Open Access Article
Open Access ArticleEffects of polyethylene oxide and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) nanofibrous substrate on omental adipose-derived mesenchymal stem cell neuronal differentiation and peripheral nerve regeneration†
Feihu Hu *a,
Ting Chenb and
Wei Wangbc
*a,
Ting Chenb and
Wei Wangbc
aSchool of Bioscience and Technology, Weifang Medical University, Weifang, Shandong, People's Republic of China. E-mail: wfmuhufeihu@hotmail.com
bDonghua University, Shanghai, People's Republic of China
cCollege of Materials Science and Engineering, Donghua University, Shanghai, People's Republic of China
First published on 5th September 2017
Abstract
Among a variety of polymers, poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) and polyethylene oxide (PEO) display biodegradable and biocompatible properties for applications in the biological, biochemical and biomedical fields. PEO incorporated with PHBV may be used to fabricate superior electrospun nanofibres. Herein, we investigated the influence of PEO content and aligned substrate topography on the viability, stemness properties and neuronal differentiation capabilities of omental adipose-derived mesenchymal stem cells (O-ASCs) in vitro. Decreasing the mass ratio of a PHBV/PEO solution from 60/40 to 90/10 is inversely correlated with increasing the fibre diameter. O-ASCs cultured on aligned and randomly-oriented nanofibrous substrates exhibit differences in cell proliferation, cell cycle distribution, and surface epitope expression. Upon induction by retinoic acid (RA), the cell morphology guidance and a higher level of Wnt signalling pathway activation of neuronally differentiated O-ASCs are achieved on the aligned substrate, wherein a combined effect of cell-selective sensitivity for the substrate topography and the external RA induction are observed. Additionally, the performance of PHBV/PEO nanofibrous conduits is evaluated for sciatic nerve regeneration in rats. Encouraging results from stained images and motor function recovery assays indicate that it is sufficient to combine aligned 90/10 PHBV/PEO nanofibrous conduits and neuronally differentiated O-ASCs for the repair of the sciatic nerve in vivo.
Introduction
Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV), a polyhydroxyalkanoate and nontoxic thermoplastic polyester produced by microorganisms, can meet the challenges for biomaterial and biomedical applications due to its thermoplasticity, biodegradability, and biocompatibility.1 Recent work demonstrated that PHBV fibrous substrates support the adhesion and proliferation of various cells, and serve as synthetic scaffolds for regeneration in the tissue engineering of nerve, bone, cartilage, and vascular tissue in vivo.2 On the other hand, it is inevitable that the shortcomings, such as high crystallinity and rigidity, poor mechanical performance and hydrophobicity, restrict its application. In this context, the polymers, gelatine, fibronectin and other components were used as additives for facilitating the biomechanical and biocompatible properties of the substrate by incorporating PHBV.3 Wu reported that the 20% bioglass could enhance the adhesion and penetration of chondrocytes into PHBV base scaffolds for bone tissue engineering.4 The calcium silicate (CS) incorporated with PHBV had a good apatite-forming ability and promoted osteogenic differentiation of MG63 cells on the scaffolds by monitoring the alkaline phosphatase (ALP) expression, which could be attributed to the electrostatic attraction of the PHBV composite.5 The curcumin-entrapped polyaniline modified PHBV for increasing the electroconductivity to the value of 5.78 × 10−5 S cm−1 and exposing the significant compatibility towards blood and fibroblast cells.6Polyethylene oxide (PEO), which is FDA certified, is a biocompatible, hydrophilic and malleable polyether and is widely applied in biomedical engineering, especially in humoural contact devices, because of its non-immunogenicity and its ability to reduce surface protein adsorption.7 Moreover, it is soluble not only in water but also in different kinds of organic solvents, which is beneficial to yield nanofibres via the electrospinning method. The electrospun fibrous substrates and scaffolds are favourable for tissue engineering, drug delivery, biosensors and wound healing by bridging gaps between truncated stumps, supporting nutrient exchange, directing regenerating cells and providing mechanical structures.8,9
The intra-abdominal omental adipose-derived mesenchymal stem cells (O-ASCs) that were tested appeared to possess good biological properties, and they could respond to specific inducing conditions and differentiate into a variety of cell lineages, including osteoblasts, lipocytes, and neurons, which reveals their notable pluripotency.10 Research exposed that substrates presenting topographical features, such as pits, pillars, and grooves, irritate the cultured cells, leading to morphological and functional change.11 Therefore, we were interested in investigating the influence of PHBV nanofibrous substrate topography on O-ASCs survival and neuronal differentiation capabilities. Meanwhile, we were devoted to elucidating signalling pathways and mechanisms caused by the impact of the substrate topography.
In this study, PEO was introduced into a PHBV-based electrospun polymer solution as a modulating element for perfecting fibres fabrication. The PHBV/PEO electrospun nanofibres with different mass ratios (90/10 and 60/40) and orientations (random-oriented and aligned) were produced to investigate the impact of composition value and arrangement on O-ASCs adhesion, survival and neural transformation. We scrutinized the characteristics of added PEO on nanofibre formation and nanofibrous diameter distributions. A cell viability assay suggested the random-oriented nanofibrous substrates are less receptive to the attachment and continued survival of O-ASCs than aligned nanofibrous substrates or the planar unpatterned substrate. In response to retinoic acid (RA), the alignment topography guides the O-ASCs and promotes elongation along the major fibre axis. The canonical Wnt signalling pathway is markedly enhanced, and a higher level of activity of this pathway was observed in the aligned group, which might further potentiate the effects of RA-inducing treatment on neuronal differentiation. These synthetic scaffolds that are often used as complemented therapies fill the injury sites and assist in tissue repair in regenerative medicine by combining the neural differentiated stem cell.12–14 Herein, the aligned and random-oriented nanofibrous conduits and RA-induced O-ASCs, as artificial nerve grafts, were implanted into a rat sciatic injury model in vivo. Animal motor function analysis, histological examination, and immunohistochemical staining analysis were used for assessing the nerve regeneration and motor function recovery.
Materials and method
Materials
Poly(3-hydroxybutyrate-co-3-hydroxyvalerate), with 12 wt% HV, 3-hydroxyvalerate, was purchased from Sigma-Aldrich, USA. PEO, poly(ethylene oxide) (MW = 1000 kDa) from Guoren Chemical Co., China, and TFE (2,2,2-trifluoroethanol) from Darui Fine Chem Ltd., China. All reagents and solvents were used as purchased without any further purification.Preparation of PEO incorporated with PHBV electrospun nanofibres
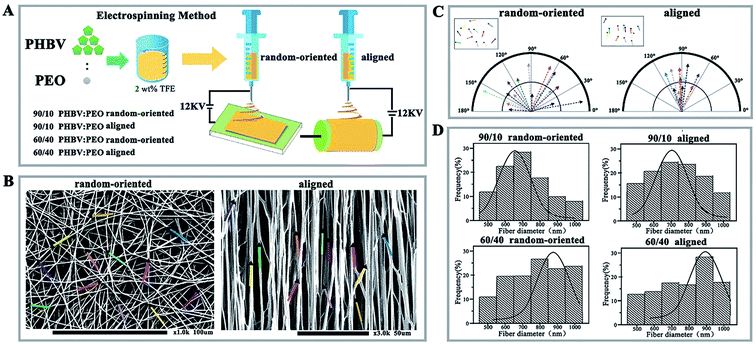
PEO was chosen as an additive (the mass ratios of PHBV to PEO were at 90/10 and 60/40, respectively), and 2 kinds of orientation nanofibres, including random-oriented and aligned, were obtained by the electrospinning method.15 In brief, different mass ratios of PHBV and PEO were dissolved in TFE to form a 2% (w/v) solution prior to electrospinning. Then, the solution was filled into a 20 mL syringe equipped with a 6# needle (inner diameter of 0.5 mm) and constantly driven by a syringe pump at a flow rate of 5 mL h−1, voltage = 12 kV high DC. A 25 cm distance was applied between the metal needle and the collector.Fibre deposition onto a stationary grounded collector resulted in randomly oriented nanofibres on the collected aluminium foil surface and formed a thin film (random-oriented group), while a rotating disc collector yielded alignment nanofibres on the aluminium foil surface (aligned group). All collected nanofibrous substrates were dried under vacuum for 24 h to remove residual solvent and stored in a desiccator for further characterization (as shown in Fig. 1A).
The morphology of nanofibrous substrates was analysed by scanning electron microscopy (SEM, ultra plus Zeiss, Germany). The angles of fibres were measured from SEM images using Matlab software (MathWorks, version 2013), and fibre diameters were measured using ImageJ software (version 6.0). A minimum of 50 fibres were measured for each sample. These different nanofibres were divided into 90/10 random-oriented group, 90/10 aligned group, 60/40 random-oriented group, and 60/40 aligned group.
O-ASCs isolation, culture and neuronal differentiation
O-ASCs were derived from Sprague Dawley rats as described previously.16 This study was performed in strict accordance with the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. This protocol was approved by the Animal Experimental Committee of Southeast University, Nanjing, China (Permit Number: 2014ZDIIKY22.1). Briefly, the intra-abdominal omental adipose tissue was sliced and digested with 0.1% Collagenase I (Sigma Aldrich, USA). The O-ASCs were isolated and propagated into T25 flasks and cultured in a basal medium composed of DMEM medium (Thermo Fisher Scientific), 5% foetal bovine serum (FBS; Gibco Lab., USA), and 1% penicillin/streptomycin (Thermo Fisher Scientific). The medium was replaced every 3 days for a period of 10 days.To assess cell adhesion and survival, the nanofibrous substrates were sterilized by a series of washing steps in 70% ethanol. Then, O-ASCs, with a density of 2 × 105 cells per cm2, were propagated onto the nanofibrous substrate surface (1 cm2 area) and allowed to attach for 48 h. The cells were propagated into 12-well flat plates and served as the planar unpatterned group (control group).
For the neuronal differentiation studies, O-ASCs were propagated on the nanofibrous substrate surface and induced with retinoic acid (RA, Sigma Aldrich, USA) for 14 days. The groups were divided into the random-oriented + RA group and aligned + RA group. The contemporaneously prepared cells on the 12-well flat plates without any treatment served as the control group (flat + un-treated group). After culturing, the cells were fixed for immunostaining and further analysis.
Fluorescence staining analysis
Hoechst 33342 (Sigma Aldrich, USA) staining was used for visualization of cell nuclei. The primary antibody was specific to the neural stem/progenitor cell marker Nestin (Santa Cruz, USA). The secondary antibody used was anti-rabbit Alexa Fluor 488 (Invitrogen). The samples were imaged using a Revolution XD confocal laser scanning microscope (Andor, Northern Ireland). The surface coverage ratio was analysed using Matlab software.Cell viability assay
The cell viability was evaluated via quantification of cell metabolic activity using the Cell Counting Kit-8 method (Dojindo, Japan) according to the manufacturer's instructions. The sample absorbance values were recorded and converted to absolute cell number by correlating with a standard curve. The number of cells in the 12-well flat plates served as a control (control group), and the experiment was repeated 3 times for each sample.Flow cytometry analysis
The surface epitope expression levels and the cell-cycle distribution were performed using an Accuri C6 Flow Cytometer (BD Bioscience, USA). Cells on the nanofibrous substrates were washed and detached with 0.25% trypsin solution, then fixed in 75% ethanol. The cells were collected and incubated with the following antibodies: CD11b-FITC (AbD serotec, USA), CD 29-FITC (Biolegend, USA), CD 44-FITC (AbD serotec, USA), CD 73-FITC (Santa Cruz, USA) and CD 106-PE (AbD serotec, USA). At least 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded and the data were analysed using CFlowPlus software and Matlab software. For the cell-cycle analysis, the cells were stained with propidium iodide (PI, Sigma Aldrich, USA) according to the manufacturer's instructions and analysed with ModFit software (Verity Software House, version 3.3).
000 events were recorded and the data were analysed using CFlowPlus software and Matlab software. For the cell-cycle analysis, the cells were stained with propidium iodide (PI, Sigma Aldrich, USA) according to the manufacturer's instructions and analysed with ModFit software (Verity Software House, version 3.3).
Western blotting and quantitative real time PCR analysis
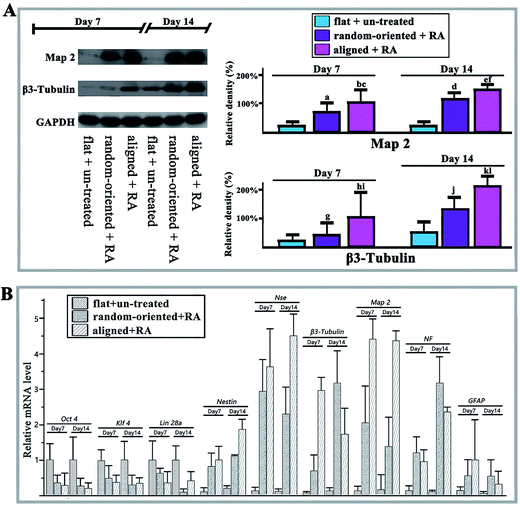
The level of stemness marker expression, the process of neuronal differentiation, and activation of Wnt signalling through the canonical Wnt/β-catenin pathway were measured via western blotting. After the cells were propagated on the nanofibrous substrates and then treated with RA for 14 days, the cellular and nuclear proteins were extracted and quantified. The protein samples were loaded on SDS-PAGE gels and electrophoresed under standard conditions. After blocking, the membranes were incubated with primary and secondary antibodies. The primary antibodies included the following: anti-Oct 4 (Santa Cruz Biotechnology, USA), anti-Sox 2 (Santa Cruz Biotechnology, USA), anti-Nanog (Santa Cruz Biotechnology, USA), anti-Map 2 (Santa Cruz Biotechnology, USA), anti-β3-Tubulin (Santa Cruz Biotechnology, USA), anti-β-Catenin (Boster Biotechnology, China), anti-Frizzled (Immunoway, USA), anti-phospho-Frizzled (Immunoway, USA), and anti-GAPDH (Cell Signaling Technology, USA).The mRNA levels of markers (Oct 4, klf 4, Lin 28a, Nestin, NSE, β3-Tubulin, Map 2, NF, GFAP, cyclin D1, c-myc, Axin 2 and c-jun) were monitored by qRT-PCR. Total RNA from the differentiated O-ASCs cultured on the 12-well flat plates (flat + un-treated group), 90/10 random-oriented nanofibrous substrates (random-oriented + RA group), and 90/10 aligned nanofibrous substrates (aligned + RA group), were isolated. One milligram of total RNA was converted to cDNA, according to the M-MLV reverse transcriptase instructions (Promega, USA). The PCR reactions were performed using SYBR Premix ExTaq (TaKaRa, China) on an ABI 7500 real time PCR system (Application Biosystems, USA). The Gapdh mRNA was used as a housekeeping gene. The primers are listed in ESI.†
Angle analysis of the RA-induced O-ASCs axon
Based on the nanofibre arrangement orientations (the direction of the fibres is the 0° line, see inset in Fig. 4C), the number of RA-induced O-ASCs with axons was measured compared to the total number of cells from the fluorescence images. The angular deviation (θ) distribution and the data were analysed with Matlab software.Preparation of nanofibrous conduits and animal surgical procedures
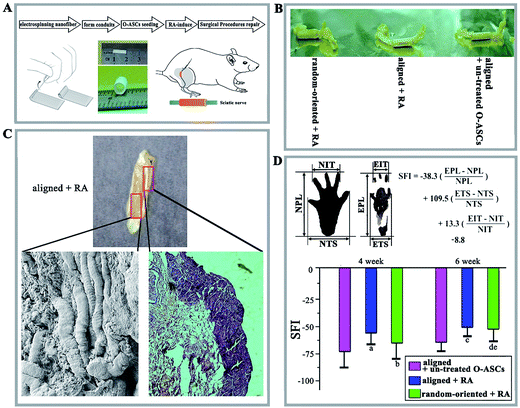
We developed a protocol for repairing rat sciatic nerve injury, and the schematic representation is shown in Fig. 7A. The nanofibrous films (random-oriented and aligned nanofibrous substrates of PHBV/PEO with a mass ratio of 90/10) were cut into rectangular shapes (15 cm length and 15 mm width) and rolled into a hollow tubular structure (15 mm length). Before use, all the nanofibrous conduits were sterilized by autoclaving at 120 °C for 20 min. The O-ASCs (1 × 106) were injected to the hollow conduits and maintained for 2 days in basal medium; then, the RA-induced medium was added and replaced every 3 days. After inducing for 15 days, the grafts containing the RA-induced O-ASCs and nanofibrous conduits were prepared for the in vivo surgical procedures.The surgical procedures were as follows: the rats were anaesthetized, and a 3 cm incision was made along the right femoral axis. The sciatic nerve was exposed, a 10 mm nerve segment was resected, the nerve segment ends were inserted into the lumen of the scaffold, the muscle and skin were sutured, and the incision was closed in layers using silk sutures. After the operation, the animals were kept in the animal facility and maintained under a 12 h light/dark period, with water and food ad libitum. The animals were divided into 3 groups, depending on which artificial nerve graft was used (n = 18): the 90/10 aligned electrospun nanofibrous conduit-containing O-ASCs without any treatment as the control group (aligned + un-treated O-ASC group), the 90/10 aligned electrospun nanofibrous conduit-containing RA-induced O-ASCs (aligned + RA group), and 90/10 random-oriented electrospun nanofibrous conduit-containing RA-induced O-ASCs (random-oriented + RA group).
Morphological assessment of regeneration nerve
At the end of the experiments (6 weeks), the rats were sacrificed and the sciatic nerve with the graft was harvested and fixed overnight in 4% paraformaldehyde (Sigma Aldrich, USA) at 4 °C for further analysis.The longitudinal section images of the grafts containing the regenerated sciatic nerve were observed using SEM. The transverse section of the grafts was stained with haematoxylin and eosin (H&E). The regenerated nerves and grafts were fixed and then dehydrated in ethanol, cleared in xylene, and embedded in paraffin. The samples were sectioned at 5 μm thickness, mounted on glass slides and observed under a light microscope.
Gait and sciatic function index (SFI) analysis
Walking track analysis was performed at the 4th week and 6th week. After pre-training, the rats were stained with black ink on their hind feet and allowed to walk across a 100 cm long passageway, and their footprints were recorded on paper that paved the bottom of the passageway. Functional assessment of sciatic nerve regeneration was expressed in term of sciatic function index (SFI).17 The lengths of the third toe to the heel (PL), the first to the fifth toe (TS) and the second to the fourth toe (IT) were measured on the experimental side (E) and the contralateral normal side (N) of each rat. The SFI value for each animal was calculated by the formula shown in Fig. 7D. Values oscillating around −100 SFI represent total dysfunction, whereas oscillation approximately 0 for normal nerve function.Statistical analysis
The results were expressed as the mean ± SD. Statistical analysis was performed using SPSS 13.0 software. Differences were compared using t-tests; statistical significance was determined as p-value < 0.05 or p-value < 0.01.Results and discussion
PEO incorporated with PHBV fabricates superior electrospun nanofibres
Electrospinning method is cost-effective and enables the manufacture of micro-size or nanoscale fibres. Under a high electrostatic force, a liquid is ejected from the apex of the so-called “Taylor cone” when the force overcomes the surface tension, and the fibres can be collected after solvent evaporation.18,19 Previous work highlighted that PHBV electrospun nanofibres are structurally analogous to the natural protein fibrils/fibres in the extracellular matrix, which has been applied for tissue engineering.20,21 However, PHBV nanofibres are typically hydrophobic and lack recognition sites for cell attachment. Therefore, it is important to introduce active biomolecules to synergistically promote cell culture. Towards this requirement, PHBV blended with PEO of different mass ratios (90/10 vs. 60/40 of PHBV/PEO) and orientations (random-oriented fibres vs. aligned fibres) was fabricated to nano-size fibres by electrospinning (Fig. 1A). Aligned nanofibres were achieved by directing the charged polymer jet towards the edge of a rapidly rotating disc collector. Conversely, deposition onto a stationary collector yielded random-oriented nanofibres due to the whipping instability of the jet caused by electrostatic repulsion on its surface.As shown in Fig. 1B, decreasing the PEO solution concentration from 60/40 to 90/10 weakly impacted the stability and morphology of nanofibres, which exhibit a typical morphology of uniform and striated line texture. Furthermore, nanofibres collected by stationary aluminium foil showed a scattered extension in the direction aligned parallel to each other in the aligned group and showed orientation angles of 90° ± 85° with a broad distribution, compared with the narrow range of 90° ± 27° from rapidly rotating collection in compass analysis (Fig. 1C). Even so, the specific average diameters of nanofibres showed an unexpected reduction, such that smaller average fibre diameters and narrower distribution profiles were observed in the 90/10 random-oriented group (Fig. 1D). Electrospinning of a 90/10 mass ratio solution yielded fibres with average values of approximately 677 nm (90/10 random-oriented group) and 713 nm (90/10 aligned group) with a 36 nm difference. The largest diameter of fibres was generated in the 60/40 aligned group with 903 nm, 26 nm more than the 877 nm from 60/40 random-oriented fibres.
As previously reported, the fibre productivity, morphology features and dimension distributions were greatly modulated by numerous regulatory factors. Schnell et al.22 compared the fibres of 0/100 poly-epsilon-caprolactone (PCL) with fibres consisting of a 25/75 collagen/PCL blend and indicated that aligned C/PCL nanofibres could work as a suitable substrate for supporting cell proliferation, and the aligned group produced better axonal guidance for Schwann and fibroblasts cells. Herein, the inverse correlation of PEO content with increasing fibre diameter was realized, and we speculated that it likely ascribes to the presence of PEO with high molecular weight and hydrophilicity, which increases the average molecular weight of the blend and reduces the surface tension of the mixed polymer solution.23
Nanofibrous substrates promote O-ASCs proliferation
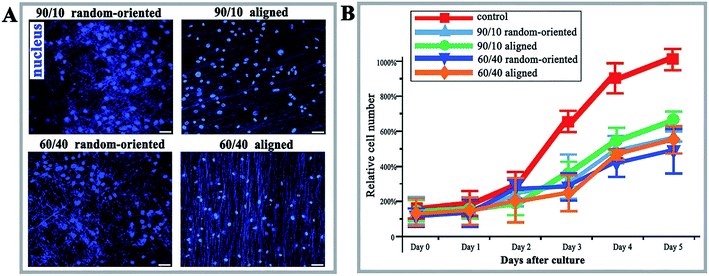
Microscopic examination of O-ASCs following propagation of the initial cells on the fibre substrates identified that the cells cultured on substrates are incapable of elongation in an unrestricted manner, while having capability on a planar unpatterned surface, and the majority of cells were characterized by a triangular or spindle-like shape and have complete nuclei (Fig. 2A). The lower number of living cells in the 4 nanofibre groups demonstrated the topographical restriction for cell survival; notwithstanding, the number is close to that observed for the control group (Fig. 2B). The 90/10 aligned group contained the highest cell relative number, with 674% and at least 1.13-fold relative to other fibre groups, with a statistical difference.Nanofibrous substrates influence the cell cycle distribution and surface epitope expression of O-ASCs
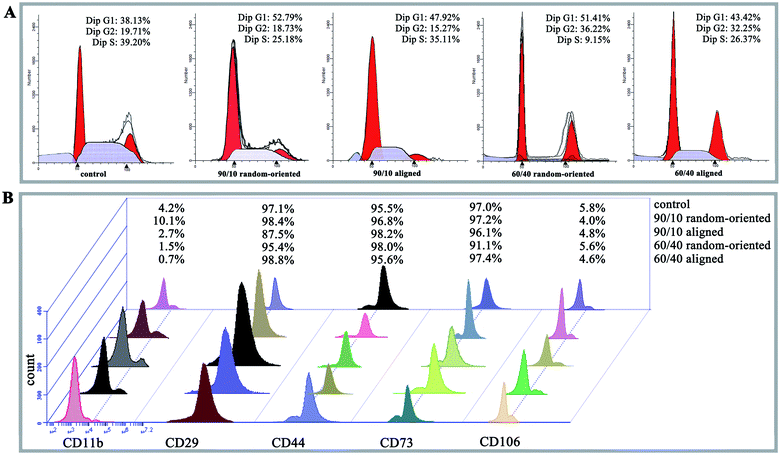
PI staining and cell cycle investigation showed that the S-phase proportion of cells cultured on 4 nanofibrous substrates was lower than that on the 12-well flat plates, regardless of the mass ratios and topography (the 4 kinds of nanofibrous groups vs. control group as 25.18%, 35.11%, 9.15%, and 26.37% vs. 39.2% in Fig. 3A). In the same mass ratio groups, such as 90/10 and 60/40, the S-phase percentage in the aligned groups was higher than that in the random-oriented groups (35.11% vs. 25.18% and 26.37% vs. 9.15%). Ramkumar et al.24 also evaluated the effect of TiO2 nanofibrous substrate on the G2/M cell cycle distribution, and the inhabitation outcomes, which verified our conclusion that the cell cycle of O-ASCs cultured on aligned nanofibres is induced from the G1-phase to the S-phase. Subsequently, surface epitope expression levels were performed, whereby the nonnegligible discrepancy was established and suggested that the nanofibrous substrates modulate cell proliferation and cell activity properties (e.g., CD11b expression in the 90/10 random-oriented group, CD 29 expression in the 90/10 aligned group, and CD 73 expression in the 60/40 random-oriented group, Fig. 3B).In general, our nanofibrous substrates are successful for O-ASCs adhesion even though the numerous nanofibres are approximately 1/40 to 1/10 of the diameter of a rounded cell. The cultured O-ASC performance revealed that the PHBV and PEO proportion participates the regulation of cell attachment in a feature size-dependent manner. On the other hand, compared to the cells on the 12-well flat plates, the topographical features of the nanofibrous substrate limits cell spread and expansion. Additionally, compared with two random-oriented groups, the aligned substrates seem to provide a superior support for cell proliferation and survival. Previous studies declared that irregular cell morphology encourages and stimulates gene expression, which ultimately decides the cells' activity and fate (survival or apoptosis).25,26 Analogously, the substrate topography cues stimulate the cell morphologic changes based on special cell sensitivity, which helps to sense the external biophysical topography. An intricate signal triggers the rearrangement of cytoskeletal components, such as actin structures and focal adhesion complexes.27,28 The above all promote cell adaptation characterized as abnormal cell proliferation, cell cycle distribution, and surface epitope expression.
Nanofibrous substrates support RA-induced O-ASCs neuronal differentiation
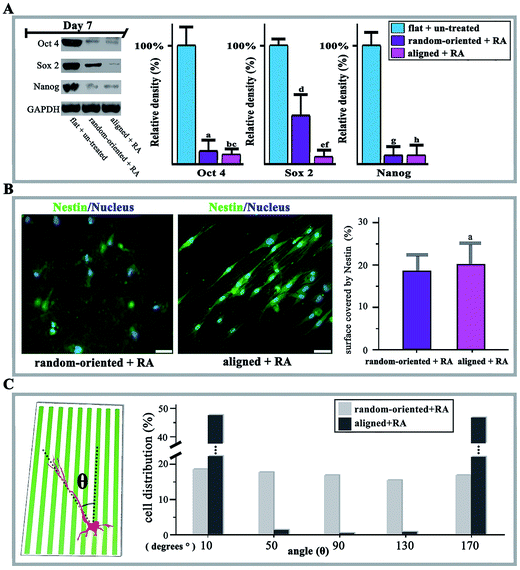
Moreover, it is also considered a question whether fibre substrates are capable of shifting stem cell responsiveness to differentiation-inducing conditions. To this end, the O-ASCs cultured on 90/10 random-oriented and aligned nanofibrous substrates were induced towards neural transformation via RA treatment for 14 days. The protein expression of Oct 4, Sox 2, Nanog, Nestin, Map 2 and Tubulin were selected for testing, since enhanced expression levels of these markers indicate O-ASCs neuronal differentiation.The cells on planar unpatterned substrates showed a basal level of stemness markers in the absence of RA treatment (flat + un-treated group). Specifically, the levels of Oct 4, Sox 2 and Nanog are lower, with 60.2% and 95.1% reduction, which indicates that fibre substrates also support the RA-induced O-ASCs at day 7. Cells on aligned fibres have dramatic decreases in stemness (random-oriented + RA vs. aligned + RA shown as 10.1% vs. 7.2% in Oct 4, 39.8% vs. 4.9% in Sox 2, and 5.9% vs. 5.6% in Nanog), pointing to the impact of substrate topography on neuronal differentiation (Fig. 4A).
Conversely, in the aligned group, over 90% of O-ASCs positively stained for Nestin and extended neurites that were several times longer than the cell bodies. The extension was guided by the axis of fibre alignment and the values of quantified Nestin coverage on the aligned substrate were higher (20.2%) than the 17.9% in the random-oriented group (Fig. 4B). As a result, in the presence of RA as an inducer, culturing O-ASCs on aligned fibres substrate significantly potentiated the yield of neural lineage cells. Axon orientation analysis is capable of exposing influences by detecting the angle of growth with respect to the direction of aligned fibres. In contrast to the random-oriented group, with an even distribution of values from 10° to 170° and without any significant trends, the value mostly gathered approximately 10° or 170° with a narrow range in the aligned group (Fig. 4C).
In addition to altering the pattern features of cell-substrate contacts, the substrate topography has also been shown to activate the neurogenesis genes towards a higher degree. Thus, we monitored higher levels of Map 2 and β3-Tubulin in aligned and random-oriented groups and they continued to increase until reaching the peak at day 14 (Fig. 5A). Notably, the expression level in the aligned group surpassed the random-oriented group. Similarly, the up-regulation of neural cell markers with simultaneous stemness marker silencing was further confirmed by q-RT-PCR (Fig. 5B).
Exogenous RA can enhance the neural transformation stimulator, NeuroD,29 and we revealed the substrate patterns (aligned and random-oriented groups) and RA treatment stimulation towards a specific contact, which facilitates the differentiation process of O-ASCs in vitro and is consistent with previous studies. Mirzaei et al.30 confirmed that the electrospun carbon nanofibres could be used as a proper substrate for human endometrial stem cell attachment and neuronal differentiation under the same RA concentrations. Aligned fibrillar fibrin hydrogel (AGF) instructed the neurogenic differentiation and rapid neurite outgrowth of human umbilical cord mesenchymal stem cells by modulating their elasticity.31 Ren et al.32 compared the aligned fibres of 600 nm and 1.6 μm diameter sensitivity for the human embryonic stem cells (ES cells) and provided an efficient protocol for ES cell-derived neural crest stem cells by combining an aligned nanofibre matrix and an optimized differentiation medium. As expected, the aligned substrate favoured the neuronally differentiated O-ASCs, and the cells propagated on the random-oriented surface were insensitive to the treatment. Therefore, we acknowledged that the RA induction and aligned substrate topography sufficiently potentiated the neuronal differentiation of O-ASCs in a possibly synergistic manner.
Aligned nanofibrous substrate facilitated Wnt signalling pathway expression
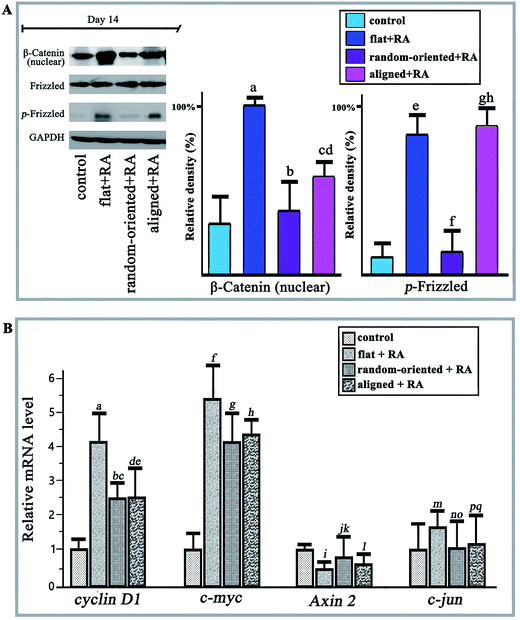
The Wnt signalling pathway (involving canonical Wnt/β-Catenin) regulates diverse cellular programmes including embryogenesis, proliferation, differentiation, and apoptosis. Activation and combination of Wnt proteins and their receptors increase the activity of glycogen synthase kinase 3β (Gsk 3β) and inhibits Axin. Consequently, cytosolic β-catenin is allowed to accumulate and translocate to the nucleus. Nuclear β-catenin binds to the TCF/LEF promoter region to activate gene expression.33 Wnt-4 and Wnt-11 (Wnt signalling pathway regulator) can promote early neural transformation by diminishing the RA-induced down-regulation of Oct 4 and Nanog and increasing the up-regulation of ASCL 1 and Neuro D1.34 Meanwhile, by combining BMP and Wnt signals, the time sequence method induced neuronal and astroglial differentiation but suppressed differentiation towards oligodendroglial lineages.35In the present work, we observed the activation of Wnt/β-catenin when cells responded to substrate topography and RA treatment. First, the RA induction stimulated nuclear β-Catenin and phosphorylated Frizzled in the flat + RA group, and the levels are at least twice greater than those of the cells on a flat substrate, which produces a basal level of activation in the absence of RA (control group, Fig. 6A). On one hand, the expression levels of nuclear β-Catenin and phosphorylated Frizzled are higher in all the RA treatment groups than in the absence of RA, which demonstrates that the RA induction significantly activates Wnt signalling. On the other hand, compared to the random-oriented fibre substrate, the aligned topographical feature markedly enhances the translocation of β-Catenin into the nucleus and phosphorylates Frizzled, in addition to guiding the differentiation of the O-ASCs. Additionally, the qRT-PCR results showed a similar function for the cyclin D1, c-myc, Axin 2, and c-jun mRNAs, while the activation of Wnt signals up-regulated the nuclear transcription factor expression and inhibited the expression of Axin (Fig. 6B).
Currently, more research has identified that the rearrangement of focal adhesion and the inhibition of cell spreading in response to substrate-presented cues are linked to activation of signalling pathways, such as MAPK,36 Runx 2,37 NF-κB,38 RhoA,39 and Wnt.40 Zhu et al.41 recognized that the scaffold nano-topography could modulate the microenvironment of mesenchymal stem cells and promote their osteogenic differentiation by regulating key ERK signalling proteins. PLLA nanofibres that mimicked the natural structure directly induced the MC3T3-E1 cell osteogenic differentiation through the RhoA-Rock signalling pathway.42 After studying the relationship of the synthetic polyamide substrate and mESC self-renewal, the fibre topography was verified as a cue for the activation of Rac and PI3K signalling pathways.43 Our results showed that the substrate topography feature produces variable amounts of signal activation; the expression levels of cells on the aligned substrates were higher than those from the random-oriented group and closer to the planar unpatterned group. Because of the dual-functional of β-catenin as a structural component of the cytoskeleton as well as a central factor in canonical Wnt signalling, it seems that extending O-ASCs morphology in response to the substrate topographical features can influence the dynamics of intracellular β-catenin bioavailability.44,45 Moreover, some research and our results confirmed that RA treatment is sufficient to up-regulate Wnt expression,46 and we suggested that the substrate topographical features can supply an additional physical signal to the cultured cells. The extracellular signal could transduce multiple intracellular biochemical signals (for instance, the Wnt signalling pathway) and change their behaviours significantly. Therefore, the RA induction and substrate topography have a combinatorial effect on stimulation of the Wnt signalling pathway. Additionally, prior work established that Wnt/β-catenin signalling, with simultaneous Sox2 silencing, also triggered NeuroD activation,29,47 which, in response to RA, was shown to promote neural transformation in adult neural stem cells.48 The intermediate steps connecting extracellular physical signal from the cell-contact surface region and the intracellular downstream signalling pathways, in response to substrate topography, need further study in the future.
PHBV/PEO nanofibrous conduits improved sciatic nerve injury regeneration and motor function recovery in vivo
Through a combination of biomaterials and stem cells, tissue engineering strategies for restoring and regenerating damaged peripheral nerves has recently been used to meet the challenges posed by nerve injury.49,50 Research groups studying nerve tissue engineering have designed a 3D nerve tubulisation and probed the mimetic structure for replacing the autologous nerve grafts in clinical practice. In this study, the regeneration of damaged sciatic nerve and the restoration of motor function were used to evaluate the electrospun nanofibres and cultured RA-induced O-ASCs in vivo. The schematic shows the experiment flow (Fig. 7A).After 6 weeks, the harvested grafts with wrapped regenerated sciatic nerves had no dislocation and inflammatory reactions. In addition, the aligned nanofibrous conduits (aligned + RA group) appeared well integrated into the host tissue with potent regeneration occurring inside the conduits (Fig. 7B). The regenerated nerve fibres and myelin sheaths proved that aligned PHBV nanofibrous conduits can facilitate nerve tissue engineering (Fig. 7C).
A tendency toward auto-restoration of motor function from 4 to 6 weeks was observed and the SFI value of RA-induced O-ASCs cultured on aligned nanofibres increased at a more rapid rate (Fig. 7D). At weeks 4 and 6, the value of the aligned + RA group was higher than other groups (−58.2 ± 9.6 vs. −63.7 ± 17.4 in the random-oriented + RA group and −73.5 ± 13.9 in the aligned + un-treated O-ASCs group at 4 weeks, and −50.4 ± 10.3 vs. −52.1 ± 12.7 and −62.1 ± 9.5 at 6 weeks).
Some studies have explored various treatment methods involving stem cells cultured on nanofibrous scaffolds stimulating peripheral nerve injury restoration.51 In the present study, histological examination showed a profound improvement in motor function, collectively suggesting a potential mechanism whereby the tissue-engineering scaffolds and stem cells encourage mixed success via the guidance of aligned topography, by which the differentiated stem cells are sufficiently transformed into nerve fibres or axons. The former provides the extracellular matrix environment between truncated stumps and prevents the apoptosis of distal neurons.52 The latter can replace lost neurons by differentiating into neural lineages or helping to secrete neurotrophic factors for inducing autologous peripheral nerve repair in vivo.53
Conclusions
Currently, PEO, as an additive, facilitates the properties of electrospun nanofibres. Herein, we investigated the influence of PEO content and aligned substrate topography on cell survival, stemness and O-ASCs neuronal differentiation in vitro. The decreasing mass ratios of the PHBV/PEO solution from 60/40 to 90/10 are inversely correlated with increasing fibre diameters. Cell viability tests indicated that the aligned nanofibres have good biocompatibility. Compared to random-oriented fibres, the alignment topography increased the neural transformation in the presence of RA with a clear guidance for cell extension. Higher Wnt signalling pathway expression levels were observed in the aligned substrate group and this enhancement might be due to the combined effect of O-ASC selective sensitivity for the substrate topography and the external RA induction. Furthermore, the animal tests in vivo showed that the 90/10 PHBV/PEO nanofibrous nerve conduits with cultured RA-induced O-ASCs could promote the regeneration of sciatic nerve deficits and the recovery of motor function. The above functionalized nanofibres and O-ASCs scaffolds provide a novel and promising platform for peripheral nerve regeneration.Conflicts of interest
The authors declare that there are no conflicts of interest.References
- M. Avella, E. Martuscelli and M. Raimo, J. Mater. Sci., 2000, 35, 523–545 CrossRef CAS.
- Y. Li, Y. Xiao and C. Liu, Chem. Rev., 2017, 117, 4376–4421 CrossRef CAS PubMed.
- L. Grondahl, A. Chandler-Temple and M. Trau, Biomacromolecules, 2005, 6, 2197–2203 CrossRef CAS PubMed.
- J. Wu, Z. Wu, Z. Xue, H. Li and J. Liu, RSC Adv., 2017, 7, 22197–22207 RSC.
- C. J. Shuai, W. Guo, C. D. Gao, Y. W. Yang, Y. Xu, L. Liu, T. Qin, H. Sun, S. Yang, P. Feng and P. Wu, Polymers, 2017, 9, 15 CrossRef.
- N. Pramanik, K. Dutta, R. K. Basu and P. P. Kundu, ACS Biomater. Sci. Eng., 2016, 2, 2365–2377 CrossRef CAS.
- L. Huang, K. Nagapudi, R. P. Apkarian and E. L. Chaikof, J. Biomater. Sci., Polym. Ed., 2001, 12, 979–993 CrossRef CAS PubMed.
- S. Megelski, J. S. Stephens, D. B. Chase and J. F. Rabolt, Macromolecules, 2002, 35, 8456–8466 CrossRef CAS.
- Y.-F. Goh, I. Shakir and R. Hussain, J. Mater. Sci., 2013, 48, 3027–3054 CrossRef CAS.
- H. Kazuaki, H. Chieko, W. Keiichi, K. Reo, I. Hiroaki and T. Yasuhiko, Nephrology, 2014, 19, 190 Search PubMed.
- W. Chen, Y. Shao, X. Li, G. Zhao and J. Fu, Nano Today, 2014, 9, 759–784 CrossRef CAS PubMed.
- M. Georgiou, J. P. Golding, A. J. Loughlin, P. J. Kingham and J. B. Phillips, Biomaterials, 2015, 37, 242–251 CrossRef CAS PubMed.
- B. G. X. Zhang, A. F. Quigley, D. E. Myers, G. G. Wallace, R. M. I. Kapsa and P. F. M. Choong, Int. J. Artif. Organs, 2014, 37, 277–291 CrossRef PubMed.
- X. S. Gu, F. Ding and D. F. Williams, Biomaterials, 2014, 35, 6143–6156 CrossRef CAS PubMed.
- W. Lu, J. Sun and X. Jiang, J. Mater. Chem. B, 2014, 2, 2369–2380 RSC.
- V. Russo, C. Yu, P. Belliveau, A. Hamilton and L. E. Flynn, Stem Cells Transl. Med., 2014, 3, 206–217 CrossRef CAS PubMed.
- J. R. Bain, S. E. Mackinnon and D. A. Hunter, Plast. Reconstr. Surg., 1989, 83, 129–138 CAS.
- M. Khamforoush and R. Agha-Moalapour, Iran. Polym. J., 2016, 25, 875–886 CrossRef CAS.
- H. S. Yoo, T. G. Kim and T. G. Park, Adv. Drug Delivery Rev., 2009, 61, 1033–1042 CrossRef CAS PubMed.
- K. Sombatmankhong, N. Sanchavanakit, P. Pavasant and P. Supaphol, Polymer, 2007, 48, 1419–1427 CrossRef CAS.
- W. Meng, S.-Y. Kim, J. Yuan, J. C. Kim, O. H. Kwon, N. Kawazoe, G. Chen, Y. Ito and I.-K. Kang, J. Biomater. Sci., Polym. Ed., 2007, 18, 81–94 CrossRef CAS PubMed.
- E. Schnell, K. Klinkhammer, S. Balzer, G. Brook, D. Klee, P. Dalton and J. Mey, Biomaterials, 2007, 28, 3012–3025 CrossRef CAS PubMed.
- A. Bianco, M. Calderone and I. Cacciotti, Mater. Sci. Eng., C, 2013, 33, 1067–1077 CrossRef CAS PubMed.
- K. M. Ramkumar, C. Manjula, G. GnanaKumar, M. A. Kanjwal, T. V. Sekar, R. Paulmurugan and P. Rajaguru, Eur. J. Pharm. Biopharm., 2012, 81, 324–333 CrossRef CAS PubMed.
- N. J. Hogrebe, J. W. Reinhardt and K. J. Gooch, J. Biomed. Mater. Res., Part A, 2017, 105, 640–661 CrossRef CAS PubMed.
- Y. Zhang, A. Gordon, W. Qian and W. Chen, Adv. Healthcare Mater., 2015, 4, 1900–1914 CrossRef CAS PubMed.
- J. M. Stukel and R. K. Willits, Tissue Eng., Part B, 2016, 22, 173–182 CrossRef CAS PubMed.
- A. Higuchi, Q.-D. Ling, Y. Chang, S.-T. Hsu and A. Umezawa, Chem. Rev., 2013, 113, 3297–3328 CrossRef CAS PubMed.
- J. Takahashi, T. D. Palmer and F. H. Gage, J. Neurobiol., 1999, 38, 65–81 CrossRef CAS PubMed.
- E. Mirzaei, J. Ai, S. Ebrahimi-Barough, J. Verdi, H. Ghanbari and R. Faridi-Majidi, Mol. Neurobiol., 2016, 53, 4798–4808 CrossRef CAS PubMed.
- S. Yao, X. Liu, S. Yu, X. Wang, S. Zhang, Q. Wu, X. Sun and H. Mao, Nanoscale, 2016, 8, 10252–10265 RSC.
- Y. J. Ren, S. Zhang, R. Mi, Q. Liu, X. Zeng, M. Rao, A. Hoke and H. Q. Mao, Acta Biomater., 2013, 9, 7727–7736 CrossRef CAS PubMed.
- N. C. Inestrosa and E. Arenas, Nat. Rev. Neurosci., 2010, 11, 77–86 CrossRef CAS PubMed.
- C. Elizalde, V. M. Campa, M. Caro, K. Schlangen, A. M. Aransay, M. dM Vivanco and R. M. Kypta, Stem Cells, 2011, 29, 141–153 CrossRef CAS PubMed.
- M. Kasai, K. Satoh and T. Akiyama, Genes Cells, 2005, 10, 777–783 CrossRef CAS PubMed.
- G. H. Kim, Y. D. Park, S. Y. Lee, A. El-Fiqi, J. J. Kim, E. J. Lee, H. W. Kim and E. C. Kim, J. Biomater. Appl., 2015, 29, 854–866 CrossRef CAS PubMed.
- M. H. Ho, C. J. Yao, M. H. Liao, P. I. Lin, S. H. Liu and R. M. Chen, Int. J. Nanomed., 2015, 10, 5941–5954 CAS.
- H. M. Yun, S. K. Kang, R. K. Singh, J. H. Lee, H. H. Lee, K. R. Park, J. K. Yi, D. W. Lee, H. W. Kim and E. C. Kim, Dent. Mater., 2016, 32, 1301–1311 CrossRef CAS PubMed.
- Y. M. Lee, H. M. Yun, H. Y. Lee, J. H. Lee, H. C. Lim, H. H. Lee, H. W. Kim and E. C. Kim, J. Biomed. Nanotechnol., 2017, 13, 180–191 CrossRef CAS.
- S. Tavakol, S. M. M. Mousavi, B. Tavakol, E. Hoveizi, J. Ai and S. M. R. Sorkhabadi, Mol. Neurobiol., 2017, 54, 2483–2496 CrossRef CAS PubMed.
- J. X. Zhu, Q. Cai, X. Zhang, X. Q. Hu, L. Li, W. P. Wang, Z. X. Shao, L. H. Dai, L. Y. Cheng, X. P. Yang, C. Y. Zhou and Y. F. Ao, J. Biomed. Nanotechnol., 2013, 9, 1757–1767 CrossRef CAS PubMed.
- J. Hu, X. H. Liu and P. X. Ma, Biomaterials, 2008, 29, 3815–3821 CrossRef CAS PubMed.
- A. Nur-E-Kamal, I. Ahmed, J. Kamal, M. Schindler and S. Meiners, Stem Cells, 2006, 24, 426–433 CrossRef PubMed.
- S. G. Sampathkumar, A. V. Li, M. B. Jones, Z. H. Sun and K. J. Yarema, Nat. Chem. Biol., 2006, 2, 149–152 CrossRef CAS PubMed.
- S. H. Lim, X. Y. Liu, H. J. Song, K. J. Yarema and H. Q. Mao, Biomaterials, 2010, 31, 9031–9039 CrossRef CAS PubMed.
- Y. Liu, Y. Z. Liu, R. X. Zhang, X. Wang, F. Huang, Z. J. Yan, M. Nie, J. Huang, Y. Z. Wang, Y. Wang, L. Chen, L. J. Yin, B. C. He and Z. L. Deng, Int. J. Biochem. Cell Biol., 2014, 47, 47–56 CrossRef CAS PubMed.
- M. Agathocleous, I. Iordanova, M. I. Willardsen, X. Y. Xue, M. L. Vetter, W. A. Harris and K. B. Moore, Development, 2009, 136, 3289–3299 CrossRef CAS PubMed.
- T. Kuwabara, J. Hsieh, A. Muotri, G. Yeo, M. Warashina, D. C. Lie, L. Moore, K. Nakashima, M. Asashima and F. H. Gage, Nat. Neurosci., 2009, 12, 1097–1107 CrossRef CAS PubMed.
- V. Chiono and C. Tonda-Turo, Prog. Neurobiol., 2015, 131, 87–104 CrossRef PubMed.
- D. Kai, G. Jin, M. P. Prabhakaran and S. Ramakrishna, Biotechnol. J., 2013, 8, 59–72 CrossRef CAS PubMed.
- S. H. Lim and H.-Q. Mao, Adv. Drug Delivery Rev., 2009, 61, 1084–1096 CrossRef CAS PubMed.
- P. Kerativitayanan, J. K. Carrow and A. K. Gaharwar, Adv. Healthcare Mater., 2015, 4, 1600–1627 CrossRef CAS PubMed.
- S. A. Hardy, D. J. Maltman and S. A. Przyborski, Curr. Stem Cell Res. Ther., 2008, 3, 43–52 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra08008e |
| This journal is © The Royal Society of Chemistry 2017 |