Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mn3O4 nanoparticles cause endoplasmic reticulum stress-dependent toxicity to Saccharomyces cerevisiae
Xiao Yi,
Weili Zhao,
Jianrong Li,
Bing Zhang,
Qilin Yu * and
Mingchun Li*
* and
Mingchun Li*
Key Laboratory of Molecular Microbiology and Technology, Ministry of Education, College of Life Science, Nankai University, Tianjin, 300071, P. R. China. E-mail: yuqilin033@163.com; nklimingchun@163.com; Tel: +86-022-23508506
First published on 28th September 2017
Abstract
Mn3O4 nanoparticles (NPs) are a significant nanomaterial (NM) due to their excellent physiochemical properties. However, little is known about their biological effects. In this study, we investigated the effect of the synthesized Mn3O4 nanoparticles (NPs) (with the size of 10–25 nm) on the important fungus model, Saccharomyces cerevisiae. Growth inhibition assays showed that Mn3O4 NPs had dose-dependent toxicity to Saccharomyces cerevisiae (IC50 = 340 ppm). The plasma membrane (PM) was not damaged by the NPs, and the addition of ROS scavengers could not attenuate growth inhibition of the NPs to yeast cells, ruling out the contribution of PM damage and oxidative stress to this toxicity. Interestingly, Mn3O4 NPs caused HAC1 mRNA splicing and remarkable up-regulation of the unfolded protein response (UPR) genes, indicating that the NPs induce severe endoplasmic reticulum (ER) stress. Moreover, treatment of the NPs severely reduced the activity of both extracellular invertase and surface ferric reductase, which might be attributable to ER stress-related disruption of the secretion pathway. This study uncovers a novel toxicity mechanism of Mn3O4 NPs against eukaryotic cells, and provides useful information for assessing the environmental impact of NMs.
1. Introduction
Oxides of manganese are suitable for super-capacitor applications due to their natural abundance, environmentally friendly nature, and the ability of manganese to exist in various states.1 The various oxidation states of manganese result in the formation of MnO2, Mn2O3, and Mn3O4.1 Mn3O4 nanoparticles (NPs) are one of the most important nanomaterials (NMs) due to their superior electrochemical properties. Besides their application in super-capacitors, they are also used in various fields, such as paints, rubbers, sensors and computer components.2 Moreover, Mn3O4 NPs have been implemented as catalysts for the oxidation of methane and carbon monoxide (CO)3 and for the abatement of volatile organic compounds and waste gases.4 Therefore, the synthesis and application of Mn3O4 NPs has become a hot spot in today's research.With the development of nanotechnology and abundant NMs being incorporated into ecosystems, it is essential and urgent to understand the potential biological impact of NMs.5 Because of their small sizes and large surface energy, NMs have much higher biological activity than bulk materials,6 they may easily enter into cells through free penetration or receptor-mediated endocytosis, and actively interact with intracellular components.7 These interactions may lead to dysfunction of protein functions, damage of DNA, interference of signalling pathways and especially accumulation of reactive oxygen species (ROS).8,9 Similar to many other NMs, Mn3O4 NPs have been demonstrated to be toxic to mammalian cells, which is associated with ROS production. Although the toxicity of Mn3O4 NPs has been revealed, the detailed toxicity mechanisms of Mn3O4 NPs remain to be investigated.
The ER, as a vital organelle, plays important roles in protein processing and modification, lipid synthesis and signalling communication.10 Owing to its significance in various cellular processes, dysfunction of the ER induces growth inhibition and even apoptosis. ER function may be disturbed by inhibition of protein glycosylation, reduction of disulfide bond formation, calcium depletion from the ER lumen, impairment of protein transport from the ER to the Golgi apparatus, and expression of misfolded proteins.11 Such dysfunction leads to ER stress.12 ER dysfunction contributes to abundant acute and chronic diseases, such as neurodegenerative diseases, diabetes and Alzheimer's disease.13 To alleviate this stress, a conserved signalling pathway, termed the unfolded protein response (UPR), was activated.14 Up to now, little is known about the contribution of ER dysfunction to the toxicity mechanism of nanomaterials.
Saccharomyces cerevisiae is a promising unicellular eukaryotic model organism for evaluating the toxicological effect of nanoparticles, as the cellular structure and functional organization of this fungus share many similarities with those of mammalian organisms.15 In this study, we for the first time investigated the potential effect of the synthesized Mn3O4 nanoparticles on this representative fungal organism, and explored possible toxicity mechanisms of these nanomaterials in the fungal cells. Interestingly, we found that ER stress, rather than the well-known plasma membrane (PM) damage and ROS accumulation, contributed to the toxicity of Mn3O4 nanoparticles.
2. Materials and methods
2.1 Preparation and characterization of Mn3O4 NPs
The Mn3O4 NPs were synthesized according to Liu's method.16 The general morphology of the products was characterized by transmission electron microscopy (TEM, Tecnai G2 F-20, FEI, USA). The crystal structure and composition of the products were characterized by X-ray diffraction (XRD, D/max-2500, Japan).2.2 Preparation of Mn3O4 NP solutions
Mn3O4 NPs stock solution was prepared in YPD medium (yeast extract 1%, peptone 2%, glucose 2%) or SC-sucrose medium (yeast nitrogen source 0.67%, sucrose 2%, amino acid mixture 0.2%) with an initial concentration of 10, 000 ppm. Then the stock solution was sonicated for 30 min (AS3120, Auto science, China) and a series of 2-fold dilutions were prepared in the corresponding medium.2.3 Yeast strains and growth conditions
Usually, the wild-type strain InvSc1 (Invitrogen, USA) was used in each experiment. In order to detect the damage to the endoplasmic reticulum of the Mn3O4 NPs, the ER stress reporting strain WT+pJC104 which contains the plasmid PFKS2-LacZ and the hac1Δ mutant to detect its sensitivity to Mn3O4 NPs.Growth inhibition was assessed by hemocytometer counting. Overnight-cultured yeast cells were suspended in fresh YPD medium to an optical density at 600 nm (OD600) of 0.2. 1 mL of cell suspension was mixed with 1 mL of diluted Mn3O4 NPs suspension in glass tubes, and the final concentrations of the NPs were 0, 100, 400, 800 ppm. After cultured with shaking at 140 rpm and 30 °C for 12 h, the cells in each tube were counted using a hemocytometer. The percent of growth was calculated as the number of cells in each group divided by the number of cells in the control ×100. Since the IC50 of the Mn3O4 NPs is 340 ppm (as described in the Results), we used this concentration in the following experiments to explore the toxicity mechanisms of the NPs. To evaluate the effect of ROS scavengers on cell growth under Mn3O4 NP treatment, 10 mM N-acetyl cysteine (NAC), 10 mM thiourea or 10 mM vitamin C (VC) were added into the co-incubation system.
2.4 Mn2+ dissolution assays
To assess Mn2+ dissolution from Mn3O4 NPs, the nanoparticles were suspended in yeast culturing supernatant with the concentrations of 400 ppm. The mixtures were subsequently cultured at 30 °C with shaking at 140 rpm for 12 h. Then the cells were harvested, washed with deionized water several times and suspended. The suspensions were repeatedly frozen and thawed, and then were vortexed violently for 20 minutes to thoroughly release the intracellular Mn2+. After centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min to pellet Mn3O4 NPs, the supernatants were digested by 10% (v/v) HNO3, and Mn2+ contents were then determined using inductively coupled plasma (ICP-AES, Thermo Elemental, USA).
000 rpm for 10 min to pellet Mn3O4 NPs, the supernatants were digested by 10% (v/v) HNO3, and Mn2+ contents were then determined using inductively coupled plasma (ICP-AES, Thermo Elemental, USA).
2.5 PM damage assays
Propidium iodide (PI) staining was used to determine the effect of Mn3O4 NPs on PM damage. Yeast cells were co-cultured with Mn3O4 NPs for 12 h, harvested, suspended in PBS buffer, and stained with 10 μg mL−1 of PI for 5 min. The cells were then harvested and observed by a fluorescence microscope (BX-51, Olympus, Japan) with the RFP filter set. The percent of PI-positive (PM damaged) cells was calculated as the number of PI-positive cells divided by the number of total cells ×100. At least 40 fields were counted.2.6 ROS detection
Cells were treated with Mn3O4 NPs shaking at 140 rpm for 12 h. The cells were then washed and suspended in PBS. 500 mL of suspensions were stained with 10 μg mL−1 of dichlorofluorescein diacetate (DCFH-DA, dissolved in ethanol, Sigma, USA) at 30 °C for 30 min. The cells were harvested, washed twice with PBS buffer, and then observed by a fluorescence microscope (BX-51, Olympus, Japan) with the RFP filter set. The percent of ROS-positive cells was calculated as the number of ROS-positive cells divided by the number of total cells ×100. At least 40 fields were counted.2.7 ER damage detection and β-galactosidase assays
The ER stress reporting strain WT+pJC104 were treated with Mn3O4 NPs as described above. Cells were then harvested and suspended in 1 mL working Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.027% (v/v) β-mercaptoethanol, pH 7.0). 150 μL of suspensions were permeabilized with 20 μL chloroform and 50 μL SDS (0.1%, m/v) at 30 °C for 5 min, mixed with 500 μL O-nitrophenyl-b-D-galactopyranoside (ONPG, 5000 ppm, BBI, USA), and incubated at 30 °C for certain time (T). Reactions were stopped by addition of 500 mL Na2CO3 (1 M) when the mixtures turned yellow. The cell number of the suspensions was also detected by hemocytometers. Suspensions were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, and the optical density of the supernatants at 420 nm (OD420) was determined. Miller units of activity were calculated as (OD420 × 1000)/(cell number × T × 10−7).
000 rpm for 10 min, and the optical density of the supernatants at 420 nm (OD420) was determined. Miller units of activity were calculated as (OD420 × 1000)/(cell number × T × 10−7).
2.8 HAC1 mRNA splicing
A pair of primers for detection of HAC1 mRNA splicing were designed on the outside of the intron, HAC1-5rt (CCGTAGACAACAACAATTTG) and HAC1-3rt (CATGAAGTGATGAAGAAATC). HAC1 products were obtained by PCR using the cDNA template. The size of the intron-containing HAC1 fragment was 433 bp and when the UPR pathway was activated, the size of the spliced HAC1 fragment was 181 bp. The PCR products were separated by 2% agarose gel electrophoresis.2.9 Quantitative real-time PCR (qRT-PCR)
The treated cells were harvested, and total RNA was extracted from the cells as previously described. An oligo (dT)-primer RT reagent Kit (Promega, Madson, USA) was used for reverse transcriptional synthesis of cDNA, and a Trans Start Green qPCR Supermix Kit (TRANSGEN, Beijing, China) was used for qRT-PCR. Transcription levels of the genes involved in the UPR genes, including PMT4 (encoding protein mannosyl transferase), YSY6 (encoding a homolog of mammalian RAMP4 protein involved in secretion), GAA1 (encoding a subunit of the GPI: protein transamidase complex), ERD2 (encoding an HDEL receptor),17 INO1 (encoding protein inositol-3-phosphate synthase) and SAC6 (encoding protein fimbrin), were normalized against the levels of ACT1 in different samples. The primers used for the reactions are listed in Table 1. The 2−ΔΔCt method was used to calculate the relative expression compared to the control.| Primer | Sequence (5′–3′) |
|---|---|
| ACT1-5rt | TGTCTTCCCATCTATCGTCG |
| ACT1-3rt | CGACGATAGATGGGAAGACA |
| PMT4-5rt | GTTGCTGAACATTGGCTCT |
| PMT4-3rt | AAGGAACCATCATAGCCACA |
| INO1-5rt | GACTTTGTCGTCTCTGGTTG |
| INO1-3rt | AAGAGGCTTCACCAAGGAC |
| SAC6-5rt | AGCAAGCAAACTGGAACAG |
| SAC6-3rt | GTGGTTTGTAATGGTGCCTT |
| GAA1-5rt | TCGGCACATTGGCATTTCCT |
| GAA1-3rt | GCTCTATCTCATCTTTCGGC |
| ERD2-5rt | AGAACCAACACCATTGCG |
| ERD2-3rt | ACACTCTCCAACCATACAGA |
| YSY6-5rt | ACAGACACCAAGACAAAGAC |
| YSY6-3rt | CCACCTACGAGAAGAAACAG |
| FKS2-5rt | CGATTGATTGGGTGAGACG |
| FKS2-3rt | AAATCTTTGTGTCGCCTTCC |
| PRM5-5rt | TCCACACAACATACCCAGT |
| PRM5-3rt | TTTCTGTTGGATTCGGATGC |
| CHS1-5rt | CAGACCCGTAAACGATGGA |
| CHS1-3rt | CGATAGAAGACACCGAGGAT |
| CHS3-5rt | CGATTGTGGCTTTCCTGAC |
| CHS3-3rt | CTTTACCAGCATCTGACCAG |
| DFG5-5rt | AGGTTGGTTGGCAATGGTA |
| DFG5-3rt | CCGCAATCTGGAACAAACA |
| CRH1-5rt | AACCAACTACAATGACGCTC |
| CRH1-3rt | AACTGCGAAATCTTCTTGGG |
| ECM33-5rt | GTTGGTGGTGGTTTCATCAT |
| ECM33-3rt | GCACCACCTCTAACAGACT |
| SUC2-5rt | ATTGCCTGGGCTTCAAACTG |
| SUC2-3rt | GACCAGGGACCAGCATTACT |
2.10 Invertase and ferric reductases assay
Invertase activity was estimated by an invertase analysis kit (Suzhou Keming Biotechnology Co., Ltd). The activity of cell-surface ferric reductases was detected using the bathophenanthroline disulfonic acid (BPS) method.182.11 Determination of residual sucrose content in culture medium
Cells were treated with or without 400 ppm Mn3O4 NPs, shaking at 140 rpm for 12 h. The cultures were then centrifuged, and then 0.1 mL of the supernatant were mixed with 0.1 mL of 30% KOH, incubated for 10 min at 100 °C. After cooled to the room temperature, 3 mL of the anthrone reagent was added into the mixture, co-incubated at 40 °C for 10–15 min. The optical density at 620 nm (OD620) was detected. Based on the standard curve, the contents of residual sucrose in the culture medium were calculated.2.12 Statistical analysis
Each text was performed with three replicates, and the values represent the means ± standard deviation (SD) of three experiments. Significant difference between the treatments was determined using the t-test (P < 0.05). All statistical analyses were performed by Statistical Packages for the Social Sciences (SPSS, Version 20).3. Results and discussion
3.1 Characterization of the synthesized Mn3O4 NPs
Transmission electron microscope (TEM) images and XRD data of the synthesized Mn3O4 nanoparticles are shown in Fig. 1. The samples displayed nanoparticle structure with cube-like morphology and with the dimension of 10–25 nm (Fig. 1a). XRD analysis revealed that the obtained nanoparticles were indexed to the pure Mn3O4 structures (Fig. 1b).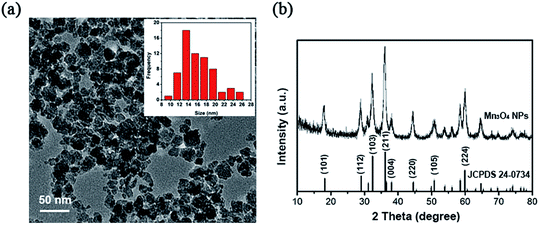 | ||
| Fig. 1 Characterization of the synthesized Mn3O4 NPs. (a) Transmission electron microscopy (TEM). (b) XRD patterns. | ||
3.2 Mn3O4 NPs show toxicity to S. cerevisiae
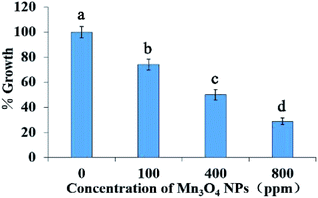
As demonstrated in Fig. 2, the NPs had inhibitory effect on yeast growth, and 340 ppm leads to approximate 50% decrease of the growth biomass (IC50 = 340 ppm). With increase of concentration of Mn3O4 NPs, the growth inhibition effect was enhanced. In the following experiments, we used 400 ppm of the NPs for exploring their toxicity mechanisms.3.3 Both PM damage and oxidative stress do not contribute to the toxicity of Mn3O4 NPs
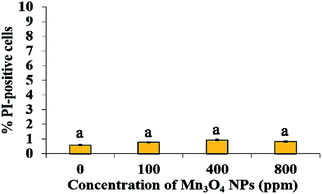
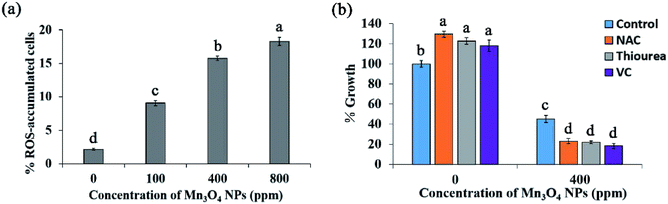
PM damage and oxidative stress are two well-known mechanisms by which NPs cause cytotoxicity.9 However, PI staining, which indicates PM damage, showed that only <1% cells were damaged by Mn3O4 NPs, even the concentrations reached to 800 ppm (Fig. 3). Hence, PM damage is not involved in the toxicity mechanism of Mn3O4 NPs.To evaluate whether ROS production and consequent oxidative stress are associated with the toxicity of Mn3O4 NPs, we measured ROS production in the Mn3O4 NP-treated yeast cells by DCFH-DA staining. Fluorescence microscopy demonstrated that the number of ROS-accumulated cells increased with the increase of the concentration of Mn3O4 NPs (Fig. 4a). The number of ROS-accumulated cells treated with 400 ppm Mn3O4 NPs is 6 to 7 times as much as control (Fig. 4a). Therefore, Mn3O4 NPs may lead to intracellular ROS accumulation. However, the addition of ROS scavengers, including NAC, thiourea and VC, could not restore the growth of yeast cells under Mn3O4 NP treatment (Fig. 4b). These results indicated that ROS accumulation caused by Mn3O4 NPs was not involved in the toxicity mechanism of these NPs.
3.4 Mn2+ dissolution only partially contribute to the toxicity
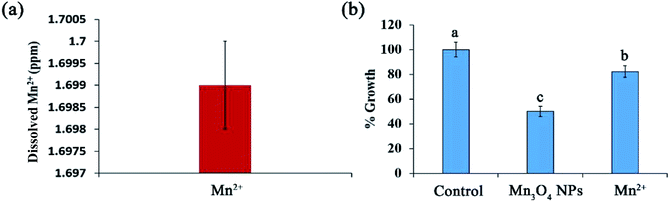
Ion dissolution is an important factor that leads to the toxicity of NMs.19 The released ions released from NPs may result in lysosomal and mitochondrial damage and ultimately cell death.20 We further determined Mn2+ dissolution from the synthesized Mn3O4 NPs. ICP assays showed that Mn2+ dissolution was enhanced with the increase of Mn3O4 NP concentrations. Approximately 1.699 ppm of Mn2+ dissolved from 400 ppm of the Mn3O4 NPs and absorbed by cells (Fig. 5a), revealing that only 0.42% of manganese released from the materials at this concentration enters the cell for toxicity.To investigate whether Mn2+ dissolution is associated with the toxicity of the NPs, we then tested the effect of the dissolved Mn2+ (1.699 ppm) on yeast growth. After 12 h of incubation, only 20% growth was inhibited by the dissolved Mn2+, while 50% growth was inhibited by Mn3O4 NPs (Fig. 5b). Therefore, Mn2+ dissolution only partially contributed to the toxicity of Mn3O4 NPs.
3.5 Mn3O4 NPs cause severe ER stress
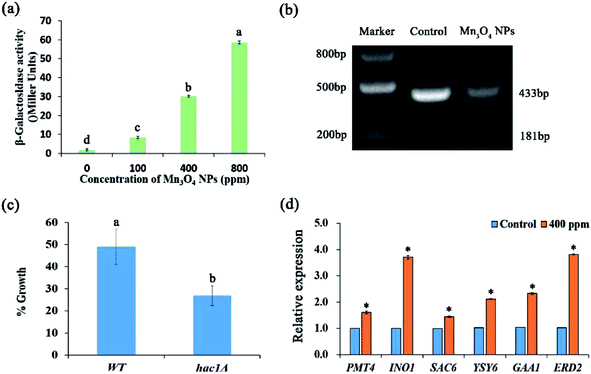
The ER plays important roles in abundant intracellular processes, such as protein synthesis, folding, modification, and transport. The protein-folding capacity of the ER is adaptable: when the capacity of the ER is exceeded and, as a result, unfolded proteins accumulate in the ER, an intracellular signalling pathway, the unfolded protein response (UPR), is induced.21 ER stress may result in dysfunction of the ER, leading to growth inhibition and apoptosis.22,23 We evaluated ER stress using the reporting strain WT+pJC104.24 β-Galactosidase assays showed that Mn3O4 NPs remarkably up-regulated the expression of the UPR promoter-governed LacZ gene, indicating that ER stress was exerted by Mn3O4 NPs (Fig. 6a).In yeast, the expression of UPR target genes is controlled by the UPR-specific transcription factor Hac1p.24 The splicing of mRNA by Ire1p is a key step in the activation of the classical UPR pathway.25 The expression of Hac1p is posttranscriptionally regulated. Dimer Irelp has endonuclease activity, splicing HAC1 mRNA introns. Haclp is a transcription factory, entering into the nucleus to activate the expression of UPR genes.26 Here, we analysed HAC1 mRNA splicing in the experimental group treated with 400 ppm Mn3O4 NPs and control group. The results showed that the size of the control group was 433 bp, with no splicing occurred. In contrast, under the treatment of NPs, HAC1 mRNA was significant spliced. This indicated the activation of ER stress with higher activity in 400 ppm NP treated strains (Fig. 6b). Moreover, we used the hac1Δ mutant to detect its sensitivity to Mn3O4 NPs. As shown in Fig. 6c, the mutant exhibited higher sensitivity to Mn3O4 NPs than the wild-type strain, suggesting the important role of UPR in tolerance to Mn3O4 NP-caused ER stress.
To confirm Mn3O4 NPs caused ER stress, we further detected expression of several UPR genes, including PMT4, INO1, SAC6, YSY6, GAA1 and ERD2. qRT-PCR analysis revealed that these UPR genes were remarkably up-regulated by Mn3O4 NP treatment (Fig. 6d). These results indicated that Mn3O4 NPs caused severe ER stress and consequent activation of the UPR pathway.
3.6 Mn3O4 NPs attenuate protein secretion
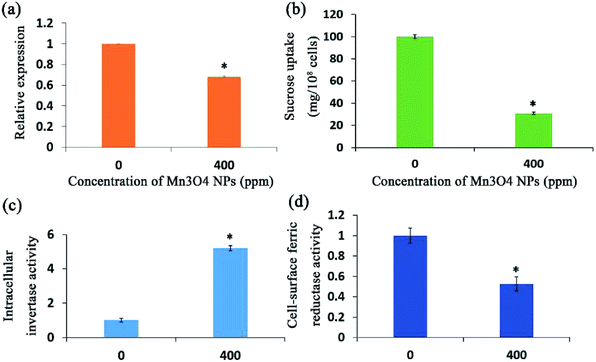
The yeast cells secrete abundant proteins from the ER to the cell surface, such as the invertase and ferric reductase, maintaining the regular absorption of essential nutrients.27 Since the ER governs protein secretion,28 we supposed that Mn3O4 NPs may have an impact on protein secretion by causing ER stress. To verify this, we first tested the activity of invertase with treatment of Mn3O4 NPs in the medium with sucrose as the sole carbon source. The yeast invertase, which is encoded by the SUC2 gene, is translated in the ER and then secreted from this organelle to the extracellular environment, functioning in utilization of extracellular sucrose.29 qRT-PCR showed that the NP-treated cells had decreased expression of SUC2 (Fig. 7a), suggesting that the NPs rendered reduction of cellular total invertase activity. However, the treated cells showed significant higher intracellular invertase activity (Fig. 7b). In addition, NP treatment led to significant decrease of sucrose uptake into the yeast cells (Fig. 7c), confirming that the NPs reduced the activity of extracellular invertase. These results indicated that the NPs severely reduced secretion of intracellular invertase.Ferric reductase is also expressed in the ER and secreted to the cell surface for mediating absorption of extracellular ferric ions. BPS staining revealed that the NP-treated cells had significant decreased activity of cell-surface ferric reductase (Fig. 7d). Hence, the NP treatment also severely reduced secretion of ferric reductase to the cell surface. Taken together, the Mn3O4 NPs attenuated protein secretion in the yeast cells.
4. Discussion
Because of the widespread applications of nanomaterials, their toxicity to the living beings, especially those exposed to emissions, is of great concern. However, as one of the main decomposers, the fungal population is usually neglected in evaluating the toxicity of nanoparticles. Fungi are the principal decomposers in the ecosystem, and play an essential role in maintaining ecological cycles and balances. In this study, we investigate the possible toxicity of Mn3O4 NPs, one kind of popular nanomaterials, on the model fungus Saccharomyces cerevisiae, and investigated the toxic mechanisms. Growth inhibition assays showed that the synthesized Mn3O4 NPs have inhibitory effect on yeast cells. Therefore, the potential hazardous effect of Mn3O4 NPs to the fungal populations should be considered when they were released into the environment.Some studies have shown that the toxicity of Mn3O4 NPs is related to ROS accumulation, mitochondrial damage, and ion dissolution.30 In many types of organisms, the mitochondria are one of the main sources of ROS, and ROS scavengers can remove free radicals reduce oxidative stress and hence attenuate the toxicity of the NPs. However, in this study, although Mn3O4 NPs caused ROS accumulation, addition of ROS scavengers, including NAC, thiourea and VC, did not restore the yeast growth under the treatment of Mn3O4 NPs. Therefore, we supposed that ROS is the by-product of Mn3O4 NPs, but is not the cause of the toxicity of the NPs.31
The ER, which functions in protein folding and assembly, lipid biosynthesis, vesicular traffic, and cellular calcium storage, is sensitive to alterations of homeostasis. Proper ER function is essential for cell survival, and perturbation of its function induces cellular damage and results in apoptosis.22 ER function can be disturbed by inhibition of protein glycosylation, a reduction in disulfide bond formation, calcium depletion from the ER lumen, impairment of protein transport from the ER to the Golgi apparatus, and the expression of misfolded proteins. Such dysfunction causes proteotoxicity in the ER, collectively termed ER stress.32 In our study, we found that the yeast strain which contain LacZ reporter plasmid pJC104 expressed high levels of β-galactosidase activity, HAC1 mRNA splicing significantly and abundant UPR genes were also up-regulated by Mn3O4 NPs. These results indicated that the Mn3O4 NPs will cause ER stress and consequent activation of the UPR pathway (Fig. 8). Since the ER function is essential for cell growth, the NP-caused ER stress may lead to decreased cell growth.
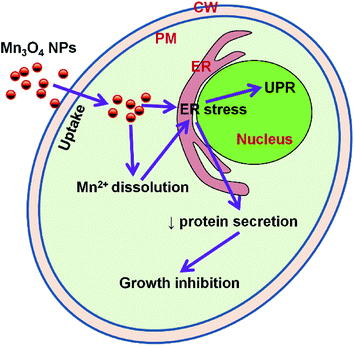 | ||
| Fig. 8 Model figure illustrating the toxicity mechanism of Mn3O4 NPs to yeast cells. ER, endoplasmic reticulum; PM, plasma membrane; CW, cell wall; UPR, unfolded protein response. | ||
One of the most important function of the ER is regulation of protein secretion. We here selected two well-known secreted proteins, invertase and ferric reductase to intestate the effect of Mn3O4 NPs on protein secretion. The extracellular invertase is responsible for the extracellular hydrolysis of sucrose33 and permits yeasts to use sucrose as a carbon and energy source. We first tested the expression of the gene and found that the expression of the gene was down-regulated. Then we examined the intracellular invertase activity. As expected, the intracellular enzyme activity increased significantly under Mn3O4 NP treatment. As the ER function is impaired, invertase secretion process is blocked, leading to invertase accumulation in the cells, followed by reduced absorption of sucrose and decreased cell growth. Ferric reductases is mainly related to the absorption of iron, Fe3+ can be reduced to cells can be used to facilitate the form of Fe2+ absorption of cells.34 In yeast cells, ferric reductases is mainly transported to the cell membrane governed by the ER. Decreased secretion of this enzyme may reduce uptake of extracellular Fe3+, which may also affect cell growth. Therefore, it can be speculated that Mn3O4 NPs cause ER stress leading to decreased protein secretion that is required for cell growth, and consequently result in decreased cell growth (Fig. 8).
5. Conclusions
This study demonstrates that Mn3O4 NPs have toxicity to the fungal model organism, Saccharomyces cerevisiae. This toxicity is not attributed to PM damage and oxidative stress, but is dependent on ER stress. This study reveals a novel toxicity mechanism of NMs in eukaryotic cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Natural Science Foundation of China (31400132, 81471923), Natural Science Foundation of Tianjin (15JCQNJC09300).References
- S. Nagamuthu, S. Vijayakumar and G. Muralidharan, Energy Fuels, 2013, 27, 3508 CrossRef CAS.
- Z.-Y. Tian, P. Mountapmbeme Kouotou, N. Bahlawane and P. H. Tchoua Ngamou, J. Phys. Chem. C, 2013, 117, 6218 CAS.
- E. Stobbe, B. De Boer and J. Geus, Catal. Today, 1999, 47, 161 CrossRef CAS.
- M. Baldi, E. Finocchio, F. Milella and G. Busca, Appl. Catal., B, 1998, 16, 43 CrossRef CAS; M. Baldi, E. Finocchio, C. Pistarino and G. Busca, Appl. Catal., A, 1998, 173, 61 CrossRef.
- R. D. Handy, F. Von der Kammer, J. R. Lead, M. Hassellöv, R. Owen and M. Crane, Ecotoxicology, 2008, 17, 287 CrossRef CAS PubMed; Y. Bai, Y. Zhang, J. Zhang, Q. Mu, W. Zhang, E. R. Butch, S. E. Snyder and B. Yan, Nat. Nanotechnol., 2010, 5, 683 CrossRef PubMed.
- L.-C. Cheng, X. Jiang, J. Wang, C. Chen and R.-S. Liu, Nanoscale, 2013, 5, 3547 RSC.
- L. Zhang, F. Gu, J. Chan, A. Wang, R. Langer and O. Farokhzad, Clin. Pharmacol. Ther. Ser., 2008, 83, 761 CrossRef CAS PubMed.
- M. S. Wason, J. Colon, S. Das, S. Seal, J. Turkson, J. Zhao and C. H. Baker, Nanomedicine, 2013, 9, 558 CrossRef CAS PubMed; E. Sanfins, J. Dairou, F. Rodrigues-Lima and J.-M. Dupret, Nanoparticle-protein interactions: from crucial plasma proteins to key enzymes, presented at Journal of Physics: Conference Series, 2011 Search PubMed.
- F. Marano, S. Hussain, F. Rodrigues-Lima, A. Baeza-Squiban and S. Boland, Arch. Toxicol., 2011, 85, 733 CrossRef CAS PubMed.
- L. Walter and G. Hajnóczky, J. Bioenerg. Biomembr., 2005, 37, 191 CrossRef CAS PubMed.
- L. Dara, C. Ji and N. Kaplowitz, Hepatology, 2011, 53, 1752 CrossRef CAS PubMed.
- R. J. Kaufman, D. Scheuner, M. Schröder, X. Shen, K. Lee, C. Y. Liu and S. M. Arnold, Nat. Rev. Mol. Cell Biol., 2002, 3, 411 CrossRef CAS PubMed; S. Oyadomari and M. Mori, Cell Death Differ., 2004, 11, 381 CrossRef PubMed.
- M. Schröder and R. J. Kaufman, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2005, 569, 29 CrossRef PubMed.
- H. P. Harding, M. Calfon, F. Urano, I. Novoa and D. Ron, Annu. Rev. Cell Dev. Biol., 2002, 18, 575 CrossRef CAS PubMed.
- K. Kasemets, S. Suppi, K. Künnis-Beres and A. Kahru, Chem. Res. Toxicol., 2013, 26, 356 CrossRef CAS PubMed; T. Nomura, J. Miyazaki, A. Miyamoto, Y. Kuriyama, H. Tokumoto and Y. Konishi, Environ. Sci. Technol., 2013, 47, 3417 CrossRef PubMed.
- M. Liu, L. Piao, L. Zhao, S. Ju, Z. Yan, T. He, C. Zhou and W. Wang, Chem. Commun., 2010, 46, 1664 RSC.
- T. T. Wimalasena, B. Enjalbert, T. Guillemette, A. Plumridge, S. Budge, Z. Yin, A. J. Brown and D. B. Archer, Fungal Genet. Biol., 2008, 45, 1235 CrossRef CAS PubMed; Q. Yu, B. Zhang, J. Li, B. Zhang, H. Wang and M. Li, Free Radicals Biol. Med., 2016, 99, 572 CrossRef PubMed.
- M. Carlson, B. C. Osmond and D. Botstein, Genetics, 1981, 98, 25 CAS.
- S. J. Klaine, P. J. Alvarez, G. E. Batley, T. F. Fernandes, R. D. Handy, D. Y. Lyon, S. Mahendra, M. J. McLaughlin and J. R. Lead, Environ. Toxicol. Chem., 2008, 27, 1825 CrossRef CAS PubMed.
- M. Pandurangan, G. Enkhtaivan, B. Venkitasamy, B. Mistry, R. Noorzai, B. Y. Jin and D. H. Kim, Biol. Trace Elem. Res., 2016, 170, 309 CrossRef CAS PubMed.
- K. J. Travers, C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman and P. Walter, Cell, 2000, 101, 249 CrossRef CAS PubMed.
- R. Chen, L. Huo, X. Shi, R. Bai, Z. Zhang, Y. Zhao, Y. Chang and C. Chen, ACS Nano, 2014, 8, 2562 CrossRef CAS PubMed.
- O. V. Tsyusko, J. M. Unrine, D. Spurgeon, E. Blalock, D. Starnes, M. Tseng, G. Joice and P. M. Bertsch, Environ. Sci. Technol., 2012, 46, 4115 CrossRef CAS PubMed.
- J. S. Cox and P. Walter, Cell, 1996, 87, 391 CrossRef CAS PubMed.
- T. Kawahara, H. Yanagi, T. Yura and K. Mori, Mol. Biol. Cell, 1997, 8, 1845 CrossRef CAS PubMed.
- U. Rüegsegger, J. H. Leber and P. Walter, Cell, 2001, 107, 103 CrossRef.
- B. Esmon, P. Novick and R. Schekman, Cell, 1981, 25, 451 CrossRef CAS PubMed; N. Xu, K. Qian, Y. Dong, Y. Chen, Q. Yu, B. Zhang, L. Xing and M. Li, Res. Microbiol., 2014, 165, 252 CrossRef PubMed; E. F. Bailão, P. S. Lima, M. G. Silvabailão, A. M. Bailão, G. R. Fernandes, D. J. Kosman and C. M. Soares, Front Microbiol., 2015, 6, 821 Search PubMed; T. Roitsch, M. E. Balibrea, M. Hofmann, R. Proels and A. K. Sinha, J. Exp. Bot., 2003, 54, 513 CrossRef PubMed.
- Y. Ma and L. M. Hendershot, J. Chem. Neuroanat., 2004, 28, 51 CrossRef CAS PubMed.
- Y. Li, W. J. Fu, N. N. Liu, M. J. Tan, G. L. Liu and Z. M. Chi, J. Mol. Catal. B: Enzym., 2015, 111, 71 CrossRef CAS.
- G. Applerot, A. Lipovsky, R. Dror, N. Perkas, Y. Nitzan, R. Lubart and A. Gedanken, Adv. Funct. Mater., 2009, 19, 842 CrossRef CAS; J. Meng, Y. Ji, J. Liu, X. Cheng, H. Guo, W. Zhang, X. Wu and H. Xu, Nanotoxicology, 2014, 8, 686 CrossRef PubMed.
- A. Deniaud, E. Maillier, D. Poncet, G. Kroemer, C. Lemaire and C. Brenner, Oncogene, 2008, 27, 285 CrossRef CAS PubMed.
- R. J. Kaufman, D. Scheuner, M. Schröder, X. Shen, K. Lee, C. Y. Liu and S. M. Arnold, Nat. Rev. Mol. Cell Biol., 2002, 3, 411 CrossRef CAS PubMed; K. Mori, Cell, 2000, 101, 451 CrossRef PubMed.
- L. F. G. De and A. Sols, Biochim. Biophys. Acta, 1962, 56, 49 Search PubMed.
- Q. Yu, Y. Dong, N. Xu, K. Qian, Y. Chen, B. Zhang, L. Xing and M. Li, FEMS Yeast Res., 2014, 14, 1037 CrossRef CAS PubMed; A. T. Mckie, G. O. Latunde-Dada, S. Miret, J. A. Mcgregor, G. J. Anderson, C. D. Vulpe, J. M. Wrigglesworth and R. J. Simpson, Biochem. Soc. Trans., 2002, 30, 722 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2017 |