Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel bioemulsifier produced by Exiguobacterium sp. strain N4-1P isolated from petroleum hydrocarbon contaminated coastal sediment†
Qinhong Caia,
Baiyu Zhang *a,
Bing Chena,
Zhiwen Zhua and
Yuming Zhao
*a,
Bing Chena,
Zhiwen Zhua and
Yuming Zhao b
b
aFaculty of Engineering and Applied Science, Memorial University of Newfoundland, St. John's, Newfoundland, Canada A1B 3X5. E-mail: bzhang@mun.ca; Tel: +1-709-864-3301
bDepartment of Chemistry, Memorial University of Newfoundland, St. John's, Newfoundland, Canada A1B 3X5
First published on 4th September 2017
Abstract
In this study, Exiguobacterium N4-1P is reported as a bioemulsifier producer for the first time. The strain was found to be closely related to E. oxidotolerans strain T-2-2T, E. antarcticum B7T, and E. antarcticum B7T with similarities of 99.47, 98.70 and 98.63%, respectively. Its phenotypic properties such as metabolic fingerprints, membrane composition, and cell morphology were determined. Different carbon sources were used for bioemulsifier production and diesel was confirmed to stimulate the yield effectively. The produced bioemulsifier is a complex mainly composed of lipopeptides with C16:0 (32.18%) and C18:0 (40.99%) as the primary fatty acids. The produced bioemulsifier could form emulsions effectively with diverse hydrocarbons. No foams were formed during the production and applications, which would facilitate its commercialization. The bioemulsifier was stable over a wide range of salinity (0–25%), pH (2–12), and temperature (below 50 °C). Exiguobacterium N4-1P and the produced bioemulsifier fills knowledge gaps and has promising application potential in diverse fields, especially in environmental engineering.
Introduction
Emulsifiers are compounds which assemble at interfaces helping in the dispersion of droplets of one immiscible liquid within another, and preventing them from coalescing.1 Their desirable characteristics such as solubility enhancement, detergency power and emulsion stabilization have enabled their applications in various industries, such as household cleaning, food processing, pharmaceuticals, petroleum, agriculture, and textiles.2,3 They have been found to have promising applications in the environmental engineering field, e.g. serving as soil washing and bioremediation enhancement agents.4–6 However, chemically synthesized emulsifiers suffer from toxicity concerns and have hazardous environmental impacts.2 Thus, safer alternatives such as bioemulsifiers are of importance. Bioemulsifiers are surface-active molecules produced by microorganisms. They are high molecular weight polymers or lipopeptides.7 They have the advantages of lower or no toxicity, high biodegradability, and high stability at extreme salinity, pH and temperature when compared to their chemically synthetic counterparts.8 Moreover, they can be produced from diverse substrates including waste streams.9,10 Despite all these advantages, the applications of bioemulsifiers have been hindered by low yields, as well as high recovery and purification costs.11 To overcome such obstacles, research efforts on developing novel bioemulsifier producers are highly important.Diverse microorganisms including algae, bacteria and fungi have been found as bioemulsifier producers. A summary of such findings is shown in Table 1, along with their identified bioemulsifier compositions. Bacterial trains belonging to genera including Acinetobacter,12 Aeribacillus,13 Alcaligenes,14 Amycolatopsis,15 Azotobacter,16 Bacillus,17 Beijerinckia,18 Corynebacterium,19 Enterobacter,20 Geobacillus,13 Halomonas,21 Klebsiella,22 Myroides,23 Pedobacter,24 Propionibacterium,25 Pseudomonas,26 Solibacillus,27 Streptomyces,28 Variovorax,29 have been reported to be bioemulsifier producers (Table 1). To date, however, no strains belong to the genus of Exiguobacterium have been reported as bioemulsifier producers.
| Phylum | Genus | Species | Bioemulsifiers composition/structure | Ref. |
|---|---|---|---|---|
| a Not available. | ||||
| Algae | Dunaliella | D. salina | Polysaccharides | 30 |
| Porphyridium | P. cruentum | Polysaccharides | 31 | |
| Archaea | Methanobacterium | M. thermoautotrophicum | Cell-associated protein with a molecular weight greater than 5000 Da | 32 |
| Halovivax | H. sp. A21 | Sugar, protein and lipid | 33 | |
| Haloarcula | H. sp. D21 | Glycoproteins | ||
| Eubacteria | Acinetobacter | A. calcoaceticus | Emulsan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lipopolysaccharide and polysaccharide lipopolysaccharide and polysaccharide |
12 |
| A. radioresistens | Alasan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein polysaccharide (1 mDa) protein polysaccharide (1 mDa) |
34 | ||
| Aeribacillus | A. pallidus YM-1 | A complex of carbohydrates (41.1%), lipids (47.6%) and proteins (11.3%) | 35 | |
| Alcaligenes | A. faecalis | Carbohydrate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein (50.93 protein (50.93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22.05) 22.05) |
14 | |
| Amycolatopsis | A. tucumanensis DSM 45259 | Lipopolysaccharides or polysaccharides based on carbon source | 15 | |
| Azotobacter | A. chroococcum | Lipid and protein (31.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 68.7) 68.7) |
16 | |
| Bacillus | B. licheniformis | Lipopeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lichenysin (1006–1034 Da) Lichenysin (1006–1034 Da) |
36 | |
| B. subtilis | Lipopeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Surfactin (1036 Da) Surfactin (1036 Da) |
37 | ||
| B. velezensis | Lipopeptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nC14-surfactin and anteisoC15-surfactin nC14-surfactin and anteisoC15-surfactin |
17 | ||
| Beijerinckia | B. indica ss lacticogenes | Polysaccharide PS-7 | 18 | |
| Corynebacterium | C. kutscheri | Carbohydrate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lipid lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein (40 protein (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 27 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29) 29) |
19 | |
| Enterobacter | E. aerogenes | Carbohydrate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein (41.79 protein (41.79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24.59) 24.59) |
14 | |
| E. cloacae TU | Polysaccharide was found to be composed of glucose and galactose with molecular weight of 12.4 ± 0.4 kDa | 20 | ||
| Geobacillus | G. Pallidus | 271, 785 Da (carbohydrates (68.6%), lipids (22.7%) and proteins (8.7%)); 526, 369 Da (carbohydrates (41.1%), lipids (47.6%) and proteins (11.3%)) | 13 | |
| Halomonas | H. eurihalina | Sulfated heteropolysaccharide | 21 | |
| Klebsiella | K. sp. Y6-1 | Lipopeptide (1000–1500 Da); polysaccharide (3.5% protein) | 22 and 38 | |
| K. oxytoca BSF-1 | Lipopolysaccharide (1700–2000 kDa); polysaccharide fraction contained L-rhamnose, D-galactose, D-glucose, and D-glucuronic acid at a molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; fatty acid content consisted of C16:0, 3-OH C12:0 and C12:0 1; fatty acid content consisted of C16:0, 3-OH C12:0 and C12:0 |
39 | ||
| Myroides | M. sp. SM1 | Mixture of L-ornithine lipids | 23 | |
| Pedobacter | Pedobacter sp. MCC-Z | 67% of carbohydrates; 30% of lipids; 3% of proteins | 24 | |
| Propionibacterium | P. acidipropionici | Polysaccharides | 40 | |
| P. freundenreichii ss shermanii | Lipopeptide | 25 | ||
| P. jensenii | Polysaccharides | 38 | ||
| P. thoenii | Polysaccharides | 38 | ||
| Pseudomonas | P. nautica | Proteins, carbohydrates and lipids (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 63 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
26 | |
| P. fluorescens | Trehaloselipid-o-dialkyl monoglycerides-protein | 41 | ||
| Solibacillus | S. silvestris AM1 | MW of 200 kDa and containing 30 kDa monomeric subunits flagellin-like protein | 27 | |
| Streptomyces | Streptomyces sp. S1 | Proteins and carbohydrates (82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18) 18) |
42 | |
| Streptomyces sp. MC1 | Glycoprotein | 28 | ||
| Variovorax | V. Paradoxus | Polysaccharides | 29 | |
| Fungi | Aspergillus | A. Niger MYA 135 | Glycolipid | 43 |
| Candida | C. lipolytica | Liposan (83% carbohydrate and 17% protein) glycoprotein | 44 | |
| C. tropicalis | Polysaccharide | 38 | ||
| C. utilis | Carbohydrate (79–98%) | 38 | ||
| Geotrichum | G. sp. CLOA40 | N/Aa | 45 | |
| Lipomyces | L. starkeyi | Polysaccharides | 46 | |
| Pichia | P. anomala | Polysaccharides | 38 | |
| Rhodospiridium | R. diobovatum | Polysaccharides | 38 | |
| Rhodotorula | R. glutinis | Carbohydrate-protein complex | 47 | |
| R. graminis | Polysaccharides | 38 | ||
| R. rubra | Polysaccharides | 38 | ||
| Saccharomyces | S. cerevisiae | Mannoprotein (90% mannose 5–10% protein) | 48 | |
| Trichosporon | T. mycotoxinivorans CLA2 | N/A | 49 | |
| T. loubieri CLV20 | N/A | 45 | ||
| T. montevideense CLOA70 | N/A | 45 | ||
| Yarrowia | Y. lipolytica | Yansan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycoprotain complex glycoprotain complex |
50 | |
In this study, a bioemulsifier producing bacterium, Exiguobacterium sp. strain N4-1P, was isolated from petroleum hydrocarbon contaminated coastal sediment in North Atlantic Canada is reported for the first time. Its genotypic and phenotypic properties were characterized, and the composition and structure of the produced bioemulsifier was investigated. The produced bioemulsifier was also tested in terms of its emulsification ability with different hydrocarbons and the stability of the formed emulsions under different pH, temperature and salinity conditions.
Experimental
Screening and isolation
Exiguobacterium sp. strain N4-1P was isolated previously from a coastal sediment sample in the vicinity of a refinery company in Northern Atlantic Canada.51 Briefly, approximately 1 g of each sediment sample was first enriched with 50 mL of medium in 125 mL conical flasks. The medium composed of (NH4)2SO4, 10 g; NaCl, 15 g; FeSO4·7H2O, 2.8 × 10−4 g; KH2PO4, 3.4 g; K2HPO4·3H2O, 4.4 g; MgSO4·7H2O, 1.02 g; yeast extract, 0.5 g and trace element solution, 0.5 mL L−1 of distilled water, with 2% (v/v) n-hexadecane as the sole carbon source. The trace element solution contained ZnSO4, 0.29 g; CaCl2, 0.24 g; CuSO4, 0.25 g; MnSO4, 0.17 g L−1 and was sterilized separately. The enrichment was shaken at 200 rpm at 30 °C for 3–5 days until observable turbidity occurred. Subsequently, the consortia were serial diluted up to 108 times, spread on agar plates with the enrichment medium and incubated at 30 °C for 3–5 days. The emerged colonies were tested with a modified drop collapsing test to screen bacterial colonies with surface activities.51 As a result, the Exiguobacterium sp. strain N4-1P was isolated along with 54 other isolates.Ribosomal DNA sequencing and phylogenetic analysis
The purified isolate was then subjected to 16S ribosomal DNA (rDNA) sequencing using universal bacterial primers F27 and R926 (position in Escherichia coli 8–27 and 926–907, respectively). An aliquot of each culture was used as DNA template in a polymerase chain reaction (PCR) using the primer pair. After gel electrophoresis confirmation of successful PCR reaction, the PCR products were subjected to a clean-up process and were measured by a NanoDrop spectrophotometer to determine the concentrations. Lastly, sequencing reactions with the last PCR products were conducted and measured with Applied Biosystems 3130 and/or 3730 systems in Core Research and Instrument Training Network (CREAIT) at Memorial University. The obtained DNA sequence was matched with Basic Local Alignment Search Tool (BLAST) database. Phylogenetic trees and distances were calculated using the software package MEGA (Molecular Evolutionary Genetics Analysis) version 5.1, after alignment of sequences with CLUSTALX. Distances were calculated using the Kimura two-parameter model.52 Trees were reconstructed using the neighbor-joining method53 with the use of ‘default settings’ and the bootstrap values were calculated based on 1000 replications.Bacterial phenotype characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to each tube. Then samples were rotated end-over-end for 10 min. After removing the aqueous (lower) phase, 3 mL of aqueous 1.2% NaOH (Reagent 4) was added and the tubes were again rotated for 5 min (Sasser, 1990). Finally, the organic (upper) phase containing FAMEs was transferred to a gas chromatography (GC) vial. The FAMEs were quantified by a GC/mass spectrometry (GC/MS) system (Agilent Technologies 7890A GC connected to an Agilent technologies 5975C inert mass spectroscopy detector) in the Northern Region Persistent Organic Pollution Control (NRPOP) laboratory. Separation of FAMEs was conducted using an Agilent BD-5MS fused-silica capillary column. The column temperature was programmed from 50 °C to 120 °C at a rate of 10 °C min−1, and then to 280 °C at a rate of 3 °C min−1. Individual compounds were identified from their mass spectra and from comparing their retention times with the standard spectra. The concentration of each individual compound was determined based on the GC/MS response relative to that of the known quantitative standards. The test was conducted in duplicate.
1, v/v) to each tube. Then samples were rotated end-over-end for 10 min. After removing the aqueous (lower) phase, 3 mL of aqueous 1.2% NaOH (Reagent 4) was added and the tubes were again rotated for 5 min (Sasser, 1990). Finally, the organic (upper) phase containing FAMEs was transferred to a gas chromatography (GC) vial. The FAMEs were quantified by a GC/mass spectrometry (GC/MS) system (Agilent Technologies 7890A GC connected to an Agilent technologies 5975C inert mass spectroscopy detector) in the Northern Region Persistent Organic Pollution Control (NRPOP) laboratory. Separation of FAMEs was conducted using an Agilent BD-5MS fused-silica capillary column. The column temperature was programmed from 50 °C to 120 °C at a rate of 10 °C min−1, and then to 280 °C at a rate of 3 °C min−1. Individual compounds were identified from their mass spectra and from comparing their retention times with the standard spectra. The concentration of each individual compound was determined based on the GC/MS response relative to that of the known quantitative standards. The test was conducted in duplicate.Bioemulsifier production with different carbon sources
A serial of production media composed of essentially the enrichment medium and four different carbon sources were tested, respectively, and compared. Three different hydrocarbon sources were applied, namely, hexadecane, diesel and benzoate. The fourth carbon source was a non-hydrocarbon one which was a mixture of 0.05% starch, 0.02% glucose, 0.02% sucrose and 0.1% citrate salt. After 7 days of incubation, the culture broths were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min to remove cells and precipitates. The surface tension and emulsification index (E24) of the culture broth were tested.4 The clear broths were concentrated to approximately 1/5 of the volume with lyophilisation, and then were mixed with three volumes of cold acetone. Subsequently, the solution was stored at 4 °C for 3 days to fully precipitate the crude bioemulsifier products. The supernatant was removed by centrifugation, and the pellet was washed twice with acetone to remove the hydrocarbon residues. Finally, the pellet was finely ground and dried first in a fume hood, and then in dissectors. When constant weight was obtained for each sample, the yield was then determined. The medium with the highest yield, E24 and lowest surface tension in the broth was selected for further characterization.
000 rpm for 5 min to remove cells and precipitates. The surface tension and emulsification index (E24) of the culture broth were tested.4 The clear broths were concentrated to approximately 1/5 of the volume with lyophilisation, and then were mixed with three volumes of cold acetone. Subsequently, the solution was stored at 4 °C for 3 days to fully precipitate the crude bioemulsifier products. The supernatant was removed by centrifugation, and the pellet was washed twice with acetone to remove the hydrocarbon residues. Finally, the pellet was finely ground and dried first in a fume hood, and then in dissectors. When constant weight was obtained for each sample, the yield was then determined. The medium with the highest yield, E24 and lowest surface tension in the broth was selected for further characterization.
Composition analysis of the bioemulsifier
The crude bioemulsifier was subjected to a dialysis process with 1 kDa MW cut dialysis tubes to remove salts and small compounds. The crude bioemulsifiers before and after dialysis were subjected to protein content determination,55 total lipid content determination56 and total carbohydrate analysis.57Further analysis of the fatty acid composition of the lipid content was conducted. The lipid content was extracted according to.58 The extracted samples were homogenized in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of ice-cold chloroform
1 mixture of ice-cold chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol with a Polytron PCU-2-110 homogenizer (Brinkmann Instruments, Rexdale, Ontario, Canada). The chloroform extracted aqueous layer was added to the sample to bring the ratio of chloroform
methanol with a Polytron PCU-2-110 homogenizer (Brinkmann Instruments, Rexdale, Ontario, Canada). The chloroform extracted aqueous layer was added to the sample to bring the ratio of chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water to 8
water to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. The sample was sonicated for 4 to 10 minutes in an ice bath and centrifuged at 5000 rpm for two minutes. The bottom organic layer was removed. Chloroform was then added back to the extraction test tube and the entire procedure was repeated 3 more times. All liquids located in the organic layers were pooled into a lipid-free vial. The sample was concentrated using a flash-evaporator (Buchler Instruments, Fort Lee, N.J.). The lipid extract was trans-esterified using methanol/sulfuric acid for 1 hour at 100 °C. The FAMEs were analyzed on a HP 6890 GC/Flame Ionized Detector (GC/FID) equipped with a 7683 Autosampler. The column temperature began at 65 °C and was held at this temperature for 0.5 minutes. The temperature was then ramped to 195 °C at a rate of 40 °C min−1, held for 15 minutes, and finally ramped to a temperature of 220 °C at a rate of 2 °C min−1. This final temperature was held for 0.75 minutes. Peaks were identified using retention times from standards purchased from Supelco, namely 37 component FAME mix, Bacterial acid methyl ester mix, polyunsaturated fatty acids (PUFA) 1 and PUFA 3. The tests were conducted in triplicate.
3. The sample was sonicated for 4 to 10 minutes in an ice bath and centrifuged at 5000 rpm for two minutes. The bottom organic layer was removed. Chloroform was then added back to the extraction test tube and the entire procedure was repeated 3 more times. All liquids located in the organic layers were pooled into a lipid-free vial. The sample was concentrated using a flash-evaporator (Buchler Instruments, Fort Lee, N.J.). The lipid extract was trans-esterified using methanol/sulfuric acid for 1 hour at 100 °C. The FAMEs were analyzed on a HP 6890 GC/Flame Ionized Detector (GC/FID) equipped with a 7683 Autosampler. The column temperature began at 65 °C and was held at this temperature for 0.5 minutes. The temperature was then ramped to 195 °C at a rate of 40 °C min−1, held for 15 minutes, and finally ramped to a temperature of 220 °C at a rate of 2 °C min−1. This final temperature was held for 0.75 minutes. Peaks were identified using retention times from standards purchased from Supelco, namely 37 component FAME mix, Bacterial acid methyl ester mix, polyunsaturated fatty acids (PUFA) 1 and PUFA 3. The tests were conducted in triplicate.
Fourier transform infrared (FT-IR) spectroscopy analysis of the bioemulsifier was conducted on a Bruker Alpha with KBr disc. A ninhydrin-based assay was applied to quantify amino acids and peptides using tryptophan as the standard.59
Emulsification ability of the bioemulsifier
The emulsification index (E24) was used for performance evaluation. The bioemulsifier solutions were mixed with equal amount of diesel and vortexed for 2 min. After settling for 24 hours, E24 was determined by the fraction height of the emulsion layer out of the total height of the mixture (Cai et al., 2015). The emulsification abilities of the bioemulsifier solution, against different hydrocarbons including hexadecane, mineral oil, crude oil, diesel, xylene, toluene, dodecane, and iso-octane were tested. The effects of concentrations of bioemulsifiers on the emulsification ability were also measured. The emulsification stability under different salinity (levels of 0, 2.5, 5, 7.5, 10, 12.5, 15, 17.5, 20 and 25%), temperature (0, 4, 30, 50, 70 and 100 °C), and pH conditions (2, 4, 6, 8, 10 and 12) were tested. The tests were duplicated.Results and discussion
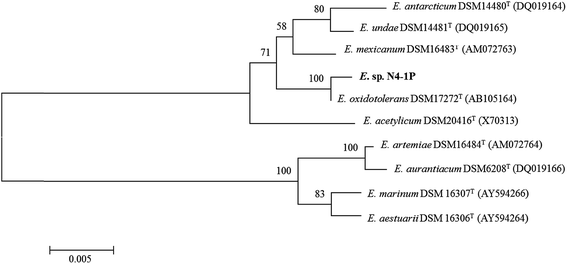
Ribosomal DNA sequencing and phylogenetic analysis
The PCR reaction obtained almost full length (1500 bp) 16S rDNA fragments for the isolate. The five replicates of the sequencing results agreed well with one another. Exiguobacterium sp. strain N4-1P (GenBank accession number: KX714225) was closely related to E. oxidotolerans strain T-2-2T with 99.47% similarity; to E. antarcticum B7T with 98.70% similarity; and to E. sibiricum 255-15T with 98.63% similarity. The phylogenetic tree of Exiguobacterium sp. strain N4-1P and the closely-related type strains are shown in Fig. 1. Exiguobacterium species were isolated from diverse habitats over a wide temperature range (−12 to 55 °C), such as glacial ice, hot springs, plant rhizospheres, Siberian permafrost, tropical soils, and temperate soil.60 Exiguobacterium strains possess interesting properties such as thermal acclimation proteins and enzymes which have proven potential for industrial applications in food processing, environmental remediation, and in the pharmaceutical manufacturing.60 No Exiguobacterium strains with emulsifying abilities have been reported so far.As shown in Fig. 2, bioemulsifier producers have been found in both Eubacteria and Archaea in the division of Proteobacteria, Firmicutes, Actinobacteria, Cytophaga–Flexibacter–Bacteroides, and Euryarchaeota. The 16S rDNA sequences of Exiguobacterium sp. strain N4-1P and the published producers with the nearly full length 16S rDNA genebank deposits were clustered and aligned. Exiguobacterium sp. strain N4-1P isolated in this study belongs to the Firmicutes divisions. Bioemulsifiers are exceptional biosurfactants with strong emulsion stabilizing ability. Reports on the discovery and characterization of new biodemulsifiers are still limited in literature.61
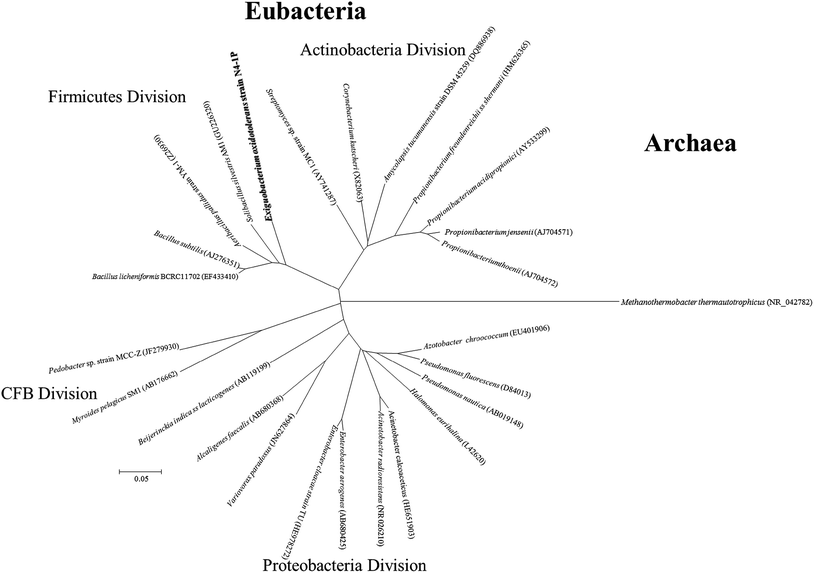 | ||
| Fig. 2 Phylogenetic tree based on 16S rDNA sequence from representative biosurfactant producers isolated in this study and those published in literature. CFB = Cytophaga–Flexibacter–Bacteroides. | ||
Phenotypic properties
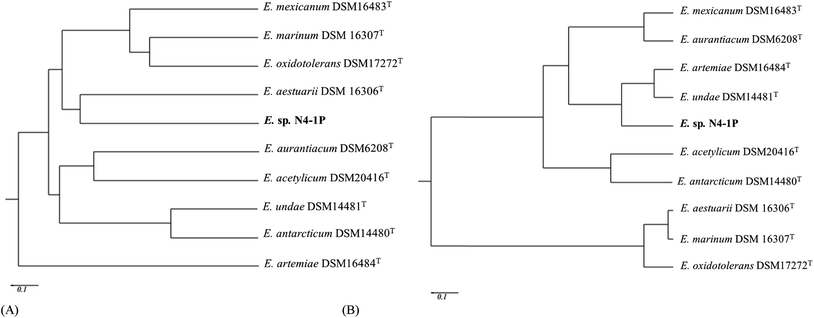
According to BIOLOG® metabolic fingerprinting results, Exiguobacterium sp. N4-1P could use 32 different carbon sources out of 95 on the GP2 plates. These include are 14 out of the 36 carbohydrates (cellobiose, fructose, glucose, sucrose, maltose, mannitol, mannose, methyl-D-glucoside, trehalose, dextrin, maltotriose, palatinose, D-ribose and pslcose); 3 out of the 16 carboxylic acids (acetic acid, D-gluconic acid and α-ketovaleric acid); 2 amides among the 4 amides (N-acety-D-galactosamine and N-acetyl mannosamine); 2 polymers (Tween 40 and 80); 2 out of the 3 esters (methyl pyruvate and methylsuccinate); 2 among the 7 alcohols (D-Sorbitol and glycerol); 6 out of 10 aromatic chemicals could be used: (arbutin, amygdalin, adenine, 2′-deoxy adenosine inosine, thymidine and uridine); and 1 out of the 9 amino acids (L-serine). No amines, phosphorylated chemicals, brominated chemicals included in the GP2 plates could be used as sole carbon sources for this strain. The strain is able to utilize the 10 substrates that other types strains in this genus can use, which defined the Exiguobacterium species.62 The carbon source utilization differentiation within the genus is shown in Table S1.† The dissimilarities among the Exiguobacterium strains are illustrated in Fig. 3A. The N4-1P strain was found to be closely related to E. aestuarii DSM 16306T and E. oxidotolerans DSM17272T with Pearson coefficients of 0.309 and 0.260, respectively, in carbon utilization.The membrane PLFA composition is shown in Tables S2.† The composition was found to be corresponded well with other members of the Exiguobacterium genus. Terminally branched-chain fatty acids are biomarkers of Gram positive bacteria.63 For Exiguobacterium sp. N4-1P, the primary PLFAs were iso-C15:0 (15.52%), iso-C17:0 (13.64%), C16:0 (12.51%), anteiso-C13:0 (11.83%), C16:1 (11.4%) and iso-C13:0 (8.92%). This pattern is very close to E. artemiae DSM16484T (ref. 60) and E. undae DSM14461T,64 which further demonstrated the assignment of the N4-1P stain as an Exiguobacterium (Fig. 3B). The SEM visualization results are shown in Fig. S1.† The Exiguobacterium sp. N4-1P was rod-shaped with a length of 2–3 μm and diameter of around 0.5 μm after 48 hours of incubation. Growing on the marine agar, the colonies were circular, entire, convex and had orange/yellow pigment. These morphological properties are similar to other members of the genus.60,62
Bioemulsifier production with different carbon sources
The general properties of the produced bacterial broth are summarized in Fig. 4. Hexadecane is a low toxicity water immiscible hydrocarbon. Diesel is composed of diverse hydrocarbons (mostly water immiscible) with moderate toxicity. It is an economic production substrate when compared with hexadecane and benzoate.65 However, its toxicity could hinder the production to some extent.66 Benzoate is a water-soluble hydrocarbon with moderate toxicity. It was found to stimulate surface-active compound production for some other biosurfactant producers,67 while eliminating the concerns of immiscible mixture during the post-processing and testing stages. The addition of all three types of hydrocarbons promoted the production of the emulsion-stabling bioemulsifier. The emulsification ability of the broths followed a sequence of diesel > hexadecane > benzoate. In terms of the yield, addition of hydrocarbon significantly increased the yield for the strain, and diesel addition generated the highest yield of the crude bioemulsifier. Interestingly, the addition of diesel produced bacterial broth can reduce surface tension significantly, indicating the simultaneous production of lower-molecular-weight biosurfactants capable of reducing surface tension.4 The obtained results indicated that the strain thrived with the moderate toxicity inherent with diesel. As a relative economic substrate (∼$0.9 per L), diesel has the potential for pilot to large scale production. Further testing on waste based substrates will be included in future studies to achieve economic production.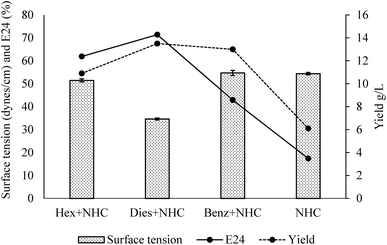 | ||
| Fig. 4 General properties of the clear broths obtained with different carbon sources (Hex = hexadecane, Dies = Diesel, Benz = Benzoate, NHC = non-hydrocarbon carbon sources; n = 3). | ||
Composition of the crude bioemulsifier
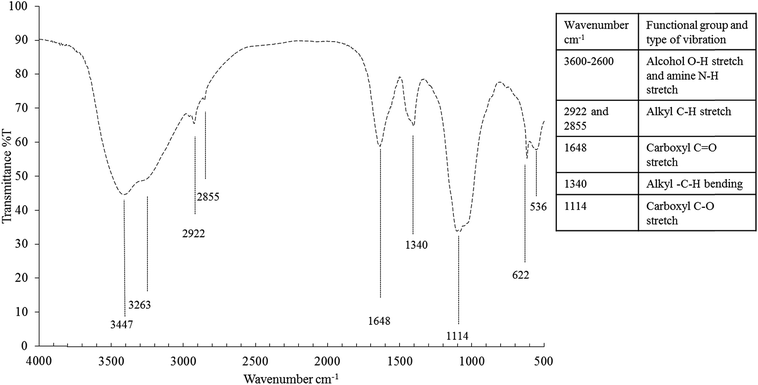
The bioemulsifier before dialysis consist of 14.64% lipid, 0.14% carbohydrate and 0.37% protein per the results of the colorimetric tests. After dialysis, the composition changed to 45.32% lipid, 5.1% carbohydrate and 0.93% protein. Eighteen fatty acids (FA) out of the 70 tested ones were detectable (Table S4†). Results showed that the hydrophobic moiety of the bioemulsifiers was primarily saturated FA (77.14%). Among which, C16:0 (32.18%) and C18:0 (40.99%) were the most abundant FA in the hydrophobic moiety. Unsaturated FA were mainly composed of monounsaturated FA (14.88) while polyunsaturated fatty acids only constituted 0.5% of total fatty acids. The hydrophobic moiety dominated by long chain FA (C16 and C18) in the bioemulsifier molecule as shown in this study is quite unique and seldom reported. The long chain FA (C16 and C18) might represent a significant improvement of emulsification activity.68 The FT-IR spectrum indicates the presence of carboxylic groups (3600–2600 cm−1, carboxyl –OH stretch; 1648 cm−1, carboxyl –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch; 1114 cm−1, carboxyl –C–O stretch) and alkyl groups (2922 cm−1 and 2855 cm−1, alkyl C–H stretch; 1340 cm−1, alkyl –C–H bending) (Fig. 5). The ninhydrin-based total amino acids/peptides test showed that the amino acids/peptides accounted for 13.11% before dialysis and 50.82% after dialysis (weight calibrated as tryptophan). When compared with the Bradford test results, the ninhydrin-based tests gave much higher readings. The Bradford test only targets proteins but not smaller peptides. The results indicate the obtained bioemulsifier is mainly a complex of lipopeptides with small fraction of glycolipids.
O stretch; 1114 cm−1, carboxyl –C–O stretch) and alkyl groups (2922 cm−1 and 2855 cm−1, alkyl C–H stretch; 1340 cm−1, alkyl –C–H bending) (Fig. 5). The ninhydrin-based total amino acids/peptides test showed that the amino acids/peptides accounted for 13.11% before dialysis and 50.82% after dialysis (weight calibrated as tryptophan). When compared with the Bradford test results, the ninhydrin-based tests gave much higher readings. The Bradford test only targets proteins but not smaller peptides. The results indicate the obtained bioemulsifier is mainly a complex of lipopeptides with small fraction of glycolipids.
Performance of the bioemulsifier
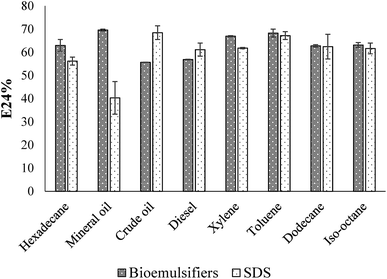
Emulsification ability of the bioemulsifier with different hydrocarbons is shown in Fig. 6. The experiment was conducted in parallel with a chemically-synthesized emulsifier, sodium dodecyl sulfate (SDS). The E24 was above 60% when using the single component aliphatic and aromatic hydrocarbons. With aromatic hydrocarbons, the bioemulsifier generally has slightly higher values of emulsification efficacy. With mixed hydrocarbons (mineral oil, crude oil and diesel), the E24 varied. High E24 values were observed with mineral oil and diesel. With crude oil, the E24 values were slightly lower than those of other hydrocarbons. The emulsification ability of the bioemulsifier is generally comparable with that of SDS. Since the bioemulsifier was capable of effectively emulsifying both aromatic and aliphatic hydrocarbons, it could be used for hydrocarbon remediation and oil recovery.69 The bioemulsifier produced by Exiguobacterium sp. N4-1P showed strikingly no-foaming characteristics even at high concentration (10%). This property would eliminate the cost of chemical defoament, sterilized pulse addition, and foam level detection during fermentation which is inevitable with common emulsifiers. In addition, the no-foaming surface-active agents were also found to have wider applications as wetting agents, rinse agents, and soil washing solutions.70 This is because the above processes rely largely on the scrubbing effect of vigorous jets or sprays of liquid which would be rendered relatively ineffective if cushioned by the action of large amounts of foam. Moreover, the foam formation will largely reduce the amount of effective liquids during applications.70As shown in Fig. 7, the E24 of the bioemulsifiers stabilized water–diesel emulsion increased when the concentrations of the bioemulsifiers increased till they reached the plateau. Before dialysis, the concentration required to reach the plateau was 15 g L−1. After the dialysis, the concentration required was reduced to 3 g L−1. Dialysis substantially improved the unit effectiveness of the bioemulsifier product. The formed oil-in-water emulsion under microscope displayed as Oil Red O-dyed oil droplets surrounded by continuous water phase with size ranging from a few microns to around 100 microns (Fig. 7). The bioemulsifier was also found able to reduce surface tension of water from 71 dynes cm−1 to 44.9 ± 0.26 dynes cm−1. The abilities of the produced bioemulsifier to stabilize oil-in-water emulsion and reduce surface tension enables its application as a soil washing agent to remove subsurface hydrocarbon plumes.4,5 Moreover, such abilities also lead to improved bioavailability of the hydrocarbon containments, thus enhance the bioremediation process.6
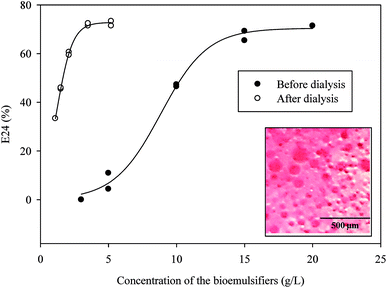 | ||
| Fig. 7 Effectiveness of the emulsifier at different concentrations (n = 2; insert: optical microscopy of cream phase of emulsions after 24 h, oil phase was dyed with Oil Red O). | ||
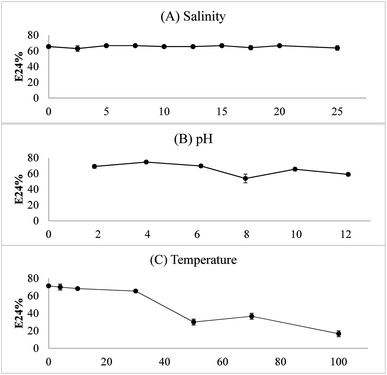
As shown in Fig. 8, the water–diesel emulsion formed with the bioemulsifier was very stable with salinity from 0 to 25%. Under acidic conditions, higher E24 values suggesting the bioemulsifiers are more effective. The temperature had significant effects on destabilization of the emulsion. When temperature was increased up to 50 °C, the emulsion started to collapse. At 100 °C, only around 20% emulsion remained. However, the emulsion was very stable and the E24 readings were the highest at or below 10 °C (>70%). Exiguobacterium sp. N4-1P is a cold-adapted bacterial strain isolated from a petroleum hydrocarbon contaminated sample in North Atlantic Canada. It, therefore, easily adapts to and thrives in the cold environment with abundant hydrocarbons. The produced bioemulsifier could perform excellent under low temperature as a soil washing agent and/or a bioremediation enhancement agent. The crude bioemulsifier when stored in an air-tight container under room temperature remained the same level of effectiveness after more than a year of storage (data not shown).
Conclusions
Exiguobacterium N4-1P is reported herein as a bioemulsifier producer for the first time. The genotypic and phenotypic properties of the strain were determined. Different carbon sources were used for bioemulsifier production and diesel stimulated the yield. The bioemulsifier is a complex mainly consist of lipopeptides with C16:0 (32.18%) and C18:0 (40.99%) as the primary FA. The produced bioemulsifier could form emulsions effectively with diverse hydrocarbons without foam generation, which facilitates the potential commercialization. The formed oil-in-water emulsions were stable upon a wide span of salinity (5–25%), pH (2–12), and temperature (below 50 °C). Exiguobacterium N4-1P and the produced bioemulsifier have promising application potential in environmental engineering, especially for soil washing and bioremediation targeting subsurface hydrocarbon pollution.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to express their gratitude to Dr Paris Georghiou (Department of Chemistry, Memorial University of Newfoundland), Research & Development Corporation (RDC) of Newfoundland and Labrador, Canada Foundation for Innovation (CFI) and Natural Sciences and Engineering Research Council of Canada (NSERC) for their support.References
- K. Holmberg, B. Jönsson, B. Kronberg and B. Lindman, Surfactants and Polymers in Aqueous Solution, 2nd edn, 2003, pp. 451–471 Search PubMed.
- A. Shete, G. Wadhawa, I. Banat and B. Chopade, J. Sci. Ind. Res., 2006, 65, 91 Search PubMed.
- C. Calvo, M. Manzanera, G. Silva-Castro, I. Uad and J. González-López, Sci. Total Environ., 2009, 407, 3634–3640 CrossRef CAS PubMed.
- Q. Cai, B. Zhang, B. Chen, X. Song, Z. Zhu and T. Cao, Environ. Monit. Assess., 2015, 187, 1–12 CrossRef CAS PubMed.
- M. Pacwa-Płociniczak, G. A. Płaza, Z. Piotrowska-Seget and S. S. Cameotra, Int. J. Mol. Sci., 2011, 12, 633–654 CrossRef PubMed.
- E. Z. Ron and E. Rosenberg, Curr. Opin. Biotechnol., 2014, 27, 191–194 CrossRef CAS PubMed.
- S. K. Satpute, A. G. Banpurkar, P. K. Dhakephalkar, I. M. Banat and B. A. Chopade, Crit. Rev. Biotechnol., 2010, 30, 127–144 CrossRef CAS PubMed.
- K. Muthusamy, S. Gopalakrishnan, T. K. Ravi and P. Sivachidambaram, Curr. Sci., 2008, 94, 736–747 CAS.
- R. S. Makkar, S. S. Cameotra and I. M. Banat, AMB Express, 2011, 1, 1–19 CrossRef PubMed.
- S. Mukherjee, P. Das and R. Sen, Trends Biotechnol., 2006, 24, 509–515 CrossRef CAS PubMed.
- I. M. Banat, A. Franzetti, I. Gandolfi, G. Bestetti, M. G. Martinotti, L. Fracchia, T. J. Smyth and R. Marchant, Appl. Microbiol. Biotechnol., 2010, 87, 427–444 CrossRef CAS PubMed.
- H. Dams-Kozlowska, M. P. Mercaldi, B. J. Panilaitis and D. L. Kaplan, Appl. Microbiol. Biotechnol., 2008, 81, 201–210 CrossRef CAS PubMed.
- C. Zheng, J. He, Y. Wang, M. Wang and Z. Huang, Bioresour. Technol., 2011, 102, 9155–9161 CrossRef CAS PubMed.
- F. Toledo, J. Gonzalez-Lopez and C. Calvo, Bioresour. Technol., 2008, 99, 8470–8475 CrossRef CAS PubMed.
- V. L. Colin, M. F. Castro, M. J. Amoroso and L. B. Villegas, J. Hazard. Mater., 2013, 261, 577–583 CrossRef CAS PubMed.
- R. Thavasi, V. S. Nambaru, S. Jayalakshmi, T. Balasubramanian and I. M. Banat, Mar. Biotechnol., 2009, 11, 551–556 CrossRef CAS PubMed.
- X. Liu, B. Ren, M. Chen, H. Wang, C. R. Kokare, X. Zhou, J. Wang, H. Dai, F. Song and M. Liu, Appl. Microbiol. Biotechnol., 2010, 87, 1881–1893 CrossRef CAS PubMed.
- F. Paul, A. Morin and P. Monsan, Biotechnol. Adv., 1986, 4, 245–259 CrossRef CAS PubMed.
- R. Thavasi, S. Jayalakshmi, T. Balasubramanian and I. M. Banat, Lett. Appl. Microbiol., 2007, 45, 686–691 CrossRef CAS PubMed.
- X. Hua, J. Wang, Z. Wu, H. Zhang, H. Li, X. Xing and Z. Liu, Biochem. Eng. J., 2010, 49, 201–206 CrossRef CAS.
- C. Calvo, F. Martinez-Checa, A. Mota, V. Bejar and E. Quesada, J. Ind. Microbiol. Biotechnol., 1998, 20, 205–209 CrossRef CAS.
- S. C. Lee, S. J. Lee, S.-H. Kim, I.-H. Park, Y. S. Lee, S. Y. Chung and Y. L. Choi, Bioresour. Technol., 2008, 99, 2288–2292 CrossRef CAS PubMed.
- S. Maneerat, T. Bamba, K. Harada, A. Kobayashi, H. Yamada and F. Kawai, Appl. Microbiol. Biotechnol., 2006, 70, 254–259 CrossRef CAS PubMed.
- T. Beltrani, S. Chiavarini, D. Cicero, M. Grimaldi, C. Ruggeri, E. Tamburini and C. Cremisini, Int. J. Biol. Macromol., 2015, 72, 1090–1096 CrossRef CAS PubMed.
- H. Hajfarajollah, B. Mokhtarani and K. A. Noghabi, Appl. Biochem. Biotechnol., 2014, 174, 2725–2740 CrossRef CAS PubMed.
- D. Husain, M. Goutx, M. Acquaviva, M. Gilewicz and J. C. Bertrand, World J. Microbiol. Biotechnol., 1997, 13, 587–590 CrossRef CAS.
- A. Markande, S. Acharya and A. Nerurkar, Process Biochem., 2013, 48, 1800–1808 CrossRef CAS.
- V. L. Colin, C. E. Pereira, L. B. Villegas, M. J. Amoroso and C. M. Abate, Chemosphere, 2013, 90, 1372–1378 CrossRef CAS PubMed.
- A. Franzetti, I. Gandolfi, C. Raimondi, G. Bestetti, I. M. Banat, T. J. Smyth, M. Papacchini, M. Cavallo and L. Fracchia, Bioresour. Technol., 2012, 108, 245–251 CrossRef CAS PubMed.
- A. Mishra, K. Kavita and B. Jha, Carbohydr. Polym., 2011, 83, 852–857 CrossRef CAS.
- J. de Jesús Paniagua-Michel, J. Olmos-Soto and E. R. Morales-Guerrero, Adv. Food Nutr. Res., 2014, 73, 221–257 Search PubMed.
- G. Trebbau de Acevedo and M. J. McInerney, J. Ind. Microbiol. Biotechnol., 1996, 16, 1–7 CrossRef CAS.
- S. Kebbouche-Gana, M. Gana, S. Khemili, F. Fazouane-Naimi, N. Bouanane, M. Penninckx and H. Hacene, J. Ind. Microbiol. Biotechnol., 2009, 36, 727–738 CrossRef CAS PubMed.
- S. Navon-Venezia, Z. Zosim, A. Gottlieb, R. Legmann, S. Carmeli, E. Ron and E. Rosenberg, Appl. Environ. Microbiol., 1995, 61, 3240–3244 CAS.
- C. Zheng, Z. Li, J. Su, R. Zhang, C. Liu and M. Zhao, J. Appl. Microbiol., 2012, 113, 44–51 CrossRef CAS PubMed.
- M. M. Yakimov, K. N. Timmis, V. Wray and H. L. Fredrickson, Appl. Environ. Microbiol., 1995, 61, 1706–1713 CAS.
- F. Peypoux, J. Bonmatin and J. Wallach, Appl. Microbiol. Biotechnol., 1999, 51, 553–563 CrossRef CAS PubMed.
- R. Shepherd, J. Rockey, I. W. Sutherland and S. Roller, J. Biotechnol., 1995, 40, 207–217 CrossRef CAS PubMed.
- P. Kim and J. H. Kim, Biotechnol. Bioprocess Eng., 2005, 10, 494–499 CrossRef CAS.
- N. Gorret, J. Maubois, J. Engasser and M. Ghoul, J. Appl. Microbiol., 2001, 90, 788–796 CrossRef CAS PubMed.
- A. Desai, K. Patel and J. Desai, Curr. Sci., 1988, 57, 500–501 CAS.
- C. Kokare, S. Kadam, K. Mahadik and B. Chopade, Indian J. Biotechnol., 2007, 6, 78 CAS.
- V. L. Colin, M. D. Baigorí and L. M. Pera, World J. Microbiol. Biotechnol., 2010, 26, 2291–2295 CrossRef CAS.
- M. C. Cirigliano and G. M. Carman, Appl. Environ. Microbiol., 1985, 50, 846–850 CAS.
- A. Monteiro, M. Bonfim, V. Domingues, A. Corrêa, E. Siqueira, C. Zani and V. Santos, Bioresour. Technol., 2010, 101, 5186–5193 CrossRef CAS PubMed.
- J. Zhang and R. Greasham, Appl. Microbiol. Biotechnol., 1999, 51, 407–421 CrossRef CAS.
- J. Oloke and B. Glick, Afr. J. Biotechnol., 2005, 4, 164 CAS.
- D. R. Cameron, D. G. Cooper and R. Neufeld, Appl. Environ. Microbiol., 1988, 54, 1420–1425 CAS.
- A. de Souza Monteiro, V. S. Domingues, M. V. Souza, I. Lula, D. B. Gonçalves, E. P. de Siqueira and V. L. dos Santos, Biotechnol. Biofuels, 2012, 5, 1 CrossRef PubMed.
- J. R. Trindade, M. G. Freire, P. F. Amaral, M. A. Z. Coelho, J. A. Coutinho and I. M. Marrucho, Colloids Surf., A, 2008, 324, 149–154 CrossRef CAS.
- Q. Cai, B. Zhang, B. Chen, Z. Zhu, W. Lin and T. Cao, Mar. Pollut. Bull., 2014, 86, 402–410 CrossRef CAS PubMed.
- M. Kimura, J. Mol. Evol., 1980, 16, 111–120 CrossRef CAS PubMed.
- N. Saitou and M. Nei, Mol. Biol. Evol., 1987, 4, 406–425 CAS.
- E. P. Ivanova, E. A. Kiprianova, V. V. Mikhailov, G. F. Levanova, A. D. Garagulya, N. M. Gorshkova, M. V. Vysotskii, D. V. Nicolau, N. Yumoto and T. Taguchi, Int. J. Syst. Evol. Microbiol., 1998, 48, 247–256 Search PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- S. Pande, R. P. Khan and T. Venkitasubramanian, Anal. Biochem., 1963, 6, 415–423 CrossRef CAS PubMed.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- C. C. Parrish, in Lipids in freshwater ecosystems, Springer, 1999, pp. 4–20 Search PubMed.
- B. Starcher, Anal. Biochem., 2001, 292, 125–129 CrossRef CAS PubMed.
- T. A. Vishnivetskaya, S. Kathariou and J. M. Tiedje, Extremophiles, 2009, 13, 541–555 CrossRef PubMed.
- C. Uzoigwe, J. G. Burgess, C. J. Ennis and P. K. Rahman, Front. Microbiol., 2015, 6, 245 Search PubMed.
- A. López-Cortés, P. Schumann, R. Pukall and E. Stackebrandt, Syst. Appl. Microbiol., 2006, 29, 183–190 CrossRef PubMed.
- D. White, J. Stair and D. Ringelberg, J. Ind. Microbiol., 1996, 17, 185–196 CrossRef CAS.
- A. Frühling, P. Schumann, H. Hippe, B. Sträubler and E. Stackebrandt, Int. J. Syst. Evol. Microbiol., 2002, 52, 1171–1176 Search PubMed.
- R. Makkar and S. Cameotra, Appl. Microbiol. Biotechnol., 2002, 58, 428–434 CrossRef CAS PubMed.
- Z. Sadouk, H. Hacene and A. Tazerouti, Oil Gas Sci. Technol., 2008, 63, 747–753 CrossRef CAS.
- T. de Sousa and S. Bhosle, Bioresour. Technol., 2012, 123, 256–262 CrossRef CAS PubMed.
- A. Johri, W. Blank and D. Kaplan, Appl. Microbiol. Biotechnol., 2002, 59, 217–223 CrossRef CAS PubMed.
- M. Ilori, C. Amobi and A. Odocha, Chemosphere, 2005, 61, 985–992 CrossRef CAS PubMed.
- Y. Hirata, M. Ryu, Y. Oda, K. Igarashi, A. Nagatsuka, T. Furuta and M. Sugiura, J. Biosci. Bioeng., 2009, 108, 142–146 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1 and Tables S1–S3. See DOI: 10.1039/c7ra07411e |
| This journal is © The Royal Society of Chemistry 2017 |