- R. Yuan and Y. Lin, Pharmacol. Ther., 2000, 86, 191–198 CrossRef CAS PubMed.
- Y. B. Jiao, Y. C. Rui, P. Y. Yang, T. J. Li and Y. Qiu, Acta Pharmacol. Sin., 2007, 42, 930–934 Search PubMed.
- L. Puchades-Carrasco and A. Pineda-Lucena, Curr. Opin. Biotechnol., 2015, 35, 73–77 CrossRef CAS PubMed.
- A. Wojakowska, M. Chekan, P. Widlak and M. Pietrowska, Int. J. Endocrinol., 2015, 2015, 258763 Search PubMed.
- H. Wu and F. Feng, RSC Adv., 2016, 6, 28279–28288 RSC.
- Y. Li, Z. Zhang, Z. Hou, L. Wang, X. Wu, L. Ju, X. Zhang and Y. Zhang, RSC Adv., 2015, 5, 202–209 RSC.
- H. G. Gika, G. A. Theodoridis, R. S. Plumb and I. D. Wilson, J. Pharm. Biomed. Anal., 2014, 87, 12–25 CrossRef CAS PubMed.
- G. A. Theodoridis, H. G. Gika, E. J. Want and I. D. Wilson, Anal. Chim. Acta, 2012, 711, 7–16 CrossRef CAS PubMed.
- C. Xu, W. Chen, S. Liu, Z. Pi, F. Song and Z. Liu, Anal. Methods, 2015, 7, 6777–6784 RSC.
- A. Scanu, F. Oliviero, R. Ramonda, P. Frallonardo, J. M. Dayer and L. Punzi, Ann. Rheum. Dis., 2012, 71, 621–624 CrossRef CAS PubMed.
- D. Krause, H. S. Suh, L. Tarassishin, Q. L. Cui, B. A. Durafourt, N. Choi, A. Bauman, M. Cosenza-Nashat, J. P. Antel, M. L. Zhao and S. C. Lee, Am. J. Pathol., 2011, 179, 1360–1372 CrossRef CAS PubMed.
- L. Zhang, O. Ovchinnikova, A. Jonsson, A. M. Lundberg, M. Berg, G. K. Hansson and D. F. Ketelhuth, Eur. Heart J., 2012, 33, 2025–2034 CrossRef CAS PubMed.
- Y. Adelibieke, M. Yisireyili, H. Y. Ng, S. Saito, F. Nishijima and T. Niwa, Nephron Exp. Nephrol., 2014, 128, 1–8 CrossRef CAS PubMed.
- N. Masai, J. Tatebe, G. Yoshino and T. Morita, Circ. J., 2010, 74, 2216–2224 CrossRef CAS PubMed.
- Z. Tumur, H. Shimizu, A. Enomoto, H. Miyazaki and T. Niwa, Am. J. Nephrol., 2010, 31, 435–441 CrossRef CAS PubMed.
- Y. Y. Zhao, H. T. Li, Y. L. Feng, X. Bai and R. C. Lin, J. Ethnopharmacol., 2013, 148, 403–410 CrossRef CAS PubMed.
- Y. Liu, X. Sun, D. Di, J. Quan, J. Zhang and X. Yang, Clin. Chim. Acta, 2011, 412, 2132–2140 CrossRef CAS PubMed.
- G. C. Mills, F. C. Schmalstieg, K. E. Newkirk and R. M. Goldblum, Clin. Chem., 1979, 25, 419–424 CAS.
- R. de Medicis, A. Lussier, A. Kahan and L. Tetreault, Eur. J. Rheumatol. Inflammation, 1983, 6, 143–148 CAS.
- M. Gaudry, C. J. Roberge, R. de Medicis, A. Lussier, P. E. Poubelle and P. H. Naccache, J. Clin. Invest., 1993, 91, 1649–1655 CrossRef CAS PubMed.
- O. Popa-Nita, L. Marois, G. Pare and P. H. Naccache, Arthritis Rheumatol., 2008, 58, 1866–1876 CrossRef CAS PubMed.
- J. Maiuolo, F. Oppedisano, S. Gratteri, C. Muscoli and V. Mollace, Int. J. Cardiol., 2016, 213, 8–14 CrossRef PubMed.
- K. D. Pfeffer, T. P. Huecksteadt and J. R. Hoidal, J. Immunol., 1994, 153, 1789–1797 CAS.
- S. J. Klebanoff, J. Leukocyte Biol., 2005, 77, 598–625 CrossRef CAS PubMed.
- F. C. Meotti, G. N. Jameson, R. Turner, D. T. Harwood, S. Stockwell, M. D. Rees, S. R. Thomas and A. J. Kettle, J. Biol. Chem., 2011, 286, 12901–12911 CrossRef CAS PubMed.
- L. K. Stamp, R. Turner, I. S. Khalilova, M. Zhang, J. Drake, L. V. Forbes and A. J. Kettle, Rheumatology, 2014, 53, 1958–1965 CrossRef CAS PubMed.
- C. Y. Sun, H. H. Hsu and M. S. Wu, Nephrol., Dial., Transplant., 2013, 28, 70–78 CrossRef CAS PubMed.
- A. Adijiang, H. Shimizu, Y. Higuchi, F. Nishijima and T. Niwa, J. Renal Nutr., 2011, 21, 105–109 CrossRef CAS PubMed.
- X. Ding, Z. Yang, Y. Han and H. Yu, Placenta, 2015, 36, 1442–1449 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08519b |
| This journal is © The Royal Society of Chemistry 2017 |