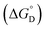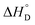Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Facile fabrication of lipase to amine functionalized gold nanoparticles to enhance stability and activity†
Sristy Shikha a,
Krishan Gopal Thakur
a,
Krishan Gopal Thakur b and
Mani Shankar Bhattacharyya
b and
Mani Shankar Bhattacharyya *a
*a
aFermentation Science and Biocatalysis Laboratory, CSIR-Institute of Microbial Technology, Sector 39 A, Chandigarh-160036, India. E-mail: manisb@imtech.res.in; Fax: +91 1722695215; Tel: +91 1726665313
bStructural Biology Laboratory, G. N. Ramachandran Protein Centre, CSIR-Institute of Microbial Technology, Chandigarh 160036, India
First published on 4th September 2017
Abstract
Among various techniques of immobilization, EDC/NHS cross linking is a simple and single step process for covalent coupling between enzymes and nanoparticles. Here we describe immobilization of lipase on amine functionalized gold nanoparticles (AuNPs-NH2) to attain enhanced activity and stability. To achieve a suitable orientation, it is necessary to understand the contribution of different functional groups on the enzyme's surface. Therefore, the crystal structure of lipase was analyzed using a computational method (PyMOL) to find the exposed acidic amino acid residues that can be exploited for conjugation. Confirmation of conjugation (AuNP-NH2-lipase) was determined by various techniques such as agarose gel electrophoresis, zeta measurement, FTIR-spectroscopy and TEM. Further, catalytic parameters (Vmax, KM,app, Kcat, and Kcat/KM,app) have been studied to establish activity enhancement upon immobilization. The data also suggested that, AuNP-NH2-lipase has desirable improved parameters such as temperature and storage stability. The thermodynamic parameters for the kinetics of deactivation (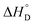 ,
, 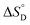 and
and 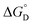 ) of the AuNP-NH2-lipase and free lipase demonstrated better stability of the conjugate. CD and fluorescence spectroscopic studies revealed minor structural rearrangements in the enzyme upon conjugation. Thus the AuNP-NH2-lipase conjugate represents a novel enzyme preparation with attributes of high activity and stability that could be an attractive choice in diverse applications ranging from catalysis to diagnostics.
) of the AuNP-NH2-lipase and free lipase demonstrated better stability of the conjugate. CD and fluorescence spectroscopic studies revealed minor structural rearrangements in the enzyme upon conjugation. Thus the AuNP-NH2-lipase conjugate represents a novel enzyme preparation with attributes of high activity and stability that could be an attractive choice in diverse applications ranging from catalysis to diagnostics.
Introduction
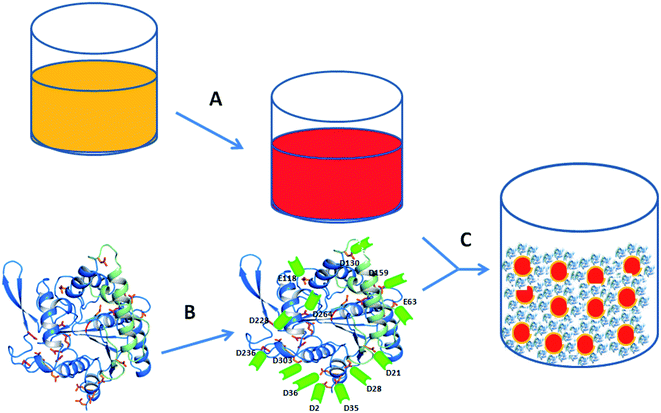
Enzymes have wide application in food, detergent, medicine, diagnostics, energy and many other industrial sectors due to their catalytic activity under mild conditions, specificity, cost-effectiveness and greener production conditions.1,2 However industrial application of free enzymes is restricted due to high lability, poor stability and problems in their recovery and reuse.3 Immobilization is the effective way to overcome these limitations and extend the horizon of their applications.4 Various methods of immobilization include physical adsorption, ionic interaction, covalent bonding between support and enzyme or entrapment within the different matrices.5 Hydrophobic interactions, ionic interactions, hydrogen bonds, van der Waals interactions and solvation are the major forces responsible for non-covalent interactions. Except for covalent attachment, the above mentioned techniques mainly rely on weak interactions, therefore enzymes are prone to detachment from the support surface with time. Covalent bonding mediated immobilization helps in improving enzyme activity at the cost of loss of conformational integrity.6 A very simple method of cross-linking biomolecules to nanoparticles relies on EDC/NHS coupling in which carbodiimides mediate formation of amide bond between carboxy and amine groups.7 High solubility in water, ease to remove byproducts, zero length cross linking molecule and single step reaction provides an edge over other methods in EDC/NHS coupling.8 Several enzymes have been conjugated using this strategy, but they lack in their structure based consideration prior to immobilization. Improper orientation after immobilization ultimately affects activity; therefore, it is highly essential to understand the structure of protein before immobilization. One of the rationale approaches is to exploit specific surface amino acids for conjugating enzymes with the nanosurface so that the catalytic properties of the enzyme do not alter significantly, but the structural integrity and stability of the enzyme improves.Nanoparticles (NPs) have emerged as the choice for matrix to immobilize enzyme due to many positive attributes such as, high dispersity and surface area, biocompatibility and availability of tools to manipulate the surface.9 Moreover, size, surface functional group available for conjugation, structure and textural attributes of the NPs has profound effect of the tethered enzyme.10 Among numerous metal nanoparticles, the property of bio-conjugation, size dependent optical and electrical properties and good biocompatibility have made gold nanoparticles a potential candidate for diverse application such as biosensor, drug delivery system, bioimaging, and cancer therapy.11–13
Lipases (triacylglycerol hydrolases, EC 3.1.1.3) are found ubiquitously from prokaryotes like bacteria to eukaryotes (yeasts, plant and animals) that catalyzes diverse reactions such as hydrolysis, and the synthesis of esters formed from glycerol and long-chain fatty acids.14 It has potential industrial applications in food, detergent, cosmetics, pharmaceutical, paper and pulp industries and biotechnological application like biodiesel production and bioremediation.15,16 Understanding its extensive applications, attempts have been made to conjugate the enzyme to improve stability and reusability of the biocatalyst. To attain this goal different techniques of immobilization have been applied using various matrices like gelatin, chitosan, mesoporous silica, magnetite nanoparticles etc.17–19 However, during conjugation aggregation of nanoparticles pose major hurdle as it reduces available surface area for enzyme immobilization or reduces availability of the substrate to the enzyme during catalysis. On the other hand, surface coupling of the enzyme to the NPs, may block the substrate binding site, or may interfere with the necessary conformational changes required for enzyme activity.20 Therefore, to address these issues; uniform distribution and proper orientation of enzyme upon immobilization with fully retained activity, we have designed our study by choosing amine functionalized gold nanoparticles as matrix and EDC/NHS as coupling agent.
In the present study, we have used rational structure based, directed immobilization approach to generate highly active and stable nanozyme conjugate. Here we have synthesized amine functionalized gold nanoparticles (AuNPs-NH2) to target surface carboxyl group (aspartic and glutamic acids) of lipase. To understand the contribution of acidic amino acid residues towards immobilization as well as structural and functional aspect, crystal structure of lipase (3-LIP) from Burkholderia cepacia, was analysed using PyMOL software. These acidic amino acids present at the surface of the lipase were identified and explored for conjugation with AuNPs-NH2 using EDC/NHS chemistry. Further, different concentrations of lipase were used to obtain maximum immobilization without aggregation. These AuNP-NH2-lipase composites have been characterized by UV-visible spectroscopy, FTIR and TEM imaging. Further, the kinetic analysis of the catalytic activity, stability parameters and thermodynamic parameters for the deactivation of the free and conjugated enzyme has been determined. CD and fluorescence spectroscopic studies were also carried out to understand the structural changes in the enzyme as an outcome of conjugation. This water-soluble, highly active and stable AuNPs-NH2-lipase nanozyme conjugate may be useful for various applications.
Methods
Material
Gold(III) chloride hydrate (50% Au basis), cysteamine–HCl, NHS were purchased from Sigma-Aldrich and NaBH4 was purchased from Merck. Lipase PS Amano (from B. cepacia), 4-nitrophenyl palmitate, Bradford reagent, EDC and NHS were procured from Sigma-Aldrich. para-Nitrophenol was purchased from Himedia, India. All chemicals and buffer components used in this study were of highest purity grade available.Structural studies
Structural analysis of B. cepacia lipase (3LIP) was carried out using PyMOL software. The number and disposition of the acidic amino acids were analyzed and finally, the surface exposed acidic amino acid residues have been explored to conjugate the enzyme with the amine functionalized gold nanoparticles.Synthesis and characterization of gold nanoparticles
Amine functionalized gold nanoparticles were synthesized following the method reported by Lee et al., with slight modification.21 Briefly, for 10 ml of HAuCl4 solution (40 ppm, pH 5.5), 100 μl of 0.213 M cysteamine–HCl solution in water was added and the mixture was stirred for 20 min at room temperature (RT) in the dark. Freshly prepared NaBH4 solution (1.25 μl, 10 mM) was further added to it and reaction mixture was again stirred at RT in the dark until the appearance of red wine colored solution. AuNPs-NH2 solution was dialysed against MQ to remove the traces of unreacted gold ions and NaBH4 and stored at 4 °C for further use.Conjugation of lipase with AuNPs-NH2, protein content measurement and immobilization efficiency
Conjugation of the lipase to AuNPs-NH2 was done by employing 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS) chemistry.22 In this method, the carboxyl group of enzyme (lipase) was activated by EDC and the intermediate was stabilized by NHS that subsequently reacted with the amino groups of the gold nanoparticles and form amide bond.In brief, 1.91 mg of EDC and 2.88 mg of NHS was added to 1 ml of 50 mM MES buffer solution containing four different concentrations of lipase, and the solution was stirred (150 rpm) for 15 min at RT for activation of carboxy group of Asp and Glu of enzyme. This mixture was transferred to 9 ml of amine functionalized gold nanoparticles solution and further incubated on stirring for next 2–3 hours. After completion of the reaction, the solution was dialyzed against MQ water to remove intermediate products, excess EDC and NHS. Conjugated AuNPs-NH2-lipase solution was kept at 4 °C for further use.
The protein content of the lipase was determined by colorimetric assay i.e., Bradford's method23 using bovine serum albumin (BSA) as standard. For estimation of actual protein content of lipase powder (lipase from PS Amano B. cepacia, Sigma), 1 mg ml−1 solution was prepared in phosphate buffer (pH 7.2), centrifuged to remove insoluble parts and then Bradford assay was performed. 1 mg of lipase powder correspond to 5.2 μg of protein content. Thereafter, all the concentration of lipase mentioned here are in terms of actual protein content. The efficiency of immobilization was expressed as % of immobilization as shown in given formula.
| % immobilization = (A − B) × 100/A |
In order to avoid the interferences of nanoparticles, the OD590 of AuNPs-NH2 supernatant was also deducted from the OD590 of the supernatant of AuNPs-NH2-lipase.
Characterizations of lipase conjugated AuNPs-NH2
Conjugation of lipase with AuNPs-NH2 was monitored by UV-visible spectrophotoscopy using Hitachi dual-beam spectrophotometer (Hitachi U-2900) operated at a resolution of 1 nm. The spectral measurement was taken in the range of 400–700 nm. Further, size measurement and the residual surface charge were measured by Malvern zetasizer. The zeta potential (ζ) was determined by laser doppler electrophoresis (LDE) using a quartz capillary electrophoresis cell. All of the measurements were performed in triplicate for a single batch of NPs, and the results shown are the average of these measurements. Confirmation of conjugation was also done by agarose gel electrophoresis (0.8%).Fourier transform infrared (FTIR) spectroscopy
FTIR spectroscopy was used to understand the conjugation lipase with AuNPs-NH2. The sample was prepared in KBr pellet. FTIR spectra of the films were recorded on instrument Vertex 70 operated in the diffuse reflectance mode at a resolution of 4 cm−1. To obtain good signal to noise ratio, 256 scans of the bio-conjugate film were taken in the range 400–4000 cm−1 and spectra was compared with free enzyme and gold nanoparticles.Transmission electron microscopy measurements
TEM measurements were performed on a JEOL Model 2100 instrument operated at an accelerating voltage of 120 kV. TEM samples were prepared by placing a drop of centrifuged and resuspended AuNPs-NH2 and AuNP-NH2-lipase on carbon-coated TEM copper grid. The film was allowed to dry for 10 min and the extra solution was removed using a blotting paper. Negative staining using 2% phosphotungstic acid (PTA) was done to visualize protein immobilized on the AuNPs-NH2 surface.Enzyme activity assay
The catalytic activity of lipase was examined by measuring the release of p-nitrophenol (p-NP) from substrate p-nitrophenyl palmitate (p-NPP) at 410 nm as described by Kuo et al., and Gupta et al., with some modifications.24,25 In brief, stock of p-NPP (25 mM) was prepared in ethanol, from which different concentration range (0–25 mM) was made. Free and immobilized enzyme solution (10 nM) was prepared in 10 mM phosphate buffer (PB, pH 7.2). 100 μl of each dilution of substrate was mixed with 100 μl of enzyme solution as mentioned and incubated at 37 °C for 5 min and then, 200 μl of 0.5 N Na2CO3 containing Triton-X100 was added to terminate the reaction. Finally, the release of p-nitrophenol (p-NP) resulting from the lipase catalyzed hydrolysis of p-NPP was measured at 410 nm with required dilution. In order to avoid the interferences of nanoparticles, similar reaction mixture for all concentrations of nanoparticles (that have been used during experiments but without substrate) was prepared, following same experimental procedure and OD410 was measured. Later this value was deducted from the experimental samples and then calculation were performed. Inferences of substrate solution were also considered and deduced from measurement. For calculation of apparent Michaelis–Menten constant (KM,app) and Vmax, the experiment was done in triplicate, average of these was plotted and data was analyzed through Prism software. One unit of enzyme activity was defined amount of enzyme required to produce μmol of product released per minute under experimental condition.Determination of the immobilized lipase stability
| (ΦMRE) = (100 × Φobs)/[d × C × (n − 1)] |
Results and discussion
Structural analysis of lipase
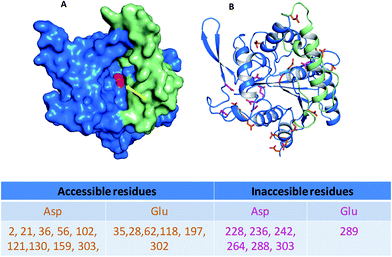
For this study, we have chosen lipase from B. cepacia. The enzyme is already proven to have versatile application26–29 and its crystal structure30 has been resolved earlier. Like other lipases it poses a catalytic triad of Ser-His-Asp buried deep in to an oxyanion hole (Ser has been indicated by yellow arrow in Fig. 2A), covered by a lid and the enzyme shares same catalytic mechanism as that of other lipases. It is also suggested that, a substantial amount of reorganization of the secondary structure is required during the activity of the enzyme.Mapping of the enzyme surface using computational methods (PyMOL) revealed the contribution of various amino acids on the structure function activity of the enzyme. The distribution of surface exposed free carboxyl group containing residues (aspartic acid, Asp and glutamic acid, Glu) has been shown in Fig. 2 along with table. The free acidic amino acid residues (categorized as accessible amino acid that does not take part in H-bond, van der Waals interactions or ionic interactions to maintain the structural integrity of the enzyme) are present on the surface on the enzyme and can be exploited for conjugation with NH2-terminated AuNPs via EDC/NHS chemistry (as shown in Fig. 1). Thus, remaining acidic amino acid residues that take part structural integrity of the enzyme through small interaction (inaccessible amino acid residues, Fig. 2) will remain undisturbed.
Synthesis, conjugation and characterization of conjugated and unconjugated gold nanoparticles
For the preparation of nanozyme composite, NH2 functionalized AuNPs were obtained from chloroauric acid solution by using NaBH4 as reducing agent and cysteamine–HCl as the capping agent. On the other side, the free carboxylate groups present on the surface of the lipase were activated by the water-soluble carbodiimide EDC in presence of NHS to provide stability to intermediate product. Thus, obtained NHS-terminated activated lipase was further mixed with NH2-functionalized AuNP solution, resulting in the displacement of NHS groups of the activated lipase by the amino groups present on the AuNP surface and formation of covalent bond. In this way, a lipase monolayer was immobilized onto the surface of NH2-functionalized AuNPs through the covalent linkage, i.e., the AuNP-NH2-lipase nanozyme composite was obtained. With the knowledge of number of available Asp and Glu on the surface of lipase, it was easy to maintain the ratio of EDC and NHS with enzyme concentration. The activating agent has been used in such a way that limited conjugation can occur with maximum proper orientation. To know the particular concentration at which complete covering of nanoparticles four different concentrations of lipase (Table 1) were used for conjugation and 1 μmol ml−1 was found to be the best for conjugation reaction. Although lowest concentration of lipase (0.5 μmol ml−1) has also shown good immobilization efficiency but this conjugate solution is not stable for longer period and precipitate. Therefore, lipase conjugated at 1 μmol ml−1 (in terms of protein content) concentration has been used for rest of the study.| Concentration of lipase used for conjugation (μmol ml−1) | % immobilization efficiency | Zeta potential (ζ) |
|---|---|---|
| 0 | No | 35 |
| 0.5 | 85 ± 5 | 14 |
| 1 | 92 ± 4 | 10 |
| 2 | 25 ± 2 | 4 |
| 4 | 15 ± 3 | 0.37 |
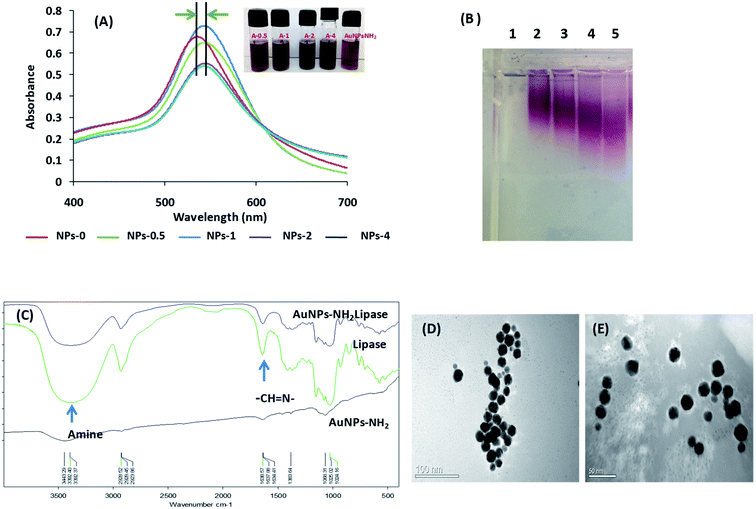
Fig. 3A shows the UV-visible spectra of unconjugated AuNPs-NH2 and conjugated AuNP-NH2-lipase, in which the peak indicates the characteristic surface plasmon resonance (SPR) of gold nanoparticles. Immobilization of lipase on the surface of AuNPs-NH2 leads to an increase in nanoparticles size, which results in the redshift of spectra (<10 nm). It is well known that larger size nanoparticles have an absorbance at a higher wavelength as compared to the smaller size nanoparticles of same shape and morphology.31 Thus, the redshift of the spectra indicated the presence of the enzyme on the AuNP-NH2 surface as observed by other groups (Fig. 4).32
Zeta potential (ζ) is a measure of surface charges carried by the nanoparticles in a suspension. Zeta potential value of conjugated nanoparticles showed that it has a lower surface charge compared to the unconjugated gold nanoparticles (as shown in Table 1 and Fig. 3B). Similar finding was also observed by Silva et al., in which they have used PEG coated polystyrene nanoparticles for conjugation of lipase.33 Lowering of zeta value indicates the masking of AuNPs-NH2 with lipase molecules that was also confirmed by gel electrophoresis (0.8% agarose). The gel shift increases with increasing concentration of protein supporting the decrease in ζ value, thus confirming the conjugation in all cases. Wangoo et al., also observed similar phenomenon using negatively charged nanoparticles.34 Immobilization efficiency (%) determined by Bradford assay indicates that maximum immobilization had occurred at the concentration of 1 μmol ml−1 of lipase. Therefore, this concentration was used for immobilization to carry rest of studies.
FTIR
Furthermore, the confirmation of immobilization of lipase to AuNPs-NH2 was characterized by FTIR spectroscopy. As shown in Fig. 3C, in the case of unconjugated gold nanoparticles (AuNPs-NH2), stretches at 1020–1250 cm−1 and at 3443 cm−1 affirms the presence of amines (1°, due to the presence of cysteamine–HCl) that capped the gold nanoparticles. These peaks are further sharpening in AuNPs-NH2-lipase and have followed pattern as of free lipase. The characteristic bands of pure lipase were at 3392 (O–H stretching and N–H stretching vibrations), 1638 (amide), and 1024 cm−1 (C–O–C stretching vibration), is consistent with the previous report.35 The AuNPs-NH2-lipase had the pattern of pure lipase, confirming the immobilization of lipase to nanoparticles. Therefore, the FTIR results provided the evidential information of modified AuNPs-NH2-lipase.TEM
The morphological characterization of conjugated/unconjugated AuNPs-NH2 was carried out by TEM. Fig. 3D shows the well-dispersed cysteamine stabilized AuNPs-NH2 with size of about 25 ± 3 nm in diameter. As peripheral protein layer cannot be imaged due to the low electron resistance of protein molecules in TEM examination, therefore nanozyme composites were stained by 2% PTA. In Fig. 3E, the faint white layer around the periphery of conjugated gold nanoparticles was visualized that indicates the presence of biomolecules and thus demonstrating immobilization of lipase to amine functionalized gold nanoparticles.36Enzyme kinetics measurement
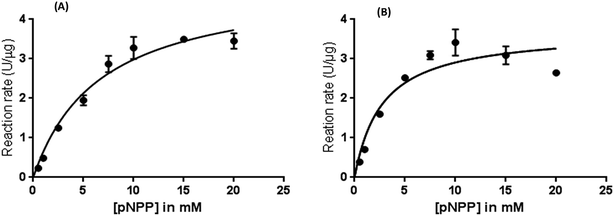
The consequences of immobilization on the enzyme kinetics were studied by determining the activity of the native enzyme and AuNPs-NH2-lipase nanozyme composite using para-nitrophenolpalmitate (p-NPP) as substrate. The apparent Michaelis–Menten constant (KM,app) of the free-lipase and AuNP-NH2-lipase was determined by plotting Michaelis–Menten saturation curve using Prism 6 software. The KM,app of the immobilized lipase was decreased by 2.43-fold over that of free lipase (Table 2). Since, KM,app is related to the binding affinity of enzyme to the substrate, lowering of KM,app of nanozyme composite indicates that the conjugation has led to the increase in substrate binding affinity. Similar findings were also observed by various groups when lipase has been conjugated to different supports.33,37–39 The Vmax value for AuNPs-NH2-lipase was lower than that for free lipase. Conjugation leads to change not only in KM,app but also in Vmax that suggested the stabilized the transition state. Moreover, 1.8-fold higher value of Vmax/KM,app for AuNP-NH2-lipase conjugate over free lipase indicated that the conjugated lipase binds to the substrate more easily and have significantly higher catalytic efficiency. The catalytic efficiencies of different enzymes can be considered by evaluating the specificity constant (Kcat/KM,app). It defines rate constant for the conversion of substrate to product and this has increased up to 80% as compared free lipase. The possible reasons of the enhanced activity of immobilized lipase are as follow: (i) maintenance of “open state” conformation of lipase due to immobilization;40–42 (ii) prevention of aggregation of free lipase due to immobilization;17,43 and (iii) change in enzyme conformation due to interfacial activation.41,44 Either one or all might be possible for enhanced activity. It suggests that covalent binding of lipase to AuNPs-NH2 might have occurred at such sites that facilitated the easy accessibility of substrate to the catalytic sites by keeping its active conformation open, leading to significant increase in catalytic efficiency.4,41,45| Parameter | Free lipase | AuNPs-NH2-lipase |
|---|---|---|
| Vmax (U μg−1) protein | 4.98 | 3.71 |
| Km,app (mM) | 6.70 | 2.76 |
| Kcat (s−1) | 2763.30 | 2059.05 |
| Kcat/KM,app (s−1 mM−1) | 412.38 | 744.95 |
Thermodynamic studies
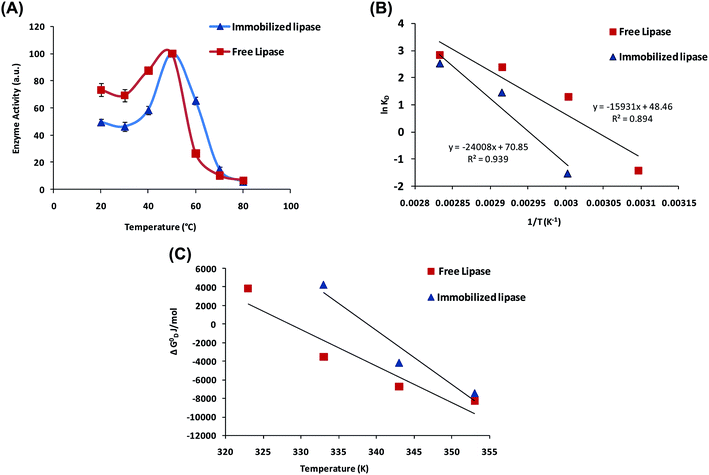
Subsequent to establishment of enhanced catalytic parameters of AuNP-NH2-lipase nanozyme composite, the enzyme activity was measured in the temperature range of 20–80 °C (an interval of 10 unit) to know temperature optima and thermodynamic stability. Free-lipase and AuNPs-NH2-lipase conjugate, both have shown maximum activity at 50 °C (assigning maximum value as 100%, from which relative activity has been calculated for different temperatures). AuNPs-NH2-lipase retains 65% of its maximum activity at 60 °C whereas free lipase has nearly 20% (Fig. 5A). It is well known that higher temperature leads to denaturation of protein, resulting in loss of activity, but the AuNPs-NH2-lipase conjugate is resistance to the denaturation of protein. In case of conjugated AuNPs-NH2-lipase, nanoparticles act as a support that either minimize the denaturation process or act as a scaffold to speed up the renaturation of protein.46 Along with the maintenance of active open conformation as mentioned earlier, immobilization generally increases the rigidity of the protein and thus increases the thermal stability of the immobilized enzyme.46Irreversible thermal deactivation can be best described by measuring the kinetic stability of the enzyme. Thermal deactivation of lipase has been found to follow the first order kinetics and can be described by the following expression:
 | (1a) |
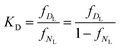 | (1b) |
ƒDL may be calculated by
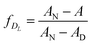 | (1c) |
The equilibrium constant KD is related to Gibbs free energy for deactivation
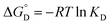 | (1d) |
Thermodynamic parameters, i.e., standard state enthalpy 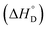 and standard state entropy
and standard state entropy 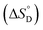 of enzyme deactivation have been calculated from Van't Hoff plot (Fig. 5B, Table 3). For this, data were selected from the range of 50 to 80 °C and compared. It was found that, the
of enzyme deactivation have been calculated from Van't Hoff plot (Fig. 5B, Table 3). For this, data were selected from the range of 50 to 80 °C and compared. It was found that, the 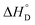 value of nanozyme composite (−199.60 kJ mol−1) is higher than free enzyme (−132.45 kJ mol−1) indicating that the nanozyme composite is more thermostable than free enzyme. On the other hand, the entropic values of the free enzyme (402.84 J mol−1 K−1) and nanozyme composite (589.04 J mol−1 K−1) indicated that the enzyme is more thermostable. However, the free energy of deactivation
value of nanozyme composite (−199.60 kJ mol−1) is higher than free enzyme (−132.45 kJ mol−1) indicating that the nanozyme composite is more thermostable than free enzyme. On the other hand, the entropic values of the free enzyme (402.84 J mol−1 K−1) and nanozyme composite (589.04 J mol−1 K−1) indicated that the enzyme is more thermostable. However, the free energy of deactivation 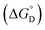 includes both the enthalpic and entropic contribution and is more accurate and reliable parameter for the prediction of enzyme stability. Therefore, the
includes both the enthalpic and entropic contribution and is more accurate and reliable parameter for the prediction of enzyme stability. Therefore, the 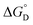 of the two enzymes was derived from eqn (1d) and plotted against the temperature and it was found that, the
of the two enzymes was derived from eqn (1d) and plotted against the temperature and it was found that, the 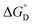 value of the nanozyme was higher than the free enzyme (Fig. 5C). A higher
value of the nanozyme was higher than the free enzyme (Fig. 5C). A higher 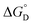 is associated with higher tolerance towards temperature deactivation, supporting the enzyme active conformation and thus confirms higher thermostability of AuNPs-NH2-lipase nanozyme conjugates as compared to free enzyme. From the same graph, when transition midpoint temperature (Tm; Table 3) of these two enzymes were compared, it was found that AuNPs-NH2-lipase nanozyme composite has higher Tm by unit of 10 °C. These results clearly show that the conjugation enhances the thermostability of enzyme consistent with previous finding obtained by Prashant et al.,47
is associated with higher tolerance towards temperature deactivation, supporting the enzyme active conformation and thus confirms higher thermostability of AuNPs-NH2-lipase nanozyme conjugates as compared to free enzyme. From the same graph, when transition midpoint temperature (Tm; Table 3) of these two enzymes were compared, it was found that AuNPs-NH2-lipase nanozyme composite has higher Tm by unit of 10 °C. These results clearly show that the conjugation enhances the thermostability of enzyme consistent with previous finding obtained by Prashant et al.,47
Time dependent thermal stability study
Further, a time course comparison of activity was performed at 40, 50 and 60 °C for free and conjugated lipase. Fig. 6 clearly demonstrates that over an hour, the AuNP-NH2-lipase nanozyme composite has 110% and 130% higher activity at 40 °C and 50 °C respectively as compared to the free lipase. In the similar condition, free lipase was able to retain only 70% and 67% of its native enzyme activity. The conjugated nanoparticles retained its nearly 80% of activity at 50 °C even after 4 hour of incubation whereas the free enzyme has nearly 30%. A very interesting finding that the nanozyme composite is able to retain its activity more than 50% after incubating at 60 °C for an hour whereas the free enzyme loses nearly all of its activity. Denaturation of enzyme (a protein) is temperature dependent and if the protein is denatured to an extent from which there is no recovery of active conformational structure that leads to activity loss. Here, fall in activity of free enzyme at all temperature suggested loss of active conformation after denaturation with respect to time. Contrary to this, immobilized lipase have shown the activity (at 40 °C and 50°) initially gain and then retain its activity (upto 80%) for longer time period. The gain of thermostability can be attributed towards covalent multipoint immobilization due to zero length crosslinker. This multipoint attachment have contributed towards structural rigidity, stability of secondary structure and active conformation of lipase, that prevents from denaturation of immobilized enzyme at higher temperature.4,48 This reinforced that immobilization of lipase on amine gold nanoparticles enhances the thermostability of enzyme that can be potentially useful for industrial applications.49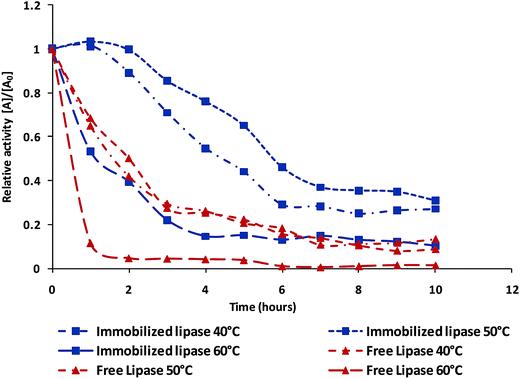 | ||
| Fig. 6 Relative activity free lipase and immobilized lipase conjugate as a function of time at three different temperatures (40, 50 and 60 °C). | ||
pH and storage stability study
For pH stability analysis, free and immobilized enzyme were kept in buffers of different pHs ranging from 4–10 for an hour at RT and the activity was analyzed at 37 °C. It was observed (ESI Fig. 1A†) that at lower pH (4–6), the activity of AuNPs-NH2-lipase is less than free enzyme that is reversed at higher pH (7–10). Lowering of pH might have disturbed the integrity of gold nanoparticles, ultimately affected the enzyme activity.AuNPs-NH2-lipase nanozyme composite can be stored for a month, and its activity was determined at an interval of 5 days. ESI Fig. 1B† clearly indicates that AuNPs-NH2-lipase can retain 60% of its activity after 25 days whereas enzyme solution lost most of its activity within 20 days of storage similar to previous finding.50 The possible reason for this stability is inhibition of aggregation of enzyme in solution whereas maintenance of open conformation of the enzyme.7 In this way, it can be said that AuNPs-NH2 facilitates durable attachment that provides the structure and long-term stability of immobilized lipase.
In order to know the stability of nanoparticles before and after conjugation, samples were withdrawn periodically at an interval of 5 days and UV spectrum was measured. It is already shown earlier that the conjugated nanoparticles have red shift in UV spectra as compared to unconjugated nanoparticles. After 5 days incubation, the SPR of the unconjugated nanoparticles (shown in ESI Fig. 2A†) have shown broadening of peak indicating the increase in size of nanoparticles. Disappearance of SPR peak at 530 nm and appearance of band after 600 nm indicated the increase in aggregation of the nanoparticles and the settlement of nanoparticles at the bottom of tube was also seen by naked eyes. However there is negligible change in UV-spectra of conjugated nanoparticles (shown in ESI Fig. 2B†) with time at different time scale suggesting no aggregation and confirming its stability. The stability of nanoparticles can be explained by Derjaguin–Landau–Verwey–Overbeek (DLVO) theory51 in which the colloidal particles are stabilized by two major forces; long range electrostatic repulsion and short range van der Waals attraction. Here it is shown in Table 1 that unconjugated nanoparticles have very high zeta potential as compared to conjugated nanoparticles, the electrostatic repulsion forces dominate over van der Waals attraction forces that led to aggregation.
Structural characterization of AuNPs-NH2-lipase
CD analysis
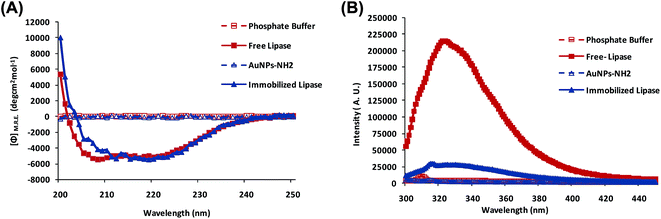
The retention of enzyme activity is the primary concern while performing immobilization especially covalent immobilization. The activity of protein mainly depends on its structural integrity (3D conformation).52 Various interactions as an outcome of conjugation may influence the structural integrity and thereby affect the activity of the enzyme (in the present enhanced activity and stability). Therefore, it was necessary to understand the structural changes occurred in the enzyme molecule due to immobilization. Thus, we conducted CD and fluorescence spectroscopic studies on both the free and AuNPs-NH2-lipase conjugates. The secondary structure profile of lipase is shown in Fig. 7A. Lipase is α-helical protein. CD spectrum of lipase shows negative peaks at 222 and 208 nm, characteristic of α-helical proteins (Fig. 7A). While the peak at 222 nm is comparable, peak at 208 nm is shifted by 3–4 nm for conjugated lipase indicating some structural rearrangements. The content of α-helix and β-sheet were calculated from K2D3 analysis server (http://cbdm-01.zdv.uni-mainz.de/%7Eandrade/cgi-bin/k2d3/k2d3_set1.pl). The α-helix and β-sheet content of free lipase were 77.44% and 1.37% whereas for AuNPs-lipase have 77.17% and 1.57% respectively. It indicates that, while there were minor secondary structural changes in the lipase due to conjugation with AuNPs-NH2 coincidentally, there was a desirable increase in the activity and stability. Here, the partial structural loss of immobilized lipase can be correlated to the involvement of surface carboxyl group of enzyme in covalent linkage that lead to alteration of hydrogen bond among functional groups to some extent and leading to smaller change in enzyme structure.53,54Fluorescence analysis
To support CD experiments, fluorescence measurements were performed to investigate the effect of AuNPs on tertiary structure of lipase. Lipase has three intrinsic fluorophores: tryptophan, tyrosine and phenylalanine that can be quenched, in which the main intrinsic fluorophore is tryptophan. The fluorescence emission maximum of tryptophan (Trp) is affected by pH of the solution and microenvironment of the particular residues. The lipase displays a fluorescence emission spectrum with a peak at 323 nm (Fig. 7B) under our experimental conditions.55 Fig. 7B shows that there is significant (nearly nine times) decrease in emission intensity of AuNP-NH2-lipase conjugate as compared to the free enzyme. Iosin et al., has shown relationship of fluorescence intensity between shape and size of gold nanoparticles to the concentration of protein.56 In which they have demonstrated that the complete binding of nanoparticles to enzyme leads to nearly complete quenching of fluorescence. Apart from this strong quenching effect, a redshift of about 2 nm (323 to 325 nm) in the fluorescence peak was also observed indicating some microenvironmental change near the Trp residues. Thus, fluorescence studies confirm the structural integrity of lipase before and after immobilization and its relation with enhanced stability.The stability of immobilized enzyme depends on various factors; nature of nanoparticles, interaction between enzyme and carrier, linking agents, binding positions of enzyme, number of bonds, properties of spacer, microenvironment of enzyme-conjugate and condition under which immobilization is performed whereas in case of native enzymes, its only intrinsic structure of protein affects the stability. Interestingly in our study, immobilization of lipase to amine functionalized gold nanoparticles contributed in gain of kinetic, thermo stability and storage stability as compared to free enzyme and that has been correlated well with structural studies. Thus, it can be said that the significance of study lie in the fact that the information obtained through structural studies helped in proceeding conjugation reaction and obtaining such immobilized lipase having minimum loss of structure with higher in various stability parameters.
Conclusion
Though, previously lipases have been conjugated with nanoparticles using different methods, but these methods involves multiple steps such as modification of nanoparticles surface to obtain desired functional group or modification of enzyme that can also alter its structure. Here, we have used cost effective method, and achieved maximum loading of lipase to nanoparticles. The elegance of this method lies mainly on; (i) obtaining amine functionalized gold nanoparticle in one step, (ii) the involvement of single step EDC/NHS chemistry for covalent conjugation and on the exploitation surface exposed acidic amino acids of the enzyme for immobilization to AuNPs-NH2. The immobilization was achieved with advantages of higher kinetic and thermal stability but, minimum alteration of the structure and this has been correlated with CD and fluorescence measurements. This study also revealed that there is little change in the secondary structural of protein and that has provided enhanced thermal and storage stability. Therefore, it can be concluded that covalent conjugation in this way is reliable in obtaining nanozyme composite having enhanced activity, stability with the minor secondary changes. Such nanozyme composite formulations will be useful for various applications in industries due to their above mentioned qualities.Author contributions
S. S. and M. S. B. designed the study. S. S. performed all the experiments, collected data. S. S. and M. S. B. analyzed results and drafted the manuscript. K. G. T. helped in analysis and interpretation of lipase structure.Conflicts of interest
The authors declare that they have no competing interest.Acknowledgements
Sristy Shikha is thankful to ICMR, New Delhi, India, for the research fellowship. The authors also thankfully acknowledge the help of Mr Shashi and Mr Randeep for obtaining the FTIR spectra and TEM images of the samples. The authors thankfully acknowledge CSIR-India for proving funds to carry out research under project no. OLP-0082.References
- R. Singh, M. Kumar, A. Mittal, P. K. Mehta, 3 Biotech, 2016, vol. 6 Search PubMed.
- M. Vellard, Curr. Opin. Biotechnol., 2003, 14, 444–450 CrossRef CAS PubMed.
- W. Tischer and F. Wedekind, Top. Curr. Chem., 1999, 200, 95–126 CrossRef CAS.
- C. Mateo, J. M. Palomo, G. Fernandez-Lorente, J. M. Guisan and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2007, 40, 1451–1463 CrossRef CAS.
- U. Hanefeld, L. Gardossi and E. Magner, Chem. Soc. Rev., 2009, 38, 453–468 RSC.
- G. A. Petkova, K. Zaruba, P. Zvatora and V. Kral, Nanoscale Res. Lett., 2012, 7, 287 CrossRef PubMed.
- D. Bartczak and A. G. Kanaras, Langmuir, 2011, 27, 10119–10123 CrossRef CAS PubMed.
- H. Shen, A. M. Jawaid and P. T. Snee, ACS Nano, 2009, 3, 915–923 CrossRef CAS PubMed.
- S. Ding, A. A. Cargill, I. L. Medintz and J. C. Claussen, Curr. Opin. Biotechnol., 2015, 34, 242–250 CrossRef CAS PubMed.
- J. Kim, J. W. Grate and P. Wang, Chem. Eng. Sci., 2006, 61, 1017–1026 CrossRef CAS.
- P. M. Tiwari, K. Vig, V. A. Dennis and S. R. Singh, Nanomaterials, 2011, 1, 31–63 CrossRef CAS PubMed.
- M. Hu, L. Qian, R. P. Brinas, E. S. Lymar, L. Kuznetsova and J. F. Hainfeld, J. Struct. Biol., 2008, 161, 83–91 CrossRef CAS PubMed.
- Y. Y. Li, H. J. Schluesener and S. Q. Xu, Gold Bull., 2010, 43, 29–41 CrossRef CAS.
- K. E. Jaeger and M. T. Reetz, Trends Biotechnol., 1998, 16, 396–403 CrossRef CAS PubMed.
- R. Sharma, Y. Chisti and U. C. Banerjee, Biotechnol. Adv., 2001, 19, 627–662 CrossRef CAS PubMed.
- F. Hasan, A. A. Shah and A. Hameed, Enzyme Microb. Technol., 2006, 39, 235–251 CrossRef CAS.
- Y. Ren, J. G. Rivera, L. He, H. Kulkarni, D. K. Lee and P. B. Messersmith, BMC Biotechnol., 2011, 11, 63 CrossRef CAS PubMed.
- J. Wang, G. Meng, K. Tao, M. Feng, X. Zhao, Z. Li, H. Xu, D. Xia and J. R. Lu, PLoS One, 2012, 7, e43478 CAS.
- Y. Wu, Y. Wang, G. Luo and Y. Dai, Bioresour. Technol., 2009, 100, 3459–3464 CrossRef CAS PubMed.
- T. Fischer and H. Hess, J. Mater. Chem., 2007, 17, 943–951 RSC.
- S. H. Lee, K. H. Bae, S. H. Kim, K. R. Lee and T. G. Park, Int. J. Pharm., 2008, 364, 94–101 CrossRef CAS PubMed.
- M. J. Fischer, Methods Mol. Biol., 2010, 627, 55–73 CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- N. Gupta, P. Rathi and R. Gupta, Anal. Biochem., 2002, 311, 98–99 CrossRef CAS PubMed.
- C. H. Kuo, Y. C. Liu, C. M. J. Chang, J. H. Chen, C. Chang and C. J. Shieh, Carbohydr. Polym., 2012, 87, 2538–2545 CrossRef CAS.
- X. W. Cao, J. K. Yang, L. Shu, B. Q. Yu and Y. J. Yan, Process Biochem., 2009, 44, 177–182 CrossRef CAS.
- P. Rathi, R. K. Saxena and R. Gupta, Process Biochem., 2001, 37, 187–192 CrossRef CAS.
- F. Sasso, A. Natalello, S. Castoldi, M. Lotti, C. Santambrogio and R. Grandori, Biotechnol. J., 2016, 11, 954–960 CrossRef CAS PubMed.
- P. R. Mello Bueno, T. F. de Oliveira, G. L. Castiglioni, M. S. Soares Junior and C. J. Ulhoa, Water Sci. Technol., 2015, 71, 957–964 CrossRef PubMed.
- J. D. Schrag, Y. G. Li, M. Cygler, D. M. Lang, T. Burgdorf, H. J. Hecht, R. Schmid, D. Schomburg, T. J. Rydel, J. D. Oliver, L. C. Strickland, C. M. Dunaway, S. B. Larson, J. Day and A. McPherson, Structure, 1997, 5, 187–202 CrossRef CAS PubMed.
- G. Raschke, S. Kowarik, T. Franzl, C. Sonnichsen, T. A. Klar, J. Feldmann, A. Nichtl and K. Kurzinger, Nano Lett., 2003, 3, 935–938 CrossRef CAS.
- S. K. Pandey, C. R. Suri, M. Chaudhry, R. P. Tiwari and P. Rishi, Mol. BioSyst., 2012, 8, 1853–1860 RSC.
- R. A. Silva, A. M. Carmona-Ribeiro and D. F. S. Petri, Molecules, 2014, 19, 8610–8628 CrossRef PubMed.
- N. Wangoo, K. K. Bhasin, S. K. Mehta and C. R. Suri, J. Colloid Interface Sci., 2008, 323, 247–254 CrossRef CAS PubMed.
- L. Z. Gao, J. M. Wu, S. Lyle, K. Zehr, L. L. Cao and D. Gao, J. Phys. Chem. C, 2008, 112, 17357–17361 CAS.
- S. K. Pandey, C. R. Suri, M. Chaudhry, R. P. Tiwari and P. Rishi, Mol. BioSyst., 2012, 8, 1853–1860 RSC.
- S. L. Cao, Y. M. Huang, X. H. Li, P. Xu, H. Wu, N. Li, W. Y. Lou and M. H. Zong, Sci. Rep., 2016, 6, 20420 CrossRef CAS PubMed.
- K. C. Badgujar and B. M. Bhanage, J. Phys. Chem. B, 2014, 118, 14808–14819 CAS.
- A. K. Singh and M. Mukhopadhyay, Korean J. Chem. Eng., 2014, 31, 1225–1232 CrossRef CAS.
- M. T. Reetz, Adv. Mater., 1997, 9, 943–954 CrossRef CAS.
- P. Adlercreutz, Chem. Soc. Rev., 2013, 42, 6406–6436 RSC.
- M. Mathesh, B. Q. Luan, T. O. Akanbi, J. K. Weber, J. Q. Liu, C. J. Barrow, R. H. Zhou and W. R. Yang, ACS Catal., 2016, 6, 4760–4768 CrossRef CAS.
- V. Sereti, M. Zoumpanioti, V. Papadimitriou, S. Pispas and A. Xenakis, J. Phys. Chem. B, 2014, 118, 9808–9816 CrossRef CAS PubMed.
- K. C. Badgujar, K. P. Dhake and B. M. Bhanage, Process Biochem., 2013, 48, 1335–1347 CrossRef CAS.
- P. Jochems, Y. Satyawali, L. Diels and W. Dejonghe, Green Chem., 2011, 13, 1609–1623 RSC.
- Y. Xie, J. An, G. Yang, G. Wu, Y. Zhang, L. Cui and Y. Feng, J. Biol. Chem., 2014, 289, 7994–8006 CrossRef CAS PubMed.
- P. Asuri, S. S. Bale, R. C. Pangule, D. A. Shah, R. S. Kane and J. S. Dordick, Langmuir, 2007, 23, 12318–12321 CrossRef CAS PubMed.
- K. A. Mahmoud, E. Lam, S. Hrapovic and J. H. Luong, ACS Appl. Mater. Interfaces, 2013, 5, 4978–4985 CAS.
- G. M. Zanin and F. F. De Moraes, Appl. Biochem. Biotechnol., 1998, 70–72, 383–394 CrossRef CAS PubMed.
- D. Andriani, C. Sunwoo, H. W. Ryu, B. Prasetya and D. H. Park, Bioprocess Biosyst. Eng., 2012, 35, 29–33 CrossRef CAS PubMed.
- N. J. Wagner and W. B. Russel, Phys. A, 1989, 155, 475–518 CrossRef.
- S. M. Kelly and N. C. Price, Curr. Protein Pept. Sci., 2000, 1, 349–384 CrossRef CAS PubMed.
- F. Secundo, Chem. Soc. Rev., 2013, 42, 6250–6261 RSC.
- P. Esmaeilnejad-Ahranjani, M. Kazemeini, G. Singh and A. Arpanaei, Langmuir, 2016, 32, 3242–3252 CrossRef CAS PubMed.
- Q. R. Jin, G. Q. Jia, Y. M. Zhang, Q. H. Yang and C. Li, Langmuir, 2011, 27, 12016–12024 CrossRef CAS PubMed.
- M. Iosin, F. Toderas, P. L. Baldeck and S. Astilean, J. Mol. Struct., 2009, 924, 196–200 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: The pH and storage stability data. See DOI: 10.1039/c7ra06075k |
| This journal is © The Royal Society of Chemistry 2017 |