Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A comparative study of microbial dynamics and phosphorus removal for a two side-stream wastewater treatment processes†
Yanyan Zhangab,
Md. Shahinoor Islamac,
Kerry N. McPhedranad,
Shimiao Donga,
Ehab M. Rashede,
Maha M. El-Shafeif,
Ahmed M. Noureldinf and
Mohamed Gamal El-Din *a
*a
aDepartment of Civil and Environmental Engineering, University of Alberta, Edmonton, Alberta, Canada. E-mail: mgamalel-din@ualberta.ca; Tel: +1-780-492-5124
bDepartment of Civil Engineering, New Mexico State University, Las Cruces, New Mexico, USA
cDepartment of Chemical Engineering, Bangladesh University of Engineering & Technology, Dhaka, Bangladesh
dDepartment of Civil, Geological and Environmental Engineering, College of Engineering, University of Saskatchewan, Saskatoon, Canada
eSanitary & Environmental Engineering, Cairo University, Giza, Egypt
fHousing and Building National Research Center (HBRC), Cairo, Egypt
First published on 26th September 2017
Abstract
In this study, a side-stream process with anoxic/aerobic tanks was designed as a denitrifying enhanced biological phosphorus removal wastewater treatment technology (denitrifying EBPR) to be evaluated for its performance in phosphorous (P) removal as compared to a modified side-stream process using contact/stabilization tanks (modified EBPR). Under variable P and ammonia–nitrogen loadings, the microbial community structure, dominant taxonomy, and community dynamics were assessed with the aid of real-time PCR analysis and MiSeq high-throughput sequencing aimed specifically at the major polyphosphate-accumulating organisms (PAOs) and glycogen-accumulating organisms (GAOs). The inclusion of an anoxic tank for the denitrifying EBPR process increased the efficiency of P removal by reducing the amount of nitrate entering the anaerobic tank, which led to increased P removal of 67.9–78.6% compared to 35.2–68.4% for the modified EBPR under various P and ammonia loading conditions. PCR analysis showed that the increased P removal performance of the denitrifying EBPR under increased ammonia loading may be attributed to higher abundances of PAO clade I organisms. MiSeq sequencing analysis indicated that the influent water composition played an important role in the microbial community structure; however, there was an increase in genetic differences between the two systems under high ammonia and P loadings. Overall, Dechloromonas and Sphingomonas were the dominant PAOs and GAOs in both systems. The denitrifying EBPR process resulted in increased concentrations of PAOs versus GAOs as compared to the modified EBPR which resulted in the improved performance of this process.
1 Introduction
Phosphorus (P) can be a growth-limiting nutrient in surface water bodies, however, excess P causes eutrophication in these waters making its removal from municipal wastewater an important aspect in wastewater management. Traditional activated sludge-type wastewater treatment plant processes were not designed specifically for P removal, and thus only remove very limited P from wastewater through cell synthesis.1 To minimize the P release in treated wastewaters, wastewater engineers developed a modified activated sludge-type process known as “enhanced biological phosphorus removal (EBPR)” for increased P removal as compared to the activated sludge process. The EBPR process utilizes cyclic anaerobic and aerobic conditions to promote the growth of polyphosphate accumulating organisms (PAOs) that can accumulate P in excess of their normal metabolic requirements and store it in the form of intracellular polyphosphate.2,3 These organisms are then settled out thereby reducing treated wastewater P concentrations. Despite this advantage of EBPRs, a competitor of PAOs often exists in these treatment systems referred as glycogen accumulating organisms (GAOs). Treatment processes with abundant GAOs result in reduced P removal given they compete with PAOs for volatile fatty acids (VFA) needed for the phosphate uptake process.EBPR processes have been developed using various reactor configurations including anaerobic and aerobic tanks. In mainstream EBPR processes, P removal is achieved by removing waste sludge microorganisms including PAOs that have accumulated polyphosphate. In contrast, side-stream EBPR processes (e.g., Phostrip process) employ an anaerobic tank that leads to the release of P from the PAOs taken from the final settling tank sludge and separates the concentrated P liquid via surface overflow.1 This anaerobic tank also provides additional readily biodegradable COD (rbCOD) from cell lysis for the synthesis of polyhydroxyalkanoates (PHAs) that promotes efficient P uptake for PAOs when it is recycled to the aerobic tank by providing a carbon and energy source. Additionally, side-stream processes have gained popularity for their ability to recover P in the form of struvite that can be sold commercially for use in fertilizer.4 Recently, a modification of the side-stream process (modified EBPR) included the addition of an aerated stabilization tank prior to the anaerobic tank to help promote the growth of PAOs and increase their P uptake from the settling tank concentrated sludge.5
Due to growing wastewater discharge standards, recent efforts have been made in the development of novel processes that enhance P removal in conjunction with improved nitrogen (N) removal. The N in municipal wastewater is in the form of ammonia that is nitrified in the aerobic tank simultaneously with P uptake by PAOs. However, the resulting nitrate is then denitrified in the anaerobic tank with the denitrifying bacteria depleting VFAs needed for PHA synthesis and P release, thus hindering the EBPR performance. Therefore, denitrifying EBPR technologies have become emerging processes for wastewater to help minimize the impact of nitrate.6 For example, an upstream anoxic tank could lead to the reduction of the nitrate entering the anaerobic tank while also increasing P removal through anaerobic–anoxic cycling with nitrate or nitrite being used as electron acceptors by denitrifying PAOs (DPAO).7 However, DPAOs have slower cell growth rates than PAOs under anaerobic/aerobic conditions.8 Other issues in the denitrifying EBPR process include denitrification intermediates such as nitrite and free nitrous acid having inhibitory effects on PAOs.9–11 Therefore, it is not clear whether denitrifying EBPR processes can efficiently remove both P and N simultaneously. Furthermore, fundamental questions remain to be answered about the distribution and abundance of the various PAOs and GAOs subgroups in denitrifying EBPR processes and their response to changes in operating conditions such as variable N and P influent loadings. It has been reported that Candidatus accumulibacter, Tetrasphaera, Rhodocyclus, Dechloromonas genera of the order of Rhodocyclales are related to PHA/PHB storage and polyphosphate accumulating from a biological P removal reactor.12–15 Among them, members of Dechloromonas have also been reported as denitrifying PAO which can uptake acetate and act as denitrifiers; and during denitrification, these species can uptake phosphorous with nitrate acting as an electron acceptor.16 Two lineages of GAOs, the alphaproteobacterial Defluviicoccus-related lineage and the gammaproteobacterial ‘Candidatus competibacter phosphatis’ lineage, have been described in EBPRs as competitor with the PAOs for available carbon source3,17 All of the GAOs identified so far have been shown capable of using nitrate as an electron acceptor in addition to oxygen.18
The main objective of the current study was to identify the side-stream EBPR that provided the best performance for the treatment of municipal wastewater with various N and P loadings while also exploring the mechanisms behind the treatment processes through identification of microbial communities. To meet this objective, two side-stream EBPR processes (Fig. 1) with contact/stabilization and anoxic/aerobic tanks were operated as modified EBPR and denitrifying EBPR processes, respectively. The modified EBPR targeted the P removal while denitrifying EBPR was used for both P and N removal. These side-stream EBPR processes were operated in parallel to compare their performance as demonstrated by chemical oxygen demand (COD), N and P removal rates. The microbial community dynamics were assessed for the various operating conditions using MiSeq high-throughput sequencing and real-time quantitative PCR (qPCR) with focus on the major PAOs and GAOs.
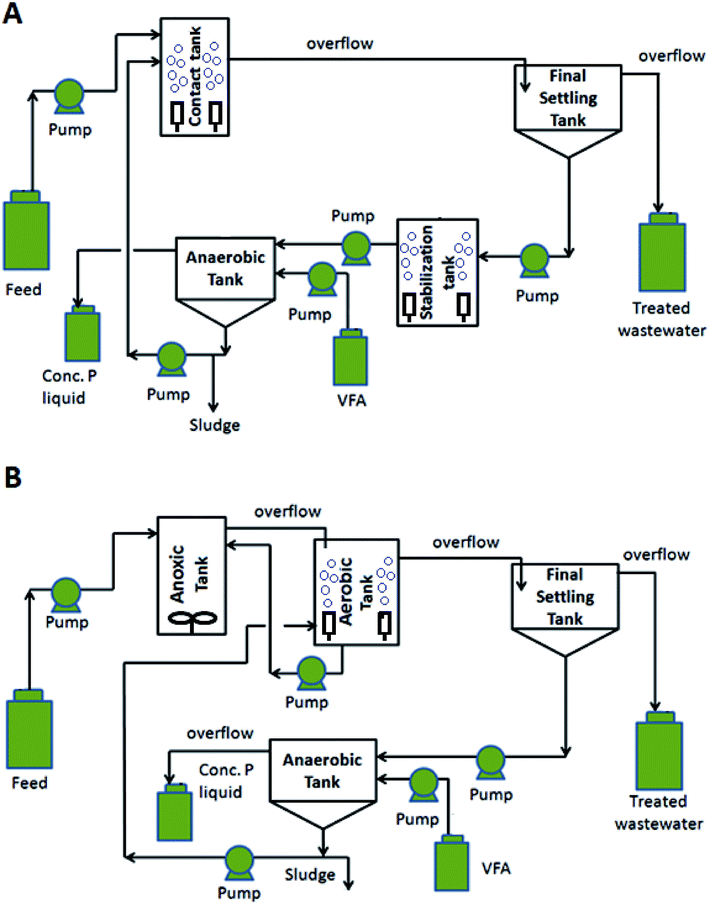 | ||
| Fig. 1 Schematics of (A) modified and (B) denitrifying side-stream EBPR treatment processes. (VFA = volatile fatty acids). | ||
2 Experimental
2.1 Wastewater collection and characterization
Raw influent wastewater was collected in two separate sampling events (March 8, 2015 and May 12, 2015) in 200 L HPDE barrels from the Goldbar Wastewater Treatment Plant (GWTP; Edmonton, AB, CA) and stored at 4 °C prior to use. Both sampling event raw wastewaters having COD concentration of 105–135 mg L−1, P (as PO43–P) concentrations of 7–8 mg L−1 and N (as NH4+–N) concentrations of 32–50 mg L−1 were used for stage 1 experiments (phase I and phase II, respectively) (Table S1†). To evaluate the system performance at various N and P concentrations, mixed wastewaters (using raw wastewater and their respective treated effluents) were prepared for stage 2 and 3 experiments. The mixed wastewater was prepared by mixing of raw and treated wastewater, instead of preparing synthetic wastewaters, which allowed for more consistent physical and chemical characteristics between the various treatment process stages. For stage 2, approximately 90 L of raw wastewater was mixed with 110 L of treated wastewater to obtain a wastewater with low P concentration (PO43−–P = 4–5 mg L−1) in the mixed wastewater. A 5.4 g NH4Cl was added into each 200 L mixed wastewater to result in a NH4+–N concentration of 22–26 mg L−1 to simulate the typical N and P concentration in domestic wastewater.19 60 g of sodium acetate (NaOAc) was added into each 200 L mixed wastewater to make the final COD concentration of 360–395 mg L−1. For stage 3, the wastewater was processed as for stage 2 with the addition of external P (14 g NaH2PO4/200 L mixed wastewater) to create a wastewater with P shock loading (PO43−–P = 21–22 mg L−1). The same amount of NaOAc and NH4Cl were added to make the final COD and NH4+–N in the same range with stage 2. The prepared mixed wastewater was stored at 4 °C prior to use in the treatment process. In addition, sodium acetate (NaOAc) was added into the feed water to increase the COD of mixed wastewater to approximately 400 mg L−1 for stages 2 and 3. The final influent COD/N ratio was 11.1, 15.7, 31.2 and 35.6 for stage 1 (phase I), stage 1 (phase II), stage 2 and 3, respectively. Before starting experiments, the collected and mixed wastewater was characterized for typical wastewater parameters after filtration using nylon membrane filters with pore size of 0.2 μm (Fisher Scientific Company, ON, CA) (Table S1†).2.2 Treatment process operation
The modified and denitrifying EBPR treatment processes are shown in Fig. 1. The modified EBPR (Fig. 1A) was the side-stream process that employed a aerated stabilization tank for the sludge in final settling tank as described previously.5 The denitrifying EBPR included an anoxic–aerobic setup for improved influent N removal, followed by a settling tank to separate treated wastewater and high P sludge due to PAO polyphosphate storage (Fig. 1B). The P release from the sludge occurred in an anaerobic tank and the concentrated P was collected via surface overflow of the supernatant for potential P recovery. Mass balance was used to calculate the flowrate of inflow and outflow for each reactor to regulate the processes. Peristaltic pumps (Cole-Parmer, QC, CA) with Masterflex tubing (Cole-Parmer, QC, CA) were used for all process flows The NaOAc (4000 mg L−1) was added into the anaerobic tanks of both systems at 0.4 mL min−1 as a carbon source to promote PHA accumulation and P release. Recirculation ratios of 0.35 were set for recycling the sludge in anaerobic tanks to the contact/aerobic tanks for both EBPRs. More details regarding tank configuration and the operation conditions can be found in both Tables 1 and S2.† An air pump with fine bubble air diffusers was used in the contact tank and stabilization tank of the modified EBPR and the aerobic tank in the denitrifying EBPR to maintain a dissolved oxygen (DO) concentration of 3 to 4 mg L−1 by adjusting air flowrate. By using a mechanical mixer in the anoxic tank of the denitrifying EBPR, the DO level was kept under 0.5 mg L−1 by changing the mixing speed. Additionally, the target solid retention time (SRT) of 10 days was maintained for both systems by wasting sludge from anaerobic tanks.| Parameter | Modified | Denitrifying |
|---|---|---|
| Purpose | Phosphorous removal | Phosphorous and nitrogen removal |
| Reactors | Aerobic (contact and stabilization) and anaerobic | Aerobic, anoxic and anaerobic |
| Wastewater feed | Aerobic (contact) | Anoxic |
| Hydraulic retention time | Settling: 2 h | Settling: 2.5 h |
| Contact: 2 h | Aerobic: 3 h | |
| Stabilization: 10 h | Anoxic: 2.5 h | |
| Anaerobic: 15 h | Anaerobic: 15 h | |
| Volume | Total: 4.4 L | Total: 8.0 L |
| Settling: 0.8 L | Settling: 0.8 L | |
| Contact: 0.7 L | Aerobic: 3.0 L | |
| Stabilization: 1.2 L | Anoxic: 2.4 L | |
| Anaerobic: 1.8 L | Anaerobic: 1.8 L | |
| Internal recycle | No | Yes (aerobic to anoxic tank: 3) |
| Sludge recycle to aerobic reactor | Yes (35% of feed) | Yes (35% of feed) |
After seeding the aerobic tank with the GWTP sludge, the treatment process was run continuously for 4 months under various loading conditions to evaluate the impact of N (as NH4+–N) and P (as PO43−–P) on the process performance. Each stage was operated until reaching steady-state as determined by PO43−–P removal rates becoming constant (at least 3 weeks) using identical operating conditions outlined above. P uptake and release rate from different tanks was determined by calculating difference between P inflow and P outflow per unit of time.
2.3 Water chemistry analysis and microbial characterization in two EBPRs
The EBPR performance for the overall process and each of the individual tanks were evaluated using various chemical and physical parameters of the feed and treated wastewaters as shown in Fig. 1. After water samples were filtered by using nylon membrane filters with pore size of 0.2 μm (Fisher Scientific Company, ON, CA), the COD, NH4+–N, P (reactive and total P), nitrite nitrogen (NO2–N), nitrate nitrogen (NO3–N) and total nitrogen (TN) were measured in duplicate two times per week using Hach kits (Hach Canada Ltd., ON, Canada). Biomass concentration (mixed liquor volatile suspended solids (MLVSS)) in the reactors, were measured in duplicate according to the standard methods.20 The redox potential, alkalinity and pH were also measured twice a week using a pH/alkalinity/redox potential measurement instrument (SevenMulti™, Mettler-Toledo, OH, USA). DO concentration in each reactor are monitored every day by using a potable DO meter (Orion Star™, Thermo Scientific, MA, USA). P uptake and release rate from different tanks was determined by calculating difference between P inflow and P outflow per unit of time. Once the steady-state in each stage was reached, the sludges from contact, stabilization and anaerobic tanks of modified EBPR and anoxic, aerobic and anaerobic tanks of denitrifying EBPR were collected in triplicates (from three consecutive days) and analyzed for microbial community structures by using real-time quantitative PCR (qPCR) and Illumina MiSeq analysis after DNA extraction (PowerSoil® DNA Isolation Kit, Mo-Bio Laboratories, CA). The data of water quality, biomass concentration and microbial abundance from qPCR was analyzed statistically using ANOVA at a significance level (α) of 0.05.The qPCR was conducted with a Bio-Rad CFX96 real-time PCR system with a C1000 Thermal Cycler (Bio-Rad Laboratories, ON, CA). A total reaction volume of 20 μL was used for each reaction including 10 μL of 2× SsoFast EvaGreen Supermix, 6 μL of sterile water, 1 μL of each primer with a concentration of 10 μM, and 2 μL of diluted sample DNA. The qPCR program consisted of an initial 3 min denaturation at 95 °C, followed by 35 cycles of denaturing at 94 °C for 30 s, annealing for 45 s and extension at 72 °C for 30 s was used to amplify the Accumulibacter ppk1 genes.21 The primer sets for ppk1 genes and annealing temperatures are listed in Table S3.† The abundance of the ppk1 genes was determined based on the qPCR using ppk1-specific primers that represent the abundance of Accumulibacter cells. Three qPCR assays were used to detect nitrifying bacteria, including the amoA gene of the ammonia oxidizing bacteria (AOB) and the 16S rDNA genes of two nitrite oxidizing bacteria (NOB) (Nitrospira spp. and Nitrobacter spp.).22,23 Four qPCR assays were used for quantification of denitrifying bacteria, targeting the nitrate reductase gene (narG), nitrite reductase genes (nirS and nirK), and the nitrous oxide reductase gene (nosZ).24 A qPCR assay for total bacteria (16S rRNA of domain bacteria) was also conducted.25 Primers for the qPCR assays of nitrifiers and denitrifers are listed in Table S4.† The corresponding qPCR amplification program for each individual gene is summarized in Table S5.† All PCR runs used plasmid standards for quantification. Standard plasmids containing target genes were constructed using a TOPO TA Cloning kit (Invitrogen Corporation, Carlsbad, California, USA). Reactions without the DNA template were used as negative controls.
Triplicated DNA samples from the steady state of each stage were pooled together for Illumina MiSeq sequencing in a commercial laboratory (Research and Testing Laboratory, Texas, USA) for identifying the microbial community structure using a previous method.26 All DNA samples were diluted to 20 ng L−1; and about 28 μL of each sample was sent to the testing laboratory for sequencing. The sequencing was performed following the manufacturers guidelines using Illumina MiSeq sequencing instruments and reagents. The sequence was performed using a forward and reverse fusion primer by amplifying the V1–V2 hypervariable regions of 16S rRNA genes prior to Illumina sequencing. Further details on the sequencing are included in the ESI.† After sequencing, the sequenced data were analyzed based on the protocol provided by the laboratory (http://www.researchandtesting.com/docs/Data_Analysis_Methodology.pdf). In brief, the raw sequenced data were denoised and processed for chimera removal (UCHIIME in de novo mode), clustered into operational taxonomic units (OTU) with 100% identity (0% divergence) using USEARCH for taxonomic identification. The Quantitative Insights Into Microbial Ecology (QIIME, http://qiime.org) pipeline with default settings was also used for processing the raw sequence data. In this process, quality checking of raw sequenced data was performed followed by multiplexed reads of samples, picking OTUs (open-reference OTU picking) through making an OTU table with threshold of 0.97, summarizing communities by taxonomic composition, and finally analysis of diversity. UCLUST consensus taxonomy classifier was used to identify representative OTU and taxonomic assignment followed by alpha diversity (diversity within a sample) and beta diversity (diversity among a group of samples) based on the OTU table. Jackknifed PCoA (principal coordinate analysis) plots with weighted UniFrac were also generated by Emperor by using OUT table.
3 Results & discussion
3.1 Performance of modified and denitrifying side-stream EBPRs
| P uptake/release (%) | P uptake/release (mg L−1 h−1) | ||||||
|---|---|---|---|---|---|---|---|
| Contact (uptake) | Stabilization (uptake) | Anaerobic (release) | Contact (uptake) | Stabilization (uptake) | Anaerobic (release) | ||
| Modified EBPR | 1, I | 43 ± 2 | 43 ± 1 | 157 ± 4 | 1.67 ± 0.09 | 0.22 ± 0.01 | 0.31 ± 0.01 |
| 1, II | 63 ± 3 | 26 ± 1 | 257 ± 6 | 2.37 ± 0.11 | 0.09 ± 0.00 | 0.46 ± 0.01 | |
| 2 | 76 ± 8 | 1 ± 0 | 617 ± 96 | 1.91 ± 0.20 | 0.00 ± 0.00 | 0.66 ± 0.10 | |
| 3 | 54 ± 5 | 19 ± 1 | 168 ± 24 | 4.89 ± 0.41 | 0.18 ± 0.01 | 0.70 ± 0.10 | |
| P uptake/release (%) | P uptake/release (mg L−1 h−1) | ||||||
|---|---|---|---|---|---|---|---|
| Anoxic (uptake) | Aerobic (uptake) | Anaerobic (release) | Anoxic (uptake) | Aerobic (uptake) | Anaerobic (release) | ||
| Denitrifying EBPR | 1, I | 28 ± 1 | 77 ± 8 | 549 ± 32 | 0.37 ± 0.01 | 0.35 ± 0.04 | 0.78 ± 0.04 |
| 1, II | 41 ± 1 | 74 ± 8 | 575 ± 34 | 0.56 ± 0.02 | 0.24 ± 0.02 | 0.82 ± 0.05 | |
| 2 | 47 ± 6 | 77 ± 14 | 895 ± 13 | 0.37 ± 0.05 | 0.21 ± 0.04 | 0.67 ± 0.01 | |
| 3 | 10 ± 8 | 76 ± 8 | 224 ± 18 | 0.38 ± 0.03 | 0.96 ± 0.10 | 0.92 ± 0.08 | |
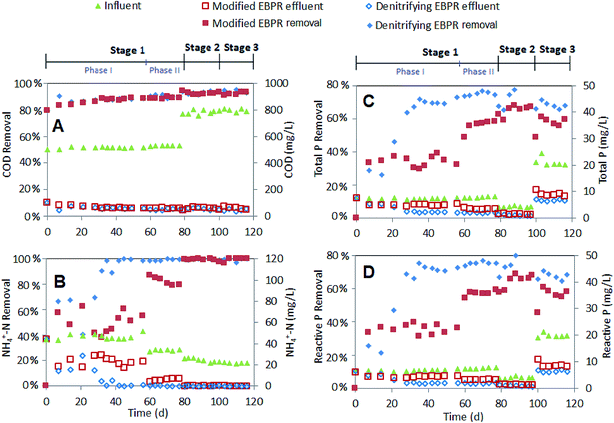
Additionally, the influent P concentration had a noticeable impact on the P removal rates. When P ‘shock’ loading of 20.7 mg L−1 was applied (stage 3), the TP removal decreased from 68.4% and 78.6% (stage 2) to 56.7% and 67.9% for the modified and denitrifying EBPR systems, respectively (Table S6†). To further analyze the P in each system, the P uptake and release rate from different tanks was determined as shown in Table 2. When P shock loading was applied, the speed of P uptake (mg L−1 h−1) increased substantially in the contact tank of the modified EBPR (from 1.91 to 4.89 mg L−1 h−1) and aerobic tank of the denitrifying EBPR (from 0.21 to 0.96 mg L−1 h−1). However, the P release (mg L−1 h−1) for both EBPRs only marginally increased (from 0.66 to 0.70 mg L−1 h−1 for modified EBPR; from 0.67 to 0.92 mg L−1 h−1 for denitrifying EBPRs) with the increased P loading (stages 2 to 3) in the anaerobic tanks, indicating that the P release limitation maybe the bottle neck in P removal at higher loadings. This result is similar to previous studies that showed that the influent P concentration negatively impacted anaerobic PHA synthesis and, subsequently, limited P release during a denitrifying P removal process.31 The decreased anaerobic P release, aerobic P uptake and sludge P content were also observed in another study when 40 mg L−1 P was present in the influent, which might be attributed to the inhibited PAO proliferation and competition from GAOs.33
3.2 Characterization of microbial community
The biomass concentrations in the various tanks are shown in Table 3. After settling tank, the concentrated sludge was observed in the stabilization tank of modified EBPR. MLVSS concentration of around 9000 mg L−1 was also witnessed in anaerobic tanks of both systems. Compared with the anoxic and aerobic tanks in denitrifying EBPR, higher biomass was maintained in the modified EBPR due to sufficient oxygen supply (DO: 3–4 mg L−1) in both contact tank and stabilization tank. These results show the advantage of denitrifying EBPR in less energy consumption and low sludge production. The change of ammonia, P and COD concentration in the influent did not affect the biomass concentration for the denitrifying EBPR. However, the biomass in contact and stabilization tanks decreased with the increase of COD/N ratio.| Stage | Modified | Denitrifying | ||||
|---|---|---|---|---|---|---|
| Contact | Stabilization | Anaerobic | Anoxic | Aerobic | Anaerobic | |
| 1 | 3185 ± 163 | 5270 ± 127 | 9390 ± 240 | 1341 ± 36 | 1730 ± 198 | 8725 ± 544 |
| 2 | 2620 ± 311 | 3440 ± 156 | 9630 ± 1768 | 1323 ± 193 | 1515 ± 332 | 9115 ± 134 |
| 3 | 2825 ± 262 | 3420 ± 156 | 8820 ± 1471 | 1557 ± 137 | 1668 ± 187 | 9200 ± 820 |
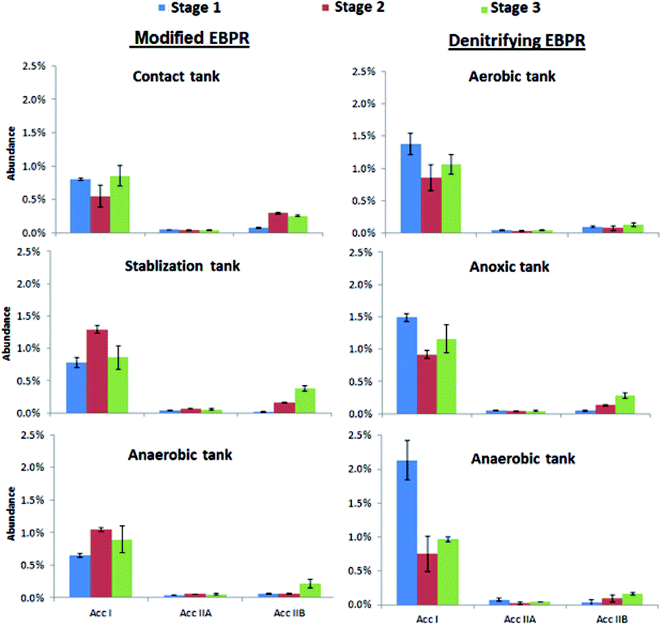
Candidatus accumulibacter has been previously identified as the dominant PAOs in laboratory-scale EBPRs with the polyphosphate kinase (ppk) gene being primarily involved in the production of polyphosphate.36 By using ppk gene sequence information, “Candidatus accumulibacter” strains can be grouped into six distinct clades, designated previously as clade I, IIA, IIB, IIC, IID, and IIF.21 In this study, no Accumulibacter clades of IIC, IID and IIF were found in either EBPR system, thus only Acc I, Acc IIA and Acc IIB were assessed for each tank of each EBPR system (Fig. 3). Clade I was the most dominant clade for all the tanks of two EBPR systems for all stages followed by Acc IIB. Previously, it was reported that “Candidatus accumulibacter” cells of clade I had nitrate reduction activity with P uptake whereas cells of clade IIA and IIB did not show similar activity.37 Therefore, the high abundance of clade I might be attributed to the denitrification activities in both systems. Interestingly, for stage 1 the abundances of clade I were significantly higher in the denitrifying EBPR compared with modified EBPR (p < 0.05; Fig. 3). During this stage the NH4+–N concentrations were higher and thus the clade I organisms might be able to couple nitrate reduction with P uptake during the denitrifying EBPR more effectively. However, no significant differences between the clade I abundances were observed between the two systems during stages 2 and 3 as the NH4+–N in the influent decreased. There was no substantial difference of clade IIA abundance in the different stages for the two systems while a substantial increase of clade IIB observed for stage 3 in most tanks may be associated with the P shock loading in the influent.
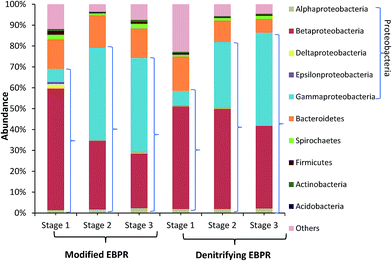 | ||
| Fig. 4 Relative abundances of different bacteria phylum/class in terms of OTUs from the anaerobic reactors of the two EBPRs during each of the stages. | ||
Further examination of the abundances of bacterial orders in both systems, it was observed that the most dominant bacteria orders during stage 1 were Burkholderiales and Rhodocyclales (Fig. 5). Burkholderiales has been reported as the main organism for the bioremediation treatment of various aromatic contaminants such as naphthalene and phenanthrene.43 The members of Rhodocyclales are widespread and abundant in wastewater treatment systems that can degrade diverse groups of aliphatic and aromatic compounds as well as nitrogen and phosphorous compounds.44 Given the diverse aromatic pollutants in the municipal wastewater, the reduced raw municipal wastewater ratio in the influent for stages 2 and 3 may contribute to the declined abundance of Burkholderiales and Rhodocyclales in stages 2 and 3 for modified EBPR as Burkholderiales and Rhodocyclales have been identified as the most dominating bacterial groups for the wastewater containing a range of aliphatic and aromatic organic compounds.45,46 In contrast, the declined trend of Burkholderiales and Rhodocyclales in denitrifying EBPR is not obvious, which might be attributed to its different system configuration. Additionally, several studies have proposed that Rhodocyclus-related bacteria are important PAO group in wastewater treatment plants.47 These bacteria could be also enriched in lab-scale P-removing systems, where they took up acetate, formed PHA anaerobically, and grew and accumulated polyphosphate aerobically. Previous study showed that the proliferation of PAOs was inhibited under high P loading.33 Thus, the decrease of Rhodocyclales abundance in stage 3 may be also attributed to the high P concentration in the influent. The increase of the abundances of Thiotrichales occurred in both the modified and denitrifying EBPR in stages 2 and 3. Thiotrichales are sulfur-oxidizing bacteria found in activated sludge systems causing sludge bulking.48,49 Thiotrichales increasing in stages 2 and 3 in our study might be the early warning of sludge bulking which could be attributed to excessive amount of acetate in mixed wastewater which favored the growth of Thiotrichales.50
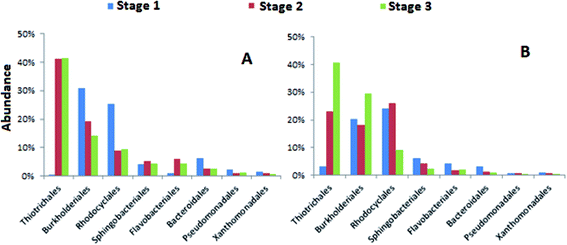 | ||
| Fig. 5 Relative abundances of bacteria orders from the anaerobic reactors during each stage of the (A) modified EBPR and (B) denitrifying EBPR processes. | ||
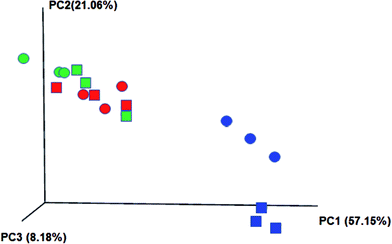
Additionally, a jackknifed PCoA plot for each sample was used to indicate the genetic differences (distances) between microorganisms (Fig. 6). It reveals that the influent water composition played an important role in the microbial community structure as the microbial communities in the same stage were clustered together. Microbial samples in stages 2 and 3 have closer genetic distance due to more similar composition of the synthetic wastewater. The larger discrepancy between two systems was discovered in stages 1 and 3, indicating that those two systems have different response to the high ammonia and P concentration in the influent.
The performance of the two EBPR systems was assessed via P removal, thus, assessment of PAO and GAO abundances is helpful in the understanding of the removal processes. Candidatus accumulibacter phosphatis have been the most frequently reported PAOs in the activated sludge-type systems at 6–22% of total abundances.3,51,52 In this study, the abundance of Candidatus accumulibacter PAOs was less than 0.2% (Table 4) which is much lower than the average reported PAO. Given the high P removal was observed in the study, it suggested that other types of PAOs played a more important role. Previously, it was reported that DPAOs capable of performing simultaneous denitrification and anoxic P uptake with advantageous in saving aeration, reducing the demand for external carbon sources and minimizing sludge yield.53 Thus, a part of phosphorous removal in the anaerobic tank may be executed by Rhodocyclus-related PAOs (denitrifying PAOs) such as Dechloromonas which reported previously.54 It was noticed that Dechloromonas was the most dominant PAO for both systems, especially during stage 1 (Table 4). Dechloromonas comprised up to 8.1% of the OTU's in anaerobic tank of denitrifying EBPR (stage 1) (Table 4). Dechloromonas-related bacteria have been detected in several EBPR processes and have been shown to accumulate PHA and polyphosphate.55–57 The higher P removal in the denitrifying EBPR compared with modified EBPR in stages 1 and 2 may be attributed to the higher percent abundances of Dechloromonas PAOs (Table 4). Specifically, the Dechloromonas denitrificans related DPAO were detected with higher abundance in the denitrifying EBPR (Table S8†). While the alternating anoxic and anaerobic conditions provided a suitable environment for DPAOs in the denitrifying EBPR, the anaerobic tank in the modified EBPR could also work as simultaneous anoxic and anaerobic conditions with both nitrate removal and P release occurring in the same tank.
| Stage | Modified | Denitrifying | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Contact | Stabilization | Anaerobic | Overall | Anoxic | Aerobic | Anaerobic | Overall | |||
| PAOs | Candidatus accumulibacter | 1 | 0.038 | 0.169 | 0.113 | 0.114 | 0.049 | 0.015 | 0.056 | 0.040 |
| 2 | 0.004 | 0.027 | 0.000 | 0.007 | 0.035 | 0.056 | 0.024 | 0.038 | ||
| 3 | 0.029 | 0.043 | 0.039 | 0.037 | 0.081 | 0.035 | 0.032 | 0.045 | ||
| Dechloromonas | 1 | 6.748 | 5.045 | 6.831 | 5.921 | 6.220 | 7.027 | 8.102 | 7.145 | |
| 2 | 0.663 | 2.423 | 2.199 | 1.642 | 4.654 | 2.773 | 5.660 | 4.429 | ||
| 3 | 1.684 | 2.758 | 3.719 | 2.812 | 3.803 | 1.689 | 1.937 | 2.352 | ||
| GAOs | Defluviicoccus | 1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| 3 | 0.012 | 0.005 | 0.004 | 0.007 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| Sphingomonas | 1 | 1.522 | 0.000 | 1.250 | 0.731 | 0.773 | 1.972 | 0.000 | 0.903 | |
| 2 | 0.407 | 0.000 | 0.000 | 0.160 | 0.000 | 0.000 | 0.000 | 0.000 | ||
| 3 | 0.012 | 0.567 | 0.204 | 0.251 | 0.000 | 0.000 | 0.106 | 0.052 | ||
The GAOs are known to compete with PAOs due to its uptake of VFA under anaerobic conditions, but no accumulation of polyphosphate under aerobic conditions.58 Therefore, the presence of GAOs in the system may interfere with the accumulation of polyphosphate by PAOs, thereby impacting the general performance of the EBPR activity.59 Candidatus competibacter phosphatis and Alphaproteobacteria GAOs are two major groups of GAOs identified in EBPRs.3 Surprisingly, no Candidatus competibacter phosphatis were detected in both systems. However, two Alphaproteobacteria GAOs including Defluviicoccus and Sphingomonas related GAOs were detected in some tanks (Table 4). Wong et al. found the deteriorated P removal performance in an acetate-fed anaerobic–aerobic membrane bioreactor was associated with Alphaproteobacteria GAOs.60 In this study, Defluviicoccus were only detected in the stage 3 of the modified EBPR, suggesting that high P shock loading may stimulate its growth. Compared with low abundance of Defluviicoccus in both systems, much more Sphingomonas were detected in the stages 1 and 3 (Table 4). Again, the increase of Sphingomonas ratio in the stage 3 may be attributed to P shocking loading. Previous study showed that long-term operation of the high phosphorus influent inhibited the proliferation of PAOs, and more importantly provided GAOs a competitive advantage over PAOs.33 The overall lower abundance of GAOs in the denitrifying EBPR than modified EBPR suggested the effectiveness of anoxic tank as a metabolic selector to inhibit the growth of GAOs.
4 Conclusions
The results showed that the high ammonia concentration in the influent could lead to the accumulation of nitrate in the anaerobic tank, which could impact the P release thereby the overall P removal. Compared with modified side-stream EBPR, the denitrifying sidestream EBPR has showed more advantages in P removal especially under high ammonia and P loading. Dechloromonas related PAOs were dominant in both systems in this study with higher abundance in denitrifying EBPR. The inclusion of an anoxic tank also resulted in reduced abundance of dominating GAOs (i.e. Defluviicoccus and Sphingomonas), which resulted in the improved P removal.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support for this project provided by House and Building National Research Centre (HBNRC), Egypt towards the inception of the project, the research activities carried out in Egypt and travel support for group meetings and interaction in Canada. The authors also acknowledge the financial support provided through an NSERC Discovery Grant (Mohamed Gamal El-Din) to support the research activities carried out in Canada.References
- Metcalf & Eddy, Inc., G. Tchobanoglous, H. D. Stensel, R. Tsuchihashi and F. Burton, Wastewater Engineering: Treatment and Resource Recovery, McGraw-Hill Education, 2014 Search PubMed.
- R. J. Seviour, T. Mino and M. Onuki, FEMS Microbiol. Rev., 2003, 27, 99–127 CrossRef CAS PubMed.
- A. Oehmen, P. C. Lemos, G. Carvalho, Z. Yuan, J. Keller, L. L. Blackall and M. A. Reis, Water Res., 2007, 41, 2271–2300 CrossRef CAS PubMed.
- S. Williams, Environ. Technol., 1999, 20, 743–747 CrossRef CAS.
- E. M. Rashed, M. M. El-Shafei, M. A. Heikal and A. M. Noureldin, HBRC Journal, 2014, 10, 92–99 CrossRef.
- N. Kishida, J. Kim, S. Tsuneda and R. Sudo, Water Res., 2006, 40, 2303–2310 CrossRef CAS PubMed.
- D. S. Lee, C. O. Jeon and J. M. Park, Water Res., 2001, 35, 3968–3976 CrossRef CAS PubMed.
- Z.-r. Hu, M. C. Wentzel and G. A. Ekama, Water Res., 2002, 36, 4927–4937 CrossRef CAS PubMed.
- T. Saito, D. Brdjanovic and M. Van Loosdrecht, Water Res., 2004, 38, 3760–3768 CrossRef CAS PubMed.
- Y. Jiang, B. Wang, L. Wang, J. Chen and S. He, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2006, 41, 2557–2570 CrossRef CAS PubMed.
- W. Zeng, A. Wang, J. Zhang, L. Zhang and Y. Peng, Chem. Eng. J., 2016, 296, 234–242 CrossRef CAS.
- M. Beer, H. M. Stratton, P. Griffiths and R. Seviour, J. Appl. Microbiol., 2006, 100, 233–243 CrossRef CAS PubMed.
- R. J. Martinez, C. H. Wu, M. J. Beazley, G. L. Andersen, M. E. Conrad, T. C. Hazen, M. Taillefert and P. A. Sobecky, PloS One, 2014, 9, e100383 Search PubMed.
- D. G. Weissbrodt, G. S. Schneiter, J.-M. Fürbringer and C. Holliger, Water Res., 2013, 47, 7006–7018 CrossRef CAS PubMed.
- O. Sanchez, Bioremediation of Wastewater: Factors and Treatment, Apple Academic Press, 2015 Search PubMed.
- B. Xie, B. Liu, Y. Yi, L. Yang, D. Liang, Y. Zhu and H. Liu, Bioresour. Technol., 2016, 207, 109–117 CrossRef CAS PubMed.
- Y. H. Ong, A. S. M. Chua, T. Fukushima, G. C. Ngoh, T. Shoji and A. Michinaka, Water Res., 2014, 64, 102–112 CrossRef CAS PubMed.
- P. H. Nielsen, A. T. Mielczarek, C. Kragelund, J. L. Nielsen, A. M. Saunders, Y. Kong, A. A. Hansen and J. Vollertsen, Water Res., 2010, 44, 5070–5088 CrossRef CAS PubMed.
- F. L. Burton, H. D. Stensel and G. Tchobanoglous, Wastewater Engineering: Treatment and Resource Recovery, McGraw-Hill, 2014 Search PubMed.
- American Public Health Association, Standard Methods for the Examination of Water and Wastewater, American Public Health Association, Washington, DC, 1998 Search PubMed.
- S. He, D. L. Gall and K. D. McMahon, Appl. Environ. Microbiol., 2007, 73, 5865–5874 CrossRef CAS PubMed.
- T. J. Mincer, M. J. Church, L. T. Taylor, C. Preston, D. M. Karl and E. F. DeLong, Environ. Microbiol., 2007, 9, 1162–1175 CrossRef CAS PubMed.
- J. M. Kim, H. J. Lee, S. Y. Kim, J. J. Song, W. Park and C. O. Jeon, Appl. Environ. Microbiol., 2010, 76, 3825–3835 CrossRef CAS PubMed.
- Y. M. Kim, D. S. Lee, C. Park, D. Park and J. M. Park, Water Res., 2011, 45, 1267–1279 CrossRef CAS PubMed.
- Y. Higashioka, H. Kojima, S. Sato and M. Fukui, J. Appl. Microbiol., 2009, 107, 126–135 CrossRef CAS PubMed.
- D. A. MacIntyre, M. Chandiramani, Y. S. Lee, L. Kindinger, A. Smith, N. Angelopoulos, B. Lehne, S. Arulkumaran, R. Brown, T. G. Teoh, E. Holmes, J. K. Nicoholson, J. R. Marchesi and P. R. Bennett, Sci. Rep., 2015, 5(8988), 1–9 Search PubMed.
- T. Müller, B. Walter, A. Wirtz and A. Burkovski, Curr. Microbiol., 2006, 52, 400–406 CrossRef PubMed.
- Y. Liu and J.-H. Tay, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2001, 36, 1027–1040 CrossRef CAS.
- S. V. Mohan, V. L. Babu, Y. V. Bhaskar and P. Sarma, Bioresour. Technol., 2007, 98, 1373–1379 CrossRef CAS PubMed.
- R. Klees and J. Silverstein, Water Sci. Technol., 1992, 26, 545–553 CrossRef CAS.
- Z. Wang, Y. Meng, T. Fan, Y. Du, J. Tang and S. Fan, Bioresour. Technol., 2015, 179, 585–594 CrossRef CAS PubMed.
- H. S. Shin, M. G. Park and J. Y. Jung, Environ. Technol., 2001, 22, 889–895 CrossRef CAS PubMed.
- C.-H. Hsu, W.-C. Chang, J.-J. Chen and J.-S. Wu, Desalin. Water Treat., 2012, 47, 42–49 CrossRef CAS.
- W. Jia, J. Zhang, H. Xie, Y. Yan, J. Wang, Y. Zhao and X. Xu, Bioresour. Technol., 2012, 113, 232–238 CrossRef CAS PubMed.
- R. Lemaire, R. Meyer, A. Taske, G. R. Crocetti, J. Keller and Z. Yuan, J. Biotechnol., 2006, 122, 62–72 CrossRef CAS PubMed.
- K. D. McMahon, S. Yilmaz, S. He, D. L. Gall, D. Jenkins and J. D. Keasling, Appl. Microbiol. Biotechnol., 2007, 77, 167–173 CrossRef CAS PubMed.
- J. J. Flowers, S. He, S. Yilmaz, D. R. Noguera and K. D. McMahon, Environ. Microbiol. Rep., 2009, 1, 583–588 CrossRef CAS PubMed.
- I. M. Head, W. D. Hiorns, T. M. Embley, A. J. McCarthy and J. R. Saunders, Microbiology, 1993, 139, 1147–1153 CrossRef CAS PubMed.
- D. Abd El Haleem, F. Von Wintzingerode, A. Moter, H. Moawad and U. Göbel, Lett. Appl. Microbiol., 2000, 31, 34–38 CrossRef CAS PubMed.
- K. Kersters, P. De Vos, M. Gillis, J. Swings, P. Vandamme and E. Stackebrandt, in The prokaryotes, Springer, 2006, pp. 3–37 Search PubMed.
- M. Bauer, M. Kube, H. Teeling, M. Richter, T. Lombardot, E. Allers, C. A. Würdemann, C. Quast, H. Kuhl and F. Knaust, Environ. Microbiol., 2006, 8, 2201–2213 CrossRef CAS PubMed.
- S. Leschine, B. J. Paster and E. Canale-Parola, in The Prokaryotes, Springer, 2006, pp. 195–210 Search PubMed.
- D. Pérez-Pantoja, R. Donoso, L. Agulló, M. Córdova, M. Seeger, D. H. Pieper and B. González, Environ. Microbiol., 2012, 14, 1091–1117 CrossRef PubMed.
- M. Hesselsoe, S. Füreder, M. Schloter, L. Bodrossy, N. Iversen, P. Roslev, P. H. Nielsen, M. Wagner and A. Loy, ISME J., 2009, 3, 1349–1364 CrossRef CAS PubMed.
- C. C. Silva, H. Hayden, T. Sawbridge, P. Mele, S. O. De Paula, L. C. Silva, P. M. Vidigal, R. Vicentini, M. P. Sousa and A. P. R. Torres, PloS one, 2013, 8, e61811 CAS.
- N. S. Abbai and B. Pillay, Mol. Biotechnol., 2013, 54, 900–912 CrossRef CAS PubMed.
- Y. Kong, J. L. Nielsen and P. H. Nielsen, Applied and Environ. Microbiol., 2004, 70, 5383–5390 CrossRef CAS PubMed.
- A. Miłobędzka and A. Muszyński, Water Sci. Technol., 2016, 71, 675–684 CrossRef PubMed.
- P. Madoni, D. Davoli and G. Gibin, Water Res., 2000, 34, 1767–1772 CrossRef CAS.
- A. M. Martins, K. Pagilla, J. J. Heijnen and M. C. van Loosdrecht, Water Res., 2004, 38, 793–817 CrossRef CAS PubMed.
- T. Fukushima, N. Uda, M. Okamoto, M. Onuki, H. Satoh and T. Mino, Microb. Environ., 2007, 22, 346–354 CrossRef.
- J. L. Zilles, J. Peccia, M.-W. Kim, C.-H. Hung and D. R. Noguera, Appl. Environ. Microbiol., 2002, 68, 2763–2769 CrossRef CAS PubMed.
- Y.-K. Wang, X.-R. Pan, Y.-K. Geng and G.-P. Sheng, Sci. Rep., 2015, 5, 1–8 CAS.
- D. Xu, H. Chen, X. Li, Q. Yang, T. Zeng, K. Luo and G. Zeng, Bioresour. Technol., 2013, 143, 204–211 CrossRef CAS PubMed.
- Y. Kong, Y. Xia, J. L. Nielsen and P. H. Nielsen, Microbiology, 2007, 153, 4061–4073 CrossRef CAS PubMed.
- J. M. Kim, H. J. Lee, D. S. Lee and C. O. Jeon, Applied and Environ. Microbiol., 2013, 79, 1969–1979 CrossRef CAS PubMed.
- X. M. Lv, M. F. Shao, C. L. Li, J. Li, X. L. Gao and F. Y. Sun, Microb. Environ., 2014, 29, 261–268 CrossRef PubMed.
- T. Mino, W.-T. Liu, F. Kurisu and T. Matsuo, Water Sci. Technol., 1995, 31, 25–34 CAS.
- J. I. Kamika, M. Coetzee, B. B. Mamba, T. Msagati and M. N. B. Momba, Int. J. Environ. Res. Public Health, 2014, 2876–2898 CrossRef PubMed.
- M.-T. Wong, F. M. Tan, W. J. Ng and W.-T. Liu, Microbiology, 2004, 150, 3741–3748 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra07610j |
| This journal is © The Royal Society of Chemistry 2017 |