Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sedimentation velocity analysis of TMPyP4-induced dimer formation of human telomeric G-quadruplex†
Yating Gaoa,
Tianlei Guanga and
Xiaodong Ye *ab
*ab
aHefei National Laboratory for Physical Sciences at the Microscale, Department of Chemical Physics, University of Science and Technology of China, Hefei, Anhui 230026, China. E-mail: xdye@ustc.edu.cn
bCAS Key Laboratory of Soft Matter Chemistry, University of Science and Technology of China, Hefei, Anhui 230026, China
First published on 5th December 2017
Abstract
5,10,15,20-Tetra-(N-methyl-4-pyridyl)porphyrin (TMPyP4), a ligand of G-quadruplex, has shown the ability to stabilize G-quadruplex structures and inhibit the activity of telomerase. Many methods have been used to study the interactions between TMPyP4 and G-quadruplex. However, many issues such as the binding number and the corresponding structural change are still controversial. Here, interactions between TMPyP4 and AGGG(TTAGGG)3 (Tel22) have been studied by a combination of analytical ultracentrifugation sedimentation velocity (AUC-SV), polyacrylamide gel electrophoresis (PAGE), circular dichroism (CD) and UV-vis absorption spectroscopy. In the presence of NaCl, the binding number of TMPyP4 per Tel22, determined by AUC-SV, increases with the increasing concentration of TMPyP4 (CTMPyP4). However, it decreases with the increasing concentration of NaCl (CNaCl). Moreover, both AUC-SV and PAGE reveal that TMPyP4 can induce the formation of the dimeric G-quadruplex–TMPyP4 complex through a medium affinity binding mode. High affinity binding modes, which may include the inner intercalation mode and end stacking, have no contribution to the formation of dimers. The weak electrostatic binding of TMPyP4 to Tel22 has a negative effect on the formation of dimers, presumably due to the instability of G-quadruplex induced by this binding mode.
Introduction
G-quadruplex, a four-stranded DNA structure consisting of π-planar G-quartets, has received much interest due to its potential application as a target in cancer therapy.1–6 Some molecules such as proteins, drugs, and ligands can bind to G-quadruplex, adjust its structure and then influence its function.7–13 For example, 5,10,15,20-tetra-(N-methyl-4-pyridyl)porphyrin (TMPyP4) (Scheme 1(a)), a well-studied ligand, has shown the abililty to stabilize the structure of human telomeric G-quadruplex and inhibit the activity of telomerase.14–38 Until now, different techniques have been used to study the interactions between G-quadruplex and this ligand, such as UV-vis absorption,17,19,23 circular dichroism (CD),17,26,36 isothermal titration calorimetry (ITC),36 fluorescence/phosphorescence spectroscopy,7,32,34 gel electrophoresis,36 molecular modeling,35,39 mass spectrometry,11 nuclear magnetic resonance (NMR) spectroscopy,40 X-ray crystallography,21 and so on. The results show that the binding mode and affinity of TMPyP4 to G-quadruplex are dependent on the structure of G-quadruplex and are sensitive to the solution conditions.23,29,35,41 Using 2-aminopurine modified Tel22, Majima et al. concluded that TMPyP4 binds to basket-type G-quadruplex, formed from human telomeric DNA in the presence of Na+, by intercalation and loop binding.42 Recently, Su et al. reported that the binding modes of TMPyP4 to the same DNA structure are end-stacking and intercalation using TMPyP4 as a triplet reporter.32 It is possible that one technique can only be sensitive to certain or several binding modes, thus it can only obtain incomplete information of the interactions.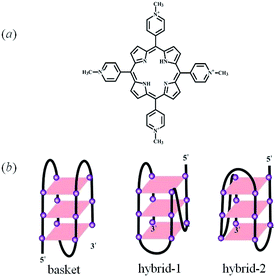 | ||
| Scheme 1 The structures of TMPyP4 (a) and G-quadruplex formed from Tel22 in a solution of Na+ (basket) or K+ (hybrid-1 and hybrid-2) (b). | ||
The binding stoichiometry, a critical parameter for understanding the interactions between TMPyP4 and DNA, is also different from previous measurements.15,16,19,23,24,26,27,37 For example, Hurley and co-workers reported a binding stoichiometry of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for TMPyP4 to the Tel22 G-quadruplex in 100 mM Na+ buffer using UV absorption spectroscopy.15 Lewis et al. reported that the binding number of TMPyP4 to Tel22 G-quadruplex is ∼4 using ITC experiments with the addition of 150 mM NaCl.37 It is obvious that the binding of TMPyP4 to Tel22 formed G-quadruplex is influenced by the ionic strength of solutions. For example, Zhang and co-workers concluded that with an increase in the concentration of K+ from 0 to 100 mM, the binding number decreases from 5 to 3.24 Until now, there is no complete understanding of the effect of salt concentration on the binding modes and binding number. Furthermore, so far, little is known about the relation between the binding and the structures of DNA–TMPyP4 complexes in aqueous solutions.
1 for TMPyP4 to the Tel22 G-quadruplex in 100 mM Na+ buffer using UV absorption spectroscopy.15 Lewis et al. reported that the binding number of TMPyP4 to Tel22 G-quadruplex is ∼4 using ITC experiments with the addition of 150 mM NaCl.37 It is obvious that the binding of TMPyP4 to Tel22 formed G-quadruplex is influenced by the ionic strength of solutions. For example, Zhang and co-workers concluded that with an increase in the concentration of K+ from 0 to 100 mM, the binding number decreases from 5 to 3.24 Until now, there is no complete understanding of the effect of salt concentration on the binding modes and binding number. Furthermore, so far, little is known about the relation between the binding and the structures of DNA–TMPyP4 complexes in aqueous solutions.
In this study, we mainly used analytical ultracentrifugation sedimentation velocity (AUC-SV) to investigate mixtures of aqueous solutions of TMPyP4 and AGGG(TTAGGG)3 (Tel22) containing 10 mM tris(hydroxymethyl)aminomethane (Tris)–HCl (pH = 7.5) and different concentrations of NaCl. The binding number of TMPyP4 increases continuously with an increase in CTMPyP4 and decreases with an increase in CNaCl. When the concentration ratio of TMPyP4 to Tel22 is 5, the binding number first decreases rapidly from 5 to 3.2 due to the competition between Na+ and weak binding TMPyP4 molecules, and then it decreases from 3.2 to 2.5 with a further increase in CNaCl, which may correspond to the competition between Na+ and TMPyP4 with medium binding affinity. Meanwhile the increase in CNa+ has a weak influence on the high affinity binding modes, as the binding number has almost no change with a further increase in CNa+ when CNa+ > 300 mM. Moreover, the results of both AUC-SV and PAGE reveal that the addition of TMPyP4 in G-quadruplex aqueous solutions can induce the formation of the dimeric G-quadruplex–TMPyP4 complex and the amount of dimers is related to the binding modes of TMPyP4. Our results show that the binding mode with medium affinity can promote the formation of the dimeric complex and weak interactions have a negative effect, presumably due to an instability of G-quadruplex caused by excessive TMPyP4 molecules.
Experimental section
Sample preparation
The human telomeric DNA fragment 5′-AGGGTTAGGGTTAGGGTTAGGG-3′ (Tel22) was purchased from Sangon Biological Engineering Technology and Services (Shanghai, China) in high performance liquid chromatography (HPLC)-purified form. The oligonucleotides were dissolved in 10 mM tris(hydroxymethyl) aminomethane (Tris)–HCl buffer solution (pH = 7.5). The DNA stock solution was heated to 95 °C for 5 min in a ThermoMixer comfort incubator (Eppendorf, Germany), slowly cooled down to room temperature and incubated at room temperature for another ∼2 h. The stock solution was stored at −20 °C for future use. The concentration of DNA was determined by measuring the absorbance at 260 nm using a UV-vis spectrophotometer (UV-2802PCS, UNICO, Shanghai) with a molar extinction coefficient of 228.5 mM−1 cm−1.19,43TMPyP4 in the form of a tetra-o-tosylate salt was purchased from Tokyo Chemical Industry (TCI) and used as received. The solid sample was dissolved in 10 mM Tris–HCl buffer solution (pH = 7.5) to obtain a stock solution with a concentration of ∼1.0 mM. The exact concentration of TMPyP4 was determined by measuring the absorbance at 424 nm with a molar extinction coefficient of 226 mM−1 cm−1.32,44 The DNA solutions containing different concentrations of TMPyP4 were heated again to 95 °C for 5 min and then incubated at room temperature for another ∼2 h. The solutions were stored at 4 °C for future use. Ultra-pure Milli-Q water with a resistivity of 18.2 MΩ cm, that was used in all experiments, was purified by filtration through a Millipore Gradient system after distillation. Tris(hydroxymethyl) aminomethane (Tris, 99%), concentrated hydrochloric acid (37.5%), sodium chloride (NaCl, 99%) and potassium chloride (KCl, 99%) from Sinopharm were used as received.
Circular dichroism (CD)
CD experiments were performed on a JASCO J-810 spectrometer at room temperature. For each sample the data were collected from 200 to 320 nm with a scanning rate of 100 nm min−1 and each final spectrum was an average of three scans using a 1 mm path length quartz cuvette. Scans of buffer solutions containing the same concentrations of salt and TMPyP4 were measured under the same experimental conditions and used as the background correction.Sedimentation velocity experiments
Sedimentation velocity (SV) experiments were carried out in a ProteomeLab XL-A analytical ultracentrifuge (Beckman Coulter Instruments) at 20.0 °C with a rotational speed of 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm using 3 mm double sector cells. The criterion to select an appropriate wavelength is that the measured absorbance should be in the range of 0.2–1.2 to ensure a good signal to noise ratio. About 200 scans were collected during each SV experiment and analyzed by Sedfit (version 14.1) using a continuous c(s) distribution model.45,46 The values of viscosity and density of the buffer solutions were obtained from Sednterp software.47
000 rpm using 3 mm double sector cells. The criterion to select an appropriate wavelength is that the measured absorbance should be in the range of 0.2–1.2 to ensure a good signal to noise ratio. About 200 scans were collected during each SV experiment and analyzed by Sedfit (version 14.1) using a continuous c(s) distribution model.45,46 The values of viscosity and density of the buffer solutions were obtained from Sednterp software.47
PAGE experiments
20 μL of DNA solution was mixed with 3 μL 10 × loading buffer (30 mM EDTA, 50% (v/v) glycerol, 0.25% (w/v) xylene cyanol FF, and 0.25% (w/v) bromophenol blue) and then loaded onto a 20% (w/v) polyacrylamide gel containing 100 mM NaCl and subjected to a constant voltage of 50 V for 2.5 h. The running buffer was 40 mM Tris–acetic acid with a pH of 7.5. Electrophoresis experiments were conducted at room temperature. After each experiment, DNA chains in the gel were stained with GelRed for 40 min and photographed by a gel image system (Tanon).Results and discussion
Binding of TMPyP4 to Tel22 G-quadruplex
The human telomeric fragment AGGG(TTAGGG)3 (Tel22) can fold into G-quadruplex in the presence of Na+ or K+ and the structural information of G-quadruplex has been determined using NMR and other techniques.1,48,49 As reported, Tel22 forms an antiparallel basket-type structure with the help of Na+, and it forms hybrid structures (hybrid 1 and hybrid 2) in the presence of K+ in the solution state, as shown in Scheme 1(b).1,49,50 In this study, first we used AUC-SV and CD to confirm and characterize G-quadruplex structures in the presence of 100 mM Na+ or K+ with 10 mM tris(hydroxymethyl) aminomethane (Tris)–HCl (pH = 7.5) as a buffer solution. The results are shown in Fig. S1.† As shown in Fig. S1(a),† the sedimentation coefficients (s) of Tel22 in the presence of 100 mM NaCl and KCl are 1.90 S and 2.02 S, respectively, which are in good agreement with earlier reports on G-quadruplex structures.43,50 Fig. S1(b)† shows that Tel22 presents a positive peak at 295 nm and a negative peak at 265 nm in the presence of 100 mM NaCl, and has a positive peak at 290 nm with a shoulder at around 270 nm and a negative peak around 235 nm with the addition of 100 mM KCl, indicating the formation of basket-type and hybrid structures in these two salt solutions, respectively.49–52Then we investigated the effect of 5,10,15,20-tetra-(N-methyl-4-pyridyl)porphyrin (TMPyP4) (Scheme 1(a)) on the structure of G-quadruplex in aqueous solutions using AUC-SV, PAGE, UV-vis spectrophotometry and CD spectroscopy. To avoid structural diversity of Tel22 in the solutions, we mainly focussed our study on the interactions between TMPyP4 and basket-type G-quadruplex in NaCl solution. The CD spectra for the titrations of TMPyP4 into Tel22 solutions with the basket-type G-quadruplex structure containing 100 mM NaCl are illustrated in Fig. 1. The concentrations of Tel22 (CTel22) and NaCl are 8.0 μM and 100 mM, respectively. The concentrations of TMPyP4 (CTMPyP4) range from 0 to 56.0 μM, and the further increase in CTMPyP4 higher than 56.0 μM leads to the formation of some precipitates. As shown in Fig. 1, the addition of TMPyP4 causes significant attenuation in the CD signals at both 265 nm and 295 nm.28,37 Besides the decrease in CD signals, TMPyP4 also causes a shift of the peaks at wavelengths of ∼265 nm and 295 nm.28,37 It should be noted that TMPyP4 has nearly no CD signal, as shown in Fig. S2,† when CTMPyP4 is 80.0 μM. Moreover, as shown in Fig. S3,† compared with the UV-vis absorption spectrum of free TMPyP4, the addition of Tel22 causes a red-shift of the maximum absorption wavelength of TMPyP4 from 424 to 435 nm and a decrease in the maximum extinction coefficient, indicating the formation of Tel22-TMPyP4 complexes.23,32 Thus, the change in the CD spectra shown in Fig. 1 should be due to the formation of the Tel22/TMPyP4 complexes. Moreover, the decreased CD intensity with the increase in CTMPyP4 when CTMPyP4 is larger than 24.0 μM indicates that the excessive TMPyP4 molecules may have a negative influence on the formation of G-quadruplex.24 The titrations of NaCl into the Tel22-ligand mixture solution (Fig. S4†) show the CD signal at 295 nm still increases when CNaCl is larger than the minimum concentration (100 mM) to fully induce the formation of G-quadruplex without the addition of TMPyP4, as reported in our previous study,43 reflecting the negative influence of TMPyP4 on the stability of G-quadruplex.
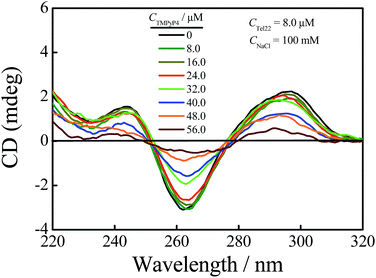 | ||
| Fig. 1 CD spectra of a Tel22 (CTel22 = 8.0 μM) solution containing 100 mM NaCl with the addition of different concentrations of TMPyP4. | ||
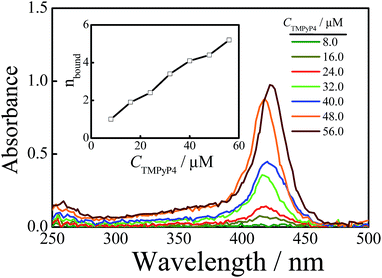
Sedimentation velocity (SV) is a powerful method for investigating the conformational change of solutes and the homogeneity of sample solutions and has been widely used in the G-quadruplex system.9,50,53–55 Herein, we used this method to investigate the structural change of DNA chains in aqueous solutions with the addition of TMPyP4. Firstly, we conducted an SV experiment on a TMPyP4 aqueous solution without DNA chains, and the result is shown in Fig. S5.† It shows that a free TMPyP4 molecule has a small sedimentation coefficient (s ∼ 0.25 S) and hardly sediments during the SV experiment. Thus the absorbance of TMPyP4 at different radial positions only slightly changes after the SV experiment. The velocity scans at the beginning and the end of one SV experiment (about 10 h) are shown in Fig. S6.† The two velocity scans have a cross-point at a radius of ∼6.8 cm, which means that the concentrations of TMPyP4 at this position before and after the SV experiment are the same. In the presence of DNA, bound TMPyP4 molecules will sediment together with DNA chains. Thus, we can determine the amount of the free TMPyP4 molecules in an aqueous solution by measuring the absorbance spectra at a radius of 6.8 cm when the rotor is still rotating at a rate of 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at the end of one SV experiment (about 10 h). For example, as shown in Fig. 2, after one SV experiment the measured concentration of free TMPyP4 (Cf) from the absorbance at a wavelength of 420 nm in the solution is 7.0 μM, where the initial concentrations of Tel22 (CDNA) and TMPyP4 (Ci) are 8.0 μM and 40.0 μM, respectively. Thus the binding number of TMPyP4 per DNA (nbound) is calculated as 4.1 using the equation of nbound = (Ci − Cf)/CDNA. The value of nbound has been widely studied.16,19,25,27,31,36,42 However, there is no exact understanding on this issue until now. Besides the structural specificities of DNA chains, the difference might result from the following reasons. Firstly, the binding number is influenced by the initial concentration ratio of TMPyP4 to Tel22 and the salt concentration in solutions.24,32 Secondly, the binding number is determined by various instruments and each instrument has its sensitivity.
000 rpm at the end of one SV experiment (about 10 h). For example, as shown in Fig. 2, after one SV experiment the measured concentration of free TMPyP4 (Cf) from the absorbance at a wavelength of 420 nm in the solution is 7.0 μM, where the initial concentrations of Tel22 (CDNA) and TMPyP4 (Ci) are 8.0 μM and 40.0 μM, respectively. Thus the binding number of TMPyP4 per DNA (nbound) is calculated as 4.1 using the equation of nbound = (Ci − Cf)/CDNA. The value of nbound has been widely studied.16,19,25,27,31,36,42 However, there is no exact understanding on this issue until now. Besides the structural specificities of DNA chains, the difference might result from the following reasons. Firstly, the binding number is influenced by the initial concentration ratio of TMPyP4 to Tel22 and the salt concentration in solutions.24,32 Secondly, the binding number is determined by various instruments and each instrument has its sensitivity.
The binding number of TMPyP4 to Tel22 increases with CTMPyP4
We used the above-mentioned method to investigate the effect of CTMPyP4 on the binding number at a constant NaCl concentration of 100 mM. Fig. 2 shows the absorbance spectra of free TMPyP4 molecules in solution. From the absorbance at the wavelength around 420 nm, we can obtain the concentration of free TMPyP4 molecules in the aqueous solution as discussed above. Then the average binding number of TMPyP4 per Tel22 chain could be obtained by nbound = (Ci − Cf)/CDNA. The inset of Fig. 2 shows that nbound increases with CTMPyP4 when the concentration of Tel22 is kept at 8.0 μM, and the binding number can become larger than 4 when the concentration of TMPyP4 is higher than 40.0 μM. However, most previous studies show that the binding stoichiometry of TMPyP4 to Tel22 ranges from 2 to 4 using Job plot and ITC.15,16,19,27,37 Herein, we also measured the binding stoichiometry using Job plot according to the reported method.27 A binding stoichiometry of ∼3 is obtained, as shown in Fig. S7.† The difference in the two experiments is presumably due to the reason that some weak binding has little contribution to the detected signal of absorbance, but it can be readily detected by our method due to the fact that both strong and weak binding TMPyP4 molecules sediment together with Tel22 chains. Using molecular dynamics simulations, Bhattacharya et al. reported that there are five modes for TMPyP4 to bind with basket-type G-quadruplex, which consist of one binding mode of end-stacking and four groove binding modes.35 Among the four groove binding modes, two of the four have stronger binding affinities, while the others have weaker binding affinities. The weak groove binding may only be due to non-specific electrostatic interactions. Previous studies have also reported that there are several specific binding modes between TMPyP4 and G-quadruplex, such as intercalation, end-stacking, and specific loop or groove binding.11,32,42,56 The binding affinities of different specific binding modes are different from each other and follow the order of intercalation > end-stacking > specific loop or groove binding.35,57 Thus, we can roughly divide them into three groups: high affinity binding modes including the inner intercalation mode and end stacking, medium affinity binding modes containing some groove binding modes, and non-specific weak electrostatic binding modes. With the increase in CTMPyP4, TMPyP4 may first bind to G-quadruplex due to high affinity binding modes and the medium affinity binding mode and then the weak non-specific electrostatic binding causes a further increase in the binding number, as shown in Fig. 2.TMPyP4 can induce the formation of a dimeric complex
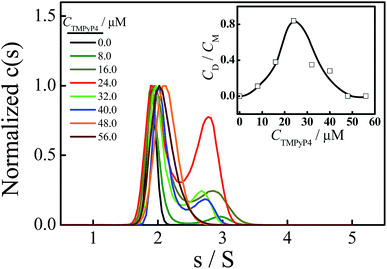
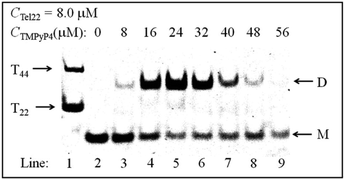
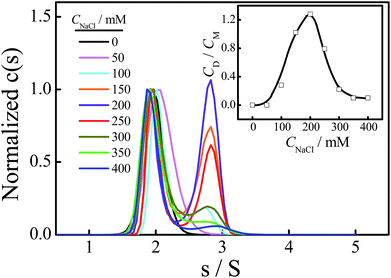
Fig. 3 shows the normalized sedimentation coefficient distributions analyzed by Sedfit using the continuous c(s) distribution model. A unimodal distribution with s = 1.90 S of Tel22 in an aqueous solution containing 100 mM NaCl and without the addition of TMPyP4 indicates an intramolecular basket-type G-quadruplex (M).43,50 With the addition of 8.0 μM TMPyP4, a second peak (D) with a sedimentation coefficient of 2.7 S appears, presumably due to the formation of a dimer between Tel22–TMPyP4 complexes. Moreover, Fig. 3 shows that the amount of dimer increases with CTMPyP4 when CTMPyP4 < 24.0 μM, and then decreases with a further increase in CTMPyP4. The effect of CTMPyP4 on the concentration ratio of the dimer to the monomer (CD/CM) is shown in the inset of Fig. 3. With an increase in CTMPyP4, TMPyP4 can gradually bind to G-quadruplex by strong interactions as illustrated in Fig. 2, and promotes the formation of the dimer. When CTMPyP4 is 24.0 μM, all the strong binding sites (∼3) are occupied. Thus CD/CM reaches its maximum value. When CTMPyP4 > 24.0 μM, TMPyP4 starts to bind to G-quadruplex through weak non-specific interactions, which may weaken the stability of G-quadruplex as illustrated in Fig. 1. The Tel22–TMPyP4 complex with a less stable G-quadruplex structure cannot form dimeric complexes. Thus, with the further increase in CTMPyP4, the amount of dimer decreases. Furthermore, we carried out two series of control SV experiments: one concerns the solution of Tel22/TMPyP4 without the addition of NaCl and the other concerns the solution of a control DNA/TMPyP4 (the sequence of control DNA is presented in Fig. S9† and the control DNA cannot form the G-quadruplex structure) with the addition of 100 mM NaCl, and the results are shown in Fig. S8 and S9.† The monodispersed distributions of the two systems indicate that the G-quadruplex structure is a critical factor for the formation of dimeric complexes.Thereafter, we conducted PAGE experiments to verify the results shown in Fig. 3. As shown in Fig. 4, line 2 represents an intramolecular G-quadruplex formed from Tel22 (M). With the addition of TMPyP4, a second band (D) appears and its content reaches a maximum when CTMPyP4 is 24.0 μM. A further increase in CTMPyP4 leads to a decrease in the content of band D, which is in good agreement with the results of SV shown in Fig. 3. Moreover, the band brightness becomes lower when CTMPyP4 is larger than 40.0 μM, presumably because the binding of TMPyP4 to Tel22 decreases the staining ability of GelRed. The gel bands shown in Fig. 4 (line: 3–9) belong to Tel22/TMPyP4 complexes and not to the ligand-free Tel22 because these bands are yellow.36
Medium affinity binding mode promotes the dimeric complex formation
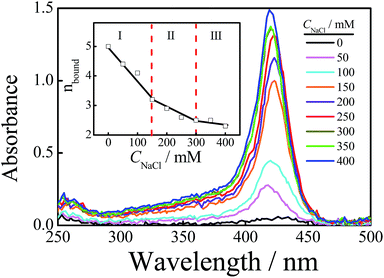
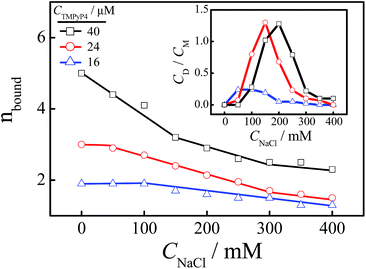
Fig. 5 shows the absorption spectra of free TMPyP4 with the addition of different concentrations of NaCl ranging from 0 to 400 mM, where the concentration of Tel22 is 8.0 μM and the concentration of TMPyP4 is 40.0 μM. The results show that the absorbance of TMPyP4 at 420 nm increases with an increase in CNaCl, indicating an increase in concentration of free TMPyP4 molecules. The effect of NaCl concentration on the calculated nbound is shown in the inset of Fig. 5. It decreases from 5 to 2.3 with increasing CNaCl to 400 mM due to the competition between Na+ and TMPyP4, and we can divide this into three regions with slopes of −0.0114, −0.0048 and −0.0020, as shown in the inset. The addition of NaCl first disturbs the weak interactions and leads to the decrease in nbound in region I when CNaCl < 150 mM. After the binding number decreases to ∼3.2 and the remaining interactions are strong, the further increase in CNaCl has a weaker effect on these strong interactions so that the binding number only slightly decreases from 3.2 to 2.3. In other words, the binding number of TMPyP4 due to the strong interactions is ∼3.2 when the concentrations of Tel22 and TMPyP4 are 8.0 and 40.0 μM, respectively, which is in good agreement with the value measured by Job plot, as shown in Fig. S7.† Note that Zhang et al. also found that the binding numbers of TMPyP4 per DNA with a similar sequence are 5 and 3 when the KCl concentrations are 0 and 100 mM, respectively.24 The increase in CNaCl has less effect on higher affinity binding modes than the binding mode with a medium affinity (possibly some groove binding). Thus we can observe two different processes (regions II and III) in Fig. 5.Fig. 6 shows the effect of CNaCl on the normalized sedimentation coefficient distributions of Tel22/TMPyP4 solutions, where the concentrations of Tel22 and TMPyP4 are 8.0 μM and 40.0 μM, respectively. As shown in Fig. 6, two peaks located around 1.9 S and 2.7 S with the addition of NaCl belong to the monomeric and dimeric complexes, respectively, as discussed above. The dependence of CNaCl on the dimeric content is plotted in the inset of Fig. 6. Firstly, CD/CM increases from 0 to 1.2 with an increase in CNaCl, presumably due to the formation of G-quadruplex and the increased stability of G-quadruplex with the increase in CNa+. A further increase in CNaCl when CNaCl > 200 mM causes a decrease in dimeric content, which may be due to the decrease in the medium affinity binding mode of specific groove binding as mentioned in Fig. 5. It is worth noting that the high affinity binding may have no contribution to the dimer formation. The small amount of dimeric complex shown in region III, where CNaCl is larger than 300 mM, may be due to the fact that a part of the medium affinity binding mode may still exist in the system.
To understand the relationship between the binding and dimeric complex formation in detail, we also studied the effects of CNaCl on the binding number and dimeric content when CTMPyP4 is 16.0 μM and 24.0 μM, as shown in Fig. 7. When CTMPyP4 is 16.0 μM, the main binding modes between TMPyP4 and G-quadruplex are high affinity binding. Thus we only found one slope of −0.0019 with the increase in CNaCl when CNaCl > 100 mM, as shown in Fig. 7. Note that there are still some TMPyP4 molecules binding to Tel22 through the medium affinity binding mode, thus a small amount of the dimeric complex can form when CNa+ < 200 mM. When CNaCl is larger than 200 mM, there is almost no dimeric complex while the high affinity binding modes still exist, indicating that the existence of the high affinity binding modes may have no contribution to dimer formation. When CTMPyp4 is 24.0 μM, nbound is almost unchanged with the addition of 50 mM NaCl in the solution, presumably due to the fact that there is no weak interaction between TMPyP4 and Tel22 at this ligand concentration. Then the further increase in CNaCl makes nbound decrease in two manners with slopes of −0.0048 and −0.0020, which are consistent with the two slopes for the medium affinity binding mode of specific groove binding and high affinity binding modes when CTMPyp4 is 40.0 μM. Due to the formation of G-quadruplex and the increased stability with the addition of NaCl, the content of the dimeric complex increases. Meanwhile, the increase in CNaCl higher than 150 mM also has a negative effect on the medium affinity binding mode, thus we observe a decrease in the dimeric content.
Potassium ions (K+) also play an important role in the formation of G-quadruplex structures. In the presence of K+, Tel22 can form a G-quadruplex with hybrid structures, as shown in Scheme 1(b).49,58 Herein, we also investigated the influence of TMPyP4 on the structural change of the hybrid-type G-quadruplex. As shown in Fig. S10,† with the addition of TMPyP4, the CD signal at 295 nm significantly decreases, and the shoulder peak at 265 nm becomes dominant. Thus, it is obvious that TMPyP4 does have a significant effect on the G-quadruplex structures. SV experiments were also carried out on these systems. As shown in Fig. S11,† the addition of TMPyP4 has no obvious effect on the sedimentation coefficient distribution of Tel22 on the first day of preparation. However, the dimeric complex appears when the solution mixture was detected on the 7th day, as shown in Fig. S11(b).† The amount of dimeric complexes with the addition of K+ is much less than that with the addition of Na+ in the same conditions, which may be due to the different conformations of G-quadruplex formed in these two systems and/or the possible different binding modes of TMPyP4 to the two kinds of G-quadruplex.
Conclusions
In conclusion, we have studied the effect of Na+ on the interactions between a human telomeric DNA fragment Tel22 and TMPyP4, and its effect on the dimeric G-quadruplex formation. The results of AUC-SV show that the binding number of TMPyP4 per Tel22 is affected by both CTMPyP4 and CNaCl. With an increase in CTMPyP4, the binding number continuously increases, and the addition of NaCl in the Tel22/TMPyP4 aqueous solution has a negative effect on the binding number. Our results show that there are three kinds of interaction between TMPyP4 and G-quadruplex: high affinity binding modes, medium affinity binding modes containing some groove binding modes, and non-specific weak electrostatic binding modes. Moreover, the addition of TMPyP4 in the basket-type G-quadruplex aqueous solutions can induce the formation of dimeric Tel22–TMPyP4 complexes, and the three kinds of interaction have different influences on the formation of dimers. Weak electrostatic interactions have a negative effect on dimer formation because they can destroy the G-quadruplex structure. Medium affinity binding modes of specific groove binding can promote the formation of the dimer, while the high affinity binding modes have no effect on its formation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The financial support of the National Natural Scientific Foundation of China (NNSFC) Projects (21674107 and 21274140) and the Fundamental Research Funds for the Central Universities (WK2340000066) is gratefully acknowledged.Notes and references
- Y. Wang and D. J. Patel, Structure, 1993, 1, 263–282 CrossRef CAS PubMed.
- C. L. Grand, H. Y. Han, R. M. Munoz, S. Weitman, D. D. Von Hoff, L. H. Hurley and D. J. Bearss, Mol. Cancer Ther., 2002, 1, 565–573 CAS.
- J. T. Davis, Angew. Chem., Int. Ed., 2004, 43, 668–698 CrossRef CAS PubMed.
- Q. Wang, J. Q. Liu, Z. Chen, K. W. Zheng, C. Y. Chen, Y. H. Hao and Z. Tan, Nucleic Acids Res., 2011, 39, 6229–6237 CrossRef CAS PubMed.
- S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discovery, 2011, 10, 261–275 CrossRef CAS PubMed.
- H. J. Kang, Y. X. Cui, H. Yin, A. Scheid, W. P. D. Hendricks, J. Schmidt, A. Sekulic, D. M. Kong, J. M. Trent, V. Gokhale, H. B. Mao and L. H. Hurley, J. Am. Chem. Soc., 2016, 138, 13673–13692 CrossRef CAS PubMed.
- J. Mohanty, N. Barooah, V. Dhamodharan, S. Harikrishna, P. I. Pradeepkumar and A. C. Bhasikuttan, J. Am. Chem. Soc., 2013, 135, 367–376 CrossRef CAS PubMed.
- K. Takahama, A. Takada, S. Tada, M. Shimizu, K. Sayama, R. Kurokawa and T. Oyoshi, Chem. Biol., 2013, 20, 341–350 CrossRef CAS PubMed.
- R. Buscaglia, M. C. Miller, W. L. Dean, R. D. Gray, A. N. Lane, J. O. Trent and J. B. Chaires, Nucleic Acids Res., 2013, 41, 7934–7946 CrossRef CAS PubMed.
- K. Takahama, A. Miyawaki, T. Shitara, K. Mitsuya, M. Morikawa, M. Hagihara, K. Kino, A. Yamamoto and T. Oyoshi, ACS Chem. Biol., 2015, 10, 2564–2569 CrossRef CAS PubMed.
- A. Marchand, A. Granzhan, K. Iida, Y. Tsushima, Y. Ma, K. Nagasawa, M. P. Teulade-Fichou and V. Gabelica, J. Am. Chem. Soc., 2015, 137, 750–756 CrossRef CAS PubMed.
- A. Chauhan, R. Paul, M. Debnath, I. Bessi, S. Mandal, H. Schwalbe and J. Dash, J. Med. Chem., 2016, 59, 7275–7281 CrossRef CAS PubMed.
- M. Debnath, S. Ghosh, R. P. Ajay Chauhan, K. Bhattacharyya and J. Dash, Chem. Sci., 2017, 8, 7448–7456 RSC.
- N. V. Anantha, M. Azam and R. D. Sheardy, Biochemistry, 1998, 37, 2709–2714 CrossRef CAS PubMed.
- R. T. Wheelhouse, D. K. Sun, H. Y. Han, F. X. G. Han and L. H. Hurley, J. Am. Chem. Soc., 1998, 120, 3261–3262 CrossRef CAS.
- I. Haq, J. O. Trent, B. Z. Chowdhry and T. C. Jenkins, J. Am. Chem. Soc., 1999, 121, 1768–1779 CrossRef CAS.
- E. Izbicka, R. T. Wheelhouse, E. Raymond, K. K. Davidson, R. A. Lawrence, D. Y. Sun, B. E. Windle, L. H. Hurley and D. D. Von Hoff, Cancer Res., 1999, 59, 639–644 CAS.
- H. Y. Han, D. R. Langley, A. Rangan and L. H. Hurley, J. Am. Chem. Soc., 2001, 123, 8902–8913 CrossRef CAS PubMed.
- C. Y. Wei, G. Q. Jia, J. L. Yuan, Z. C. Feng and C. Li, Biochemistry, 2006, 45, 6681–6691 CrossRef CAS PubMed.
- H. Mita, T. Ohyama, Y. Tanaka and Y. Yamamoto, Biochemistry, 2006, 45, 6765–6772 CrossRef CAS PubMed.
- G. N. Parkinson, R. Ghosh and S. Neidle, Biochemistry, 2007, 46, 2390–2397 CrossRef CAS PubMed.
- C. Wei, G. Han, G. Jia, J. Zhou and C. Li, Biophys. Chem., 2008, 137, 19–23 CrossRef CAS PubMed.
- A. Arora and S. Maiti, J. Phys. Chem. B, 2008, 112, 8151–8159 CrossRef CAS PubMed.
- H. J. Zhang, X. F. Wang, P. Wang, X. C. Ai and J. P. Zhang, Photochem. Photobiol. Sci., 2008, 7, 948–955 CAS.
- G. Q. Jia, Z. C. Feng, C. Y. Wei, J. Zhou, X. L. Wang and C. Li, J. Phys. Chem. B, 2009, 113, 16237–16245 CrossRef CAS PubMed.
- L. Martino, B. Pagano, I. Fotticchia, S. Neidle and C. Giancola, J. Phys. Chem. B, 2009, 113, 14779–14786 CrossRef CAS PubMed.
- C. Y. Wei, G. Q. Jia, J. Zhou, G. Y. Han and C. Li, Phys. Chem. Chem. Phys., 2009, 11, 4025–4032 RSC.
- R. D. Gray, J. Li and J. B. Chaires, J. Phys. Chem. B, 2009, 113, 2676–2683 CrossRef CAS PubMed.
- A. Arora and S. Maiti, J. Phys. Chem. B, 2009, 113, 8784–8792 CrossRef CAS PubMed.
- A. Kabir, M. Hossain and G. S. Kumar, J. Chem. Thermodyn., 2013, 57, 445–453 CrossRef CAS.
- A. G. Kudrev, Talanta, 2013, 116, 541–547 CrossRef CAS PubMed.
- D. Song, W. Yang, T. X. Qin, L. D. Wu, K. H. Liu and H. M. Su, J. Phys. Chem. Lett., 2014, 5, 2259–2266 CrossRef CAS PubMed.
- L. P. Bai, J. Liu, L. Han, H. M. Ho, R. X. Wang and Z. H. Jiang, Anal. Bioanal. Chem., 2014, 406, 5455–5463 CrossRef CAS PubMed.
- D. D. Le, M. Di Antonio, L. K. M. Chan and S. Balasubramanian, Chem. Commun., 2015, 51, 8048–8050 RSC.
- A. Ali, M. Bansal and S. Bhattacharya, J. Phys. Chem. B, 2015, 119, 5–14 CrossRef CAS PubMed.
- M. Bončina, C. Podlipnik, I. Piantanida, J. Eilmes, M. P. Teulade-Fichou, G. Vesnaver and J. Lah, Nucleic Acids Res., 2015, 43, 10376–10386 Search PubMed.
- J. I. DuPont, K. L. Henderson, A. Metz, V. H. Le, J. P. Emerson and E. A. Lewis, Biochim. Biophys. Acta, Gen. Subj., 2016, 1860, 902–909 CrossRef CAS PubMed.
- A. G. Kudrev, Russ. J. Gen. Chem., 2016, 86, 1353–1363 CrossRef CAS.
- F. S. Di Leva, E. Novellino, A. Cavalli, M. Parrinello and V. Limongelli, Nucleic Acids Res., 2014, 42, 5447–5455 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, H. Y. Gaw and D. J. Patel, Nat. Chem. Biol., 2005, 1, 167–173 CrossRef CAS PubMed.
- P. Murat, Y. Singh and E. Defrancq, Chem. Soc. Rev., 2011, 40, 5293–5307 RSC.
- T. Kimura, K. Kawai, M. Fujitsuka and T. Majima, Tetrahedron, 2007, 63, 3585–3590 CrossRef CAS.
- Y. Gao, S. Wu and X. Ye, Soft Matter, 2016, 12, 5959–5967 RSC.
- R. F. Pasternack, E. J. Gibbs and J. J. Villafranca, Biochemistry, 1983, 22, 2406–2414 CrossRef CAS PubMed.
- P. Schuck, Biophys. J., 2000, 78, 1606–1619 CrossRef CAS PubMed.
- P. H. Brown and P. Schuck, Biophys. J., 2006, 90, 4651–4661 CrossRef CAS PubMed.
- T. M. Laue, B. D. Shah, T. M. Ridgeway and S. L. Pelletier, ed. S. E. Harding, A. J. Rowe and J. C. Horton, Computer-aided interpretation of analytical sedimentation data for proteins, Analytical Ultracentrifugation in Biochemistry and Polymer Science, Royal Society of Chemistry, Cambridge, UK, 1992, pp. 90–125 Search PubMed.
- K. N. Luu, A. T. Phan, V. Kuryavyi, L. Lacroix and D. J. Patel, J. Am. Chem. Soc., 2006, 128, 9963–9970 CrossRef CAS PubMed.
- A. Ambrus, D. Chen, J. Dai, T. Bialis, R. A. Jones and D. Yang, Nucleic Acids Res., 2006, 34, 2723–2735 CrossRef CAS PubMed.
- J. Li, J. J. Correia, L. Wang, J. O. Trent and J. B. Chaires, Nucleic Acids Res., 2005, 33, 4649–4659 CrossRef CAS PubMed.
- M. Vorlíčková, J. Chládková, I. Kejnovská, M. Fialová and J. Kypr, Nucleic Acids Res., 2005, 33, 5851–5860 CrossRef PubMed.
- P. Tóthová, P. Krafčíková and V. Viglaský, Biochemistry, 2014, 53, 7013–7027 CrossRef PubMed.
- L. M. Hellman, D. W. Rodgers and M. G. Fried, Eur. Biophys. J., 2010, 39, 389–396 CrossRef CAS PubMed.
- Z. L. Luo and G. Z. Zhang, Macromolecules, 2010, 43, 10038–10044 CrossRef CAS.
- J. B. Chaires, W. L. Dean, H. T. Le and J. O. Trent, Methods Enzymol., 2015, 562, 287–304 Search PubMed.
- I. Lubitz, N. Borovok and A. Kotlyar, Biochemistry, 2007, 46, 12925–12929 CrossRef CAS PubMed.
- M. H. Li, Q. Luo and Z. S. Li, J. Phys. Chem. B, 2010, 114, 6216–6224 CrossRef CAS PubMed.
- J. Dai, M. Carver, C. Punchihewa, R. A. Jones and D. Yang, Nucleic Acids Res., 2007, 35, 4927–4940 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra07758k |
| This journal is © The Royal Society of Chemistry 2017 |