Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Insights into the detection of mutations and epigenetic markers using diamondoid-functionalized sensors
Ganesh Sivaramana,
Rodrigo G. Amorimbc,
Ralph H. Scheicherb and
Maria Fyta *a
*a
aInstitute for Computational Physics, Universität Stuttgart, Stuttgart, Germany. E-mail: mfyta@icp.uni-stuttgart.de
bDivision of Materials Theory, Department of Physics and Astronomy, Uppsala University, Sweden
cDepartamento de Física, ICEx, Universidade Federal Fluminense – UFF, Volta Redonda, RJ, Brazil
First published on 6th September 2017
Abstract
Nanogaps functionalized with small diamond-like particles, diamondoids, have been shown to effectively distinguish between different DNA nucleotides. Here, we focus on the detection of mutations and epigenetic markers using such devices. Based on quantum mechanical simulations within the density functional theory approach coupled with the non-equilibrium Green’s function scheme, we provide deeper insight into the inherent differences in detecting modified nucleotides. Our results strongly underline the influence of the type of functionalization molecule of the nanogap, as well its conformational details within the nanogap, on the sensing efficiency of the device. The electronic features for the mutations and epigenetic markers are compared to those for the respective canonical nucleotides that are detected by different devices. The calculations directly correlate the structural and electronic properties of the different nucleotides with the electronic transmission across the diamondoid-based device. The latter was found to be controlled by the functionalizing molecule and its binding to the nucleotides. We report on the direct connection of these characteristics to the sensitivity of the diamondoid-functionalized nanogaps, which could eventually be embedded in a nanopore device, and discuss the implications for DNA sensing.
I. Introduction
The sequencing of DNA using nanopores has been the aim of a series of studies in the past 20 years.1–5 The process is based on the electrophoretic motion of a biomolecule, such as DNA, through a nanometer-sized pore within a salt solution6 and allows the realization of single-molecule experiments. The use of solid-state nanopores,7 including silicon-nitride8 and graphene,9 has shown a high potential for low-cost and ultra-fast DNA sequencing.10 Different protocols for reading-out the DNA sequence have been proposed. One of them is the use of transverse tunneling currents.11,12 In that case, metallic electrodes are embedded into the nanopore and an additional electric field drives the tunneling current from one electrode to the other. This current can be electrically read-out and used to distinguish between the DNA nucleotides translocating through the nanopore. A common drawback of this scheme is the fact that often the tunneling signals for the four nucleotides overlap, leading to a low signal-to-noise ratio.13,14 A possible way to enhance the tunneling current and improve the signal-to-noise ratio is to use functionalized nanopores.1 An enhancement by at least one order of magnitude compared to the signals from a non-functionalized nanopore15 would be possible for all four DNA nucleotides. In the functionalized nanopores, the metallic electrodes embedded in the nanopore are functionalized by a small molecule of the size of the nucleotides. The functionalizing molecule binds through hydrogen bonds to each translocating DNA unit in a very specific way.Different molecules have been chosen for the functionalization of nanopores in order to increase their ability to read-out the nucleotides.16–18 We have recently shown that using small diamond-like functionalizations for the electrodes can very much improve the read-out process of DNA nucleotides.19,20 The functionalizing molecules are derivatives of diamondoids,21 which are tiny diamond-caged molecules22,23 forming a family of nanoscale building blocks with a variety of sizes and modifications.24–27 They have a size close to that of a nucleotide and tunable properties.28,29 Diamondoid derivatives are prone to form hydrogen bonds with DNA nucleobases.30,31 In diamondoid-functionalized electrodes embedded in a nanopore, the respective tunneling signals are nucleotide-specific and tuning the gating voltage in the diamondoid-based sensing device can improve the detection of DNA sequences.19
As a part of improving technologies for sequencing DNA, the development of more efficient techniques for also detecting DNA mutations32–35 is a very active field of research. Being able to read-out mutations or modifications in DNA molecules can be a keystone in revolutionizing cancer therapies. As a representative example, using two-dimensional molecular electronics spectroscopy to measure differential conductance is able to lead to distinguishable signatures from different types of DNA nucleobases and methylated nucleobases.36 Within this scope, we further investigate the sensitivity of diamondoid-functionalized electrodes for the detection of mutations and epigenetic markers. The aim is to provide a better understanding of the inherent characteristics of the detection properties of canonical and modified DNA units. Accordingly, the paper is organized as follows: the methodology is first presented in Section II, followed by the comparative analysis from different sensors in Section III. Finally, Section IV draws conclusions from our work.
II. Methodology
In this work, we set up a sensing device made up of gold electrodes functionalized using three different amine-modified diamondoid derivatives:20,21 rimantadine (‘rim’), amantadine (‘ama’), and memantine (‘mem’). The diamondoid is attached on the gold (111) surface of one of the electrodes. Two natural DNA nucleotides, deoxycytidine monophosphate (dAMC, in the following abbreviated simply as C) and deoxyguanosine monophosphate (dAMG, in the following abbreviated simply as G), and their modifications, a mutation (the 8-oxodeoxyguanosine monophosphate – 8-oxo-Gua, in the following abbreviated simply as d8oG37), and an epigenetic marker (the 5-methyl deoxycytidine monophosphate – 5-mdCMP, in the following abbreviated simply as d5mc38) are separately placed between the functionalized electrodes. An extensive analysis of amantadine-functionalized gold electrodes and the natural nucleotides was carried out previously.19,20 Here, we extend the investigation to study the efficiency of such diamondoid-functionalized electrodes in detecting mutations and epigenetic markers.The study is carried out through density functional theory based (DFT)39,40 simulations as implemented in the code SIESTA.41 The generalized gradient approximation of Perdew–Burke–Ernzerhof (PBE-GGA),42 and the norm-conserving Troullier–Martins pseudopotentials43 were used. A double-ζ polarized basis-set (DZP) for the nucleotides, and diamondoid derivatives and a single-ζ, with polarized (SZP), with (5d10, 6s1) orbital states for the gold atoms were considered.15,19 An energy shift of 0.01 Ry with a real space sampling grid (mesh cutoff) of 200 Ry, and 5 × 5 × 1 k-points using the Monkhorst–Pack scheme were also used. The ionic relaxations were performed until the net atomic forces of each atomic component are smaller than 0.01 eV Å−1. The gold (111) unit cell was fully relaxed and we obtained a lattice constant of 4.186 Å, which compares well with the data reported in literature.44 For the device functionalization, a supercell of 14.8 × 14.8 × 40.8 Å for rim and ama and 14.8 × 14.8 × 39.8 Å for mem was used. Five unit cells in the x and y directions and ten along the z direction with a gap of 19 Å for rim and ama and 18 Å for mem were repeated to build the supercell. Semi-infinite electrodes were used for the calculations. The different gap sizes were taken due to the different conformations of the diamondoids, which, in the case of rim and ama, required a larger gap. However, this has no effect on the consistency of our results. A thiolated amine-modified diamondoid was then placed close to the left gold layers and relaxed until the thiol group of the diamondoid was bonded to the gold surface. Finally, a nucleotide was added close to the diamondoid with an orientation favoring the formation of hydrogen bonds with the diamondoid. The sugar–phosphate group is pointing towards the right inner gold layer. The choice of the distance between the gold layers is based on the condition that even the largest nucleotide (G) could fit inside the electrode gap.
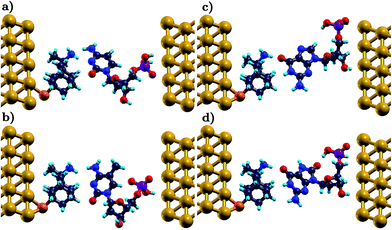
Each nucleotide interacts with the functionalized electrodes through hydrogen bonds with the functionalizing diamondoid.31,45 In this binding, amantadine and memantine are always donors. In the case of a rimantadine functionalization, the diamondoid can be a donor or an acceptor in the hydrogen bonding with the nucleotides. This depends on the relative arrangements of these two molecules. For the rimantadine case, we account for the two different arrangements in which rimantadine acts as (a) an acceptor (in the following referred to as rim-1) or (b) a donor (in the following referred to as rim-2) to the hydrogen bonding. A schematic representation of the respective two rimantadine-based devices is shown in Fig. 1 for two of the natural nucleotides, a mutation, and an epigenetic marker. The variation of the arrangements of the molecules is clearly seen in this figure. Comparison of panels (a) and (b) with (c) and (d) reveals the different role of the functionalization in bonding with the nucleotides, as the amine group of the rimantadine molecule can act either as the acceptor or the donor.
An assessment of the efficiency of the diamondoid-based devices in detecting modifications in DNA was made with the aid of electronic transport calculations. These are performed using DFT combined with the Non-Equilibrium Green’s Function (NEGF) formalism, as implemented in TranSIESTA.46 Details on the exact setup can be found elsewhere.19 The electronic transmission through the sensing device, including a left (L), a right (R) electrode, and a scattering region, can be written as:
T(E) = Tr[ΓL(E)![[capital G, script]](https://www.rsc.org/images/entities/i_char_e112.gif) (E)ΓR(E) (E)ΓR(E)![[capital G, script]](https://www.rsc.org/images/entities/i_char_e112.gif) †(E)], †(E)],
| (1) |
![[capital G, script]](https://www.rsc.org/images/entities/i_char_e112.gif) (E) Green’s function can be written as:
(E) Green’s function can be written as:
![[capital G, script]](https://www.rsc.org/images/entities/i_char_e112.gif) (E) = [E × SS − HS[ρ] − ΣL(E) − ΣR(E)]−1, (E) = [E × SS − HS[ρ] − ΣL(E) − ΣR(E)]−1,
| (2) |
III. Results and discussion
A. Conductance
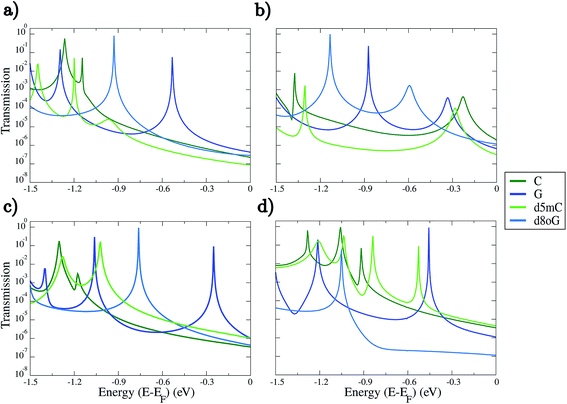
Our analysis will first focus on the transport properties of the devices shown in Fig. 1, as well as the ama- and mem-functionalized electrodes. These properties are directly mapped through the electronic transmission described in eqn (1) and are summarized in Fig. 2 for the two modified nucleotides, d5mC and d8oG, and their natural counterparts, C and G, respectively. The results directly correlate to the gating conductance, g, across the functionalized electrodes. The gating conductance as a function of the gate voltage, Vg, can be defined as:50| g(Vg) = G0T(μ), | (3) |
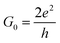 . T is the electronic transmission function in eqn (1), where the energy has been replaced by the chemical potential in order to account for the gating. Note, that all calculations are performed at zero bias.
. T is the electronic transmission function in eqn (1), where the energy has been replaced by the chemical potential in order to account for the gating. Note, that all calculations are performed at zero bias.
Inspection of Fig. 2 reveals some interesting trends. As a first note, each functionalized device seems to be able to sense in a quite different way, as the trends in the electronic transmission significantly vary within the energy range analyzed. In most of the cases, strong and clear peaks are evident down to energies of about −0.9 eV for all panels in Fig. 2. The first transmission peak, i.e., the peak closest to the Fermi energy, is the one corresponding to guanine (G). This holds in all cases in which the diamondoid-functionalization acts as an acceptor to the hydrogen bonding with the nucleotide (panels (a), (c), and (d) of Fig. 2). The peak for G closest to the Fermi energy was found in the case of the amantadine-functionalization at −0.25 eV in Fig. 2(c). Similarly, the first peak for G in the rim-1 and mem cases was found at −0.53 eV and −0.46 eV in panels (a) and (d) of Fig. 2, respectively. For the single case (rim-2) in which the diamondoid is a donor to the hydrogen bond with the nucleotide, the peak for G is shifted to −0.33 eV from the Fermi level (see Fig. 2(b)). In this case, for G, the electrons can be less efficiently transmitted across the device. Although G shows a stronger transmission peak at smaller energies for the rim-1, ama, and mem devices (Fig. 2(a), (c) and (d)), it decreases by about 3-fold in strength in the rim-2 arrangement (Fig. 2(b)). More specifically, the most prominent peak for G is also strongly shifted further away from the Fermi level (at −0.87 eV) when it becomes an acceptor to the hydrogen bonding with the electrodes, as is evident from Fig. 2(b). Apparently, for G the role of an acceptor is not that efficient for electronic transport. A possible physical explanation for the shifts in the peaks for the different devices and the various nucleotides could be given in terms of the coupling. A strong coupling between the nucleotide and the diamondoid corresponds to a strong coupling between the DNA (or nucleotide) electronic states and those of the device. This exactly manifests the role of the functionalization.
Further analysis of the transmission spectra in Fig. 2 shows that the hierarchy in the conductance varies for the four different devices. The rim-1, ama, and mem cases show similar trends; the guanine-based nucleotides are better resolved at energies closer to the Fermi level than the cytosine-based nucleotides (see Fig. 2(a), (c) and (d)). For the rim-2 and mem cases in Fig. 2(b) and (d), d5mC is the second nucleotide with the prominent peak closer to the Fermi level, while d8oG is the nucleotide with its prominent transmission peak further away from the Fermi energy. The order of the transmission signals for each nucleotide with respect to the position of the first transmission peak would be: G > d8oG > C > d5mC for rim-1, C > d5mC > G > d8oG for rim-2, G > d8oG > d5mC > C for ama, and G > d5mC > C > d8oG for mem. In this trend, the energy for each nucleotide peak is different. The respective trends for the zero-bias conductance, i.e., the transmission at the Fermi energy, are the same for rim-1, but for the other cases these are: C > d8oG > G > d5mC for rim-2, G > d5mC > d8oG > C for ama, and d5mC > C > G > d8oG for mem. Here, the energy for all nucleotides corresponds to the Fermi level. Note, that the transmission peaks for all nucleotides begin to overlap at low energies, leading to the recommendation that experimental setups should use a gate that operates at a low voltage.
Distinguishing the natural from the modified nucleotides, that is C from d5mC and G from d8oG, cannot be done with a similar efficiency for all diamondoid functionalizations. Again, panels (a) and (c) in Fig. 2 strongly denote that for rim-1 and ama the two peaks corresponding to the natural and modified nucleotides (compare the light and dark blue (green) lines) are strongly distinguishable. A read-out with rim-2 or mem might not be as efficient, as the corresponding transmission peaks are closer to each other (Fig. 2(b) and (d)). For rim-1, the two peaks for C and d5mC are very close and only ≈0.1 eV apart (Fig. 2(a) and (d)). Depending on the bandwidth with which a real device would work, this difference might be buried in the noise inherent in the electronic measurements. For the ama case, the peaks for G and d8oG are further apart (Fig. 2(c)). Note, that though the first transmission peaks below the Fermi energy are well resolved in most of the devices, there are also cases like d5mC in the mem-based device, which show more transmission peaks in the same energy window. This might strongly influence the sensitivity of the device, and this will be analyzed in the following section. The fact that for some energies no clear trends are observed indicates that the structural arrangements of the molecules (diamondoid and nucleotide) do not provide clear pathways for the electrons to tunnel across the devices. According to the transmission data presented here, we propose that experiments should work in the gating windows [−0.5, −1.3] V, [−0.8, −1.4] V, [−0.7, −1.4] V, and [−0.4, −1.3] V for the rim-1, rim-2, ama, and mem devices, respectively. This choice was made based on the energy position of the most prominent transmission peaks for all nucleotides (see Fig. 2), which will be considered in the analysis of the device sensitivity.
B. Electronic properties
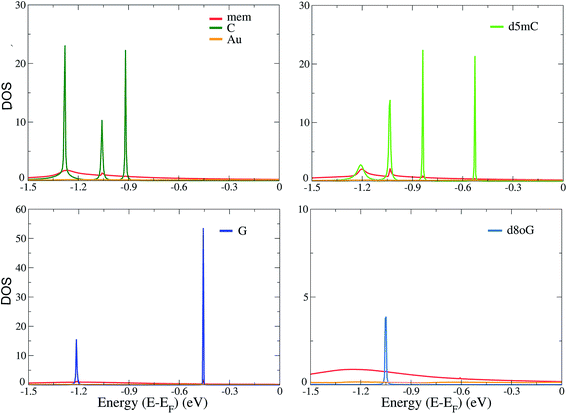
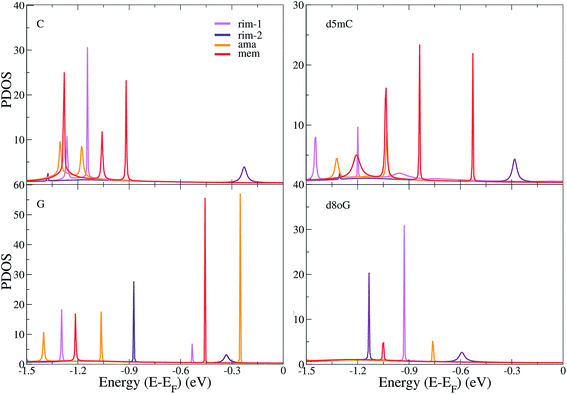
In order to understand the underlying physics in all devices and nucleotides, we turn to the analysis of the electronic properties, such as the total electronic density of states (DOS) and the projected density of states (PDOS). The total DOS for the memantine-based device are shown in Fig. 3 for the canonical and modified nucleotides studied here. The figure includes the contributions from the gold electrodes, the diamondoid, and the nucleotide. It appears that the gold electrodes do not contribute significantly to the PDOS as they add only to the background and do not influence the peaks. It is also clear from all panels of this figure that the nucleotides are strongly influencing the structure of the DOS, as the corresponding peaks arise predominantly from the contribution of the nucleotides. This was evident for all cases (data not shown for all). Another important finding when comparing the total DOS with the transmission data from Fig. 2 is that all transmission peaks can be seen as a result of an increased occupancy level at that same energy level as seen in the DOS plot (Fig. 3). In that sense, inspection of the DOS, i.e. the available electronic levels, can directly give an insight into the energy at which a peak in the transmission is expected. Although the diamondoids did not play an explicit role in determining these levels, they inherently influence the electronic behavior of the devices through their specific interaction with the nucleotides.In order to further understand the differences among the diamondoid-functionalizations, we summarize the results of the PDOS for all functionalized electrodes and the four canonical and modified nucleotides in Fig. 4. The contribution of the gold atoms and the diamondoids are added to the nucleotide part and the total DOS data is shown in Fig. 4. A comparison of the PDOS for all functionalized electrodes (rim-1, rim-2, ama, mem) reveals a different order of the peaks, as is also indicated by the transmission curves. For C (top-left panel), the rim-2 case shows the first peak (at −0.23 eV) closer to the Fermi energy, while for rim-1 and ama, the first PDOS peaks are further away and at similar energies (−1.14 eV and −1.18 eV, respectively). For d5mC (top-right panel), again rim-2 shows the first PDOS peak (at −0.28 eV), while the rim-1 device develops a peak almost 1 eV lower. For G (left-bottom panel), the peaks closer to the Fermi level are observed in the case of rim-2 and ama at −0.33 eV and −0.25 eV, respectively. The fact that these peaks are very close denotes that these two devices are similarly efficient in detecting G. This, though, should be confirmed by the strength of the corresponding transmission peaks in Fig. 2(b) and (c). Inspection of that figure reveals that for the ama-device the peak is about 3 orders of magnitude stronger. Hence, an ama-based device would be more efficient than a rim-2 one for reading-out G, as is also suggested by the following sensitivity analysis.
For the mutation d8oG (right-bottom panel), the peaks are again shifted to lower energies, further away from the Fermi level, with the first peak closest to the Fermi level occurring for the rim-2 case and the next for the ama case, at −0.59 eV and −0.76 eV, respectively. Summarizing these results shows (a) the significance of the structural arrangement of the two important ingredients of the sensing devices, the nucleotide and the diamondoid, alternating the donor and acceptor role in their binding and (b) the structural specificity in this binding, through the different chemical structures of the nucleotides and diamondoids. According to all observations made above, the different trends in the peak order in the PDOS indicate a variability in the sensing efficiency of each diamondoid-based device.
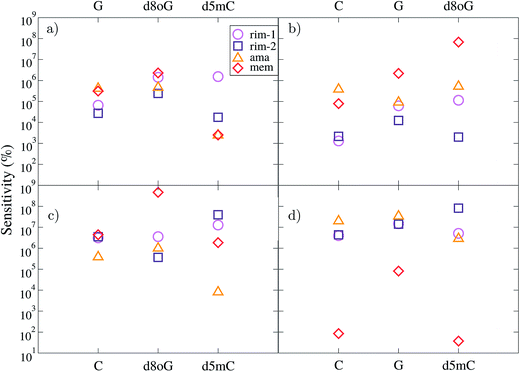
C. Sensing characteristics
Taking into account the specific electronic features of the natural and modified nucleotides, we compare these features within the context of the device sensitivity. The sensitivity provides an alternative quantitative view of the transmission spectra in Fig. 2, related to the differences in sensing the various nucleotides, and the importance of the chosen gating voltage. Based on the description of the conductance from eqn (3) we can define the sensitivity S(Vg)19 of the device in detecting a certain nucleotide as:
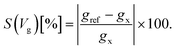 | (4) |
In this equation, gref is the reference zero-bias conductance under a certain gating voltage, which corresponds to the transmission peak of a specific nucleotide at a gating voltage Vg, and gx is the gating conductance at the same gate voltage for any other nucleotide. The sensitivity reveals how well resolved the conductance of a reference nucleotide will be with respect to other nucleotides.
Here, we take separately the nucleotides, C, d5mC, G, and d8oG as references. The results for all diamondoid-based devices are summarized in Fig. 5. Each panel of this figure reveals how well each device can resolve the reference nucleotide with respect to the other three nucleotides. Within the gating window described previously we take the energy (or gating voltage) for each resonance peak, which is different for each device and each nucleotide. For example, for C taken as a reference in panel (a), its resonance peaks were found at −1.26 eV, −1.38 eV, −1.30 eV, and −1.06 eV for rim-1, rim-2, ama, and mem, respectively. These are the energies at which the sensitivities in panel (a) are given for each device. Similarly, for d5mC (panel (b)), these peaks are found at −1.20 eV, −1.30 eV, −1.02 eV, and −0.84 eV for the rim-1, rim-2, ama, and mem devices, respectively. For G (panel (c)) these peaks are at −0.53 eV, −0.87 eV, −1.06 eV, and −0.46 eV for the rim-1, rim-2, ama, and mem devices, respectively. For d8oG (panel (d)) these peaks are at −0.93 eV, −1.13 eV, −0.76 eV, and −1.05 eV for the rim-1, rim-2, ama, and mem devices, respectively. The features described next are all related to these first resonance peak energies for each reference nucleotide.
According to panel (a) of Fig. 5, the rim-1 device can resolve C 5–6 orders of magnitude better than the other four nucleotides, while mem can resolve C about 5–6 orders of magnitude better than G and d8oG and about 3 orders of magnitude better than d5mC. In a similar way, panel (b) reveals that the mem-device has quite a different sensitivity for separating among the nucleotides, as it can resolve d5mC 5, 6, and 8 orders of magnitude better than C, G, and d8oG, respectively. Panel (c) shows that mem is again the most efficient (almost 9-fold) in distinguishing G from its mutation d8oG, while the other 3 devices are about 3-fold less efficient than mem for this. This comparisons are made at different reference energies, as the reference nucleotide has a resonance peak at different energies for each device. However, they clearly underline the variations among the devices and the importance of a careful choice of the gating voltage at which sensing is taking place.
A comparison at the same gating voltage (i.e. transmission energy) shows the difference between the two arrangements in the rim-1 and rim-2 devices. According to panel (a), in the former setup, C is resolved 5, 6, and 6 orders of magnitude better than G, d8oG, and d5mC, respectively. In the rim-2 setup, C is 4, 5, and 4 times better resolved than G, d8oG, and d5mC, respectively. C can be resolved from d5mC in the same way using the ama and mem devices. Panel (b) implies that the rim-1 device can resolve d5mC about 3, 5, and 5 times better than C, G, and d8oG, respectively. This sensitivity changes to 3-, 4-, and 3-fold for the rim-2 case. However, panel (d) implies that a rimantadine functionalization can better distinguish d8oG from the other molecules, with an efficiency of 6, 7, and 6 for rim-1 and 6, 7, and 8 for rim-2 for C, G, and d5mC, respectively.
Inspection of Fig. 5(b) and (d), shows clearly, that the ama-based device can resolve d5mC about 5–6 orders of magnitude better than C, G, and d8oG, respectively and d8oG about 7-fold better than C, G, and d5mC, respectively. The mem device can resolve d5mC about 5, 6, and 8 orders of magnitude better than C, G, and d8oG, respectively and d8oG about 2, 5, and 2 times better than C, G, and d5mC, respectively. This figure also implies that some of the devices cannot distinguish between certain nucleotides at certain gate voltages. For example, the rim-1-based device seems to not be efficient in distinguishing between d8oG and d5mC when C is taken as the reference (corresponding to a gate voltage of −1.26 eV). It also cannot separate C from d8oG when G is the reference (gate voltage of −0.53 eV). Similarly, for the ama-based device, when d8oG is taken as the reference (gate voltage of −0.76 eV), C and G cannot be recognized. For the same device working at a gate voltage (−1.30 eV) corresponding to C as the reference, G and d8oG give similar signals.
IV. Conclusions and relevance to DNA sequencing
An insight into the possibilities of detecting nucleotides and their modifications was provided in this work. For this, we focused on two of the canonical nucleotides, cytosine (C) and guanine (G), a mutation (d8oG), and an epigenetic marker (d5mC). For their detection we used four different diamondoid-functionalized electrodes. Transport calculations can provide information on the electronic transmission across each of these devices. Our results successfully revealed the differences and the efficiency of the sensing devices in distinguishing the canonical nucleotides from their modified counterparts. In most of the cases, the guanine nucleotide was the one better resolved, meaning that a strong transmission peak was found closer to the Fermi energy than for the other nucleotides. This implies that such a peak would be more easily detected in experiments. It was also shown that the most distinguishable conductance peaks are found close to the Fermi level, since further below −0.9 eV the electronic signals for the different nucleotides start to overlap. This suggests that potential experiments should tune the gating voltage of the devices in order to sense in a certain energy window below the Fermi level and up to around −1 eV.Another important finding involves the detection of modified nucleotides from their canonical counterparts. Although these are structurally very close, they show a different interaction with the sensing device through the diamondoid functionalization. These differences alter the pathways that the electrons can follow to tunnel in the functionalized device from one electrode to the other. Accordingly, depending on the exact conformational arrangement of the two molecules (diamondoid and nucleotide), the electronic transmission can show quite distinguishable features for some devices. Our results have shown, for example, that in the rim-1 and ama devices, the mutations or epigenetic markers can be strongly distinguished from the canonical nucleotides. For the other devices, there still exist clear peaks, which are closer in energy and might become broader for devices not working within a certain bandwidth. Additionally, we have shown that the resonance peaks in the electronic transmission are directly related to the available electronic states in the devices and can indicate the gating voltage that needs to be used to detect a certain nucleotide. In the end, our results strongly underline the fact that the explicit interaction and conformational arrangement of the functionalizing molecule and DNA significantly control the sensing aspect.
These results are based on an investigation that neglects certain elements, such as the solvent and structural and thermal conformational variability, which are important in a real sensing device. It is expected that these effects would influence the electronic transmission and the sensing efficiency of the devices. Nevertheless, a proof of concept was given here, which is expected to be very important in designing biosensors. In essence, our results propose the importance of a very careful design of functionalized nanogaps. At the edges of such nanogaps, functionalized electrodes are placed. This setup is very relevant to DNA sensing with nanopores. The nanogaps investigated in this work can potentially be embedded in a nanopore platform threading DNA molecules. Nanogaps in nanopores have already been experimentally realized.51 Such a design should seriously consider the specific type and interaction of the functionalization with DNA translocating through the nanopore. The DNA will in any case fluctuate within the nanopore, and there will be a large configurational space it will visit while interacting with the functionalizing molecule. However, through a strong interaction between the two essential units, the functionalizing molecule and the DNA nucleotides, it should be possible to reduce the noise and consequently increase the sensitivity of the device.
Note that the idea behind the functionalization of a nanogap would be to modify both electrodes.1 With such an approach, the molecules on both sides could alternately switch their roles, one of them being the ‘backbone grabber’ for slowing down the translocating DNA, while the second is the ‘nucleobase reader’, which identifies the nucleobase. This work is a first step towards that approach. We have started the investigation with one functionalization for simplicity, in order to show a proof of the principles of the sensitivity of the diamondoid-functionalization. A next step would be the use of a double functionalization, i.e. functionalization on both electrodes and should be the subject of a separate study. This work has an additional strong impact in nanopore sensing, as it signals the path through which mutations and epigenetic markers could be detected. The choice of the device and the bandwidth it works at are crucial in being able to distinguish these modified bases from the canonical nucleotides. With the clear input from our theoretical results, it remains to be shown in relevant experiments how other factors need to be tuned in order to bring diamondoid-functionalized nanopores closer to real DNA sequencing techniques and to efficiently scan for chemical alterations in the DNA chain. Finally, this work is the first step of a long and detailed investigation. In order to provide a proof of principle, we have first followed the simplest approach and plan to increase the complexity stepwise by considering additional factors. These include additional functionalization (see above), the influence of conformational variability (also including longer sequences), the inclusion of a solvent, possible contamination of the gold electrodes, the influence of an applied bias, the fluctuation of the functionalizing molecule, etc. It is not possible to tackle all factors simultaneously, which urged us to start from the simplest case and resort in our follow-up studies to other computational methods (ab initio and/or atomistic molecular dynamics). In this way, we plan to increase the complexity by investigating all additional factors and their influence on the electronic transmission spectra in order to move closer to a realistic functionalized nanogap for detecting biomolecules.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
G. S. and M. F. acknowledge financial support from the collaborative network SFB 716 “Dynamic simulations of systems with large particle numbers” funded by the German Funding Agency (Deutsche Forschungsgemeinschaft-DFG). This work was performed on the computational resource ForHLR Phase I funded by the Ministry of Science, Research and the Arts Baden-Württemberg and DFG. The initial benchmark calculations where performed on the computational resource bwUniCluster funded by the Ministry of Science, Research and the Arts Baden-Württemberg and the Universities of the State of Baden-Württemberg, Germany, within the framework program bwHPC. R. G. A. acknowledges financial support from the Carl Tryggers Stiftelse and R. H. S. thanks the Swedish Research Council. MF acknowledges support from the Juniorprofessorenprogramm funded by the Ministry of Science, Research and the Arts Baden-Württemberg (MWK).References
- D. Branton, D. W. Deamer, A. Marziali, H. Bayley, S. A. Benner and T. Butler, et al., The potential and challenges of nanopore sequencing, Nat. Biotechnol., 2008, 26(10), 1146 CrossRef CAS PubMed.
- B. M. Venkatesan and R. Bashir, Nanopore sensors for nucleic acid analysis, Nat. Nanotechnol., 2011, 6(10), 615–624 CrossRef CAS PubMed.
- R. H. Scheicher, A. Grigoriev and R. Ahuja, DNA sequencing with nanopores from an ab initio perspective, J. Mater. Sci., 2012, 47(21), 7439–7446 CrossRef CAS.
- M. Fyta, Threading DNA through nanopores for biosensing applications, J. Phys.: Condens. Matter, 2015, 27(27), 273101 CrossRef PubMed.
- S. Agah, M. Zheng, M. Pasquali and A. B. Kolomeisky, DNA sequencing by nanopores: advances and challenges, J. Phys. D: Appl. Phys., 2016, 49(41), 413001 CrossRef.
- C. Dekker, Solid-state nanopores, Nat. Nanotechnol., 2007, 2(4), 209–215 CrossRef CAS PubMed.
- J. Li, D. Stein, C. McMullan, D. Branton, M. J. Aziz and J. A. Golovchenko, Ion-beam sculpting at nanometre length scales, Nature, 2001, 412(6843), 166–169 CrossRef CAS PubMed.
- J. Li, M. Gershow, D. Stein, E. Brandin and J. A. Golovchenko, DNA molecules and configurations in a solid-state nanopore microscope, Nat. Mater., 2003, 2(9), 611 CrossRef CAS PubMed.
- S. J. Heerema and C. Dekker, Graphene nanodevices for DNA sequencing, Nat. Nanotechnol., 2016, 11(2), 127 CrossRef CAS PubMed.
- M. Wanunu, Nanopores: A journey towards DNA sequencing, Phys. Life Rev., 2012, 9(2), 125–158 CrossRef PubMed.
- M. Zwolak and M. Di Ventra, Colloquium: Physical approaches to DNA sequencing and detection, Rev. Mod. Phys., 2008, 80(1), 141 CrossRef.
- M. Di Ventra, M. Taniguchi and D. N. A. Decoding, RNA and peptides with quantum tunnelling, Nat. Nanotechnol., 2016, 11(2), 117 CrossRef CAS PubMed , review.
- M. Zwolak and M. Di Ventra, Electronic signature of DNA nucleotides via transverse transport, Nano Lett., 2005, 5(3), 421–424 CrossRef CAS PubMed.
- J. Lagerqvist, M. Zwolak and M. Di Ventra, Fast DNA sequencing via transverse electronic transport, Nano Lett., 2006, 6(4), 779–782 CrossRef CAS PubMed.
- H. He, R. H. Scheicher, R. Pandey, A. Reily Rocha, S. Sanvito and A. Grigoriev, et al., Transmission eigenchannels from nonequilibrium Green’s functions, J. Phys. Chem. C, 2008, 112, 3456 CAS.
- S. Huang, J. He, S. Chang, P. Zhang, F. Liang and S. Li, et al., Identifying single bases in a DNA oligomer with electron tunnelling, Nat. Nanotechnol., 2010, 5(12), 868–873 CrossRef CAS PubMed.
- V. Mussi, P. Fanzio, L. Repetto, G. Firpo, P. Scaruffi and S. Stigliani, et al., DNA-functionalized solid state nanopore for biosensing, Nanotechnology, 2010, 21(14), 145102 CrossRef CAS PubMed.
- B. Pathak, H. Löfås, J. Prasongkit, A. Grigoriev, R. Ahuja and R. H. Scheicher, Double-functionalized nanopore-embedded gold electrodes for rapid DNA sequencing, Appl. Phys. Lett., 2012, 100(2), 023701 CrossRef.
- G. Sivaraman, R. G. Amorim, R. H. Scheicher and M. Fyta, Diamondoid-functionalized gold nanogaps as sensors for natural, mutated, and epigenetically modified DNA nucleotides, Nanoscale, 2016, 8, 10105 RSC.
- G. Sivaraman, R. G. Amorim, R. H. Scheicher and M. Fyta, Benchmark investigation of diamondoid-functionalized electrodes for nanopore DNA sequencing, Nanotechnology, 2016, 27(41), 414002 CrossRef PubMed.
- A. Spasov, T. Khamidova, L. Bugaeva and I. Morozov, Adamantane derivatives: Pharmacological and toxicological properties (review), Pharm. Chem. J., 2000, 34(1), 1–7 CrossRef CAS.
- J. Dahl, S. Liu and R. Carlson, Isolation and structure of higher diamondoids, nanometer-sized diamond molecules, Science, 2003, 299(5603), 96–99 CrossRef CAS PubMed.
- H. Schwertfeger, A. A. Fokin and P. R. Schreiner, Diamonds are a chemist’s best friend: diamondoid chemistry beyond adamantane, Angew. Chem., Int. Ed., 2008, 47(6), 1022–1036 CrossRef CAS PubMed.
- A. A. Fokin, B. A. Tkachenko, P. A. Gunchenko, D. V. Gusev and P. R. Schreiner, Functionalized nanodiamonds part I. An experimental assessment of diamantane and computational predictions for higher diamondoids, Chem.–Eur. J., 2005, 11(23), 7091–7101 CrossRef CAS PubMed.
- M. A. Gunawan, J. C. Hierso, D. Poinsot, A. A. Fokin, N. A. Fokina and B. A. Tkachenko, et al., Diamondoids: functionalization and subsequent applications of perfectly defined molecular cage hydrocarbons, New J. Chem., 2014, 38(1), 28–41 RSC.
- Y. Zhou, A. D. Brittain, D. Kong, M. Xiao, Y. Meng and L. Sun, Derivatization of diamondoids for functional applications, J. Mater. Chem. C, 2015, 3, 6947 RSC.
- G. C. McIntosh, M. Yoon, S. Berber and D. Tománek, Diamond fragments as building blocks of functional nanostructures, Phys. Rev. B: Condens. Matter Mater. Phys., 2004, 70(4), 045401 CrossRef.
- A. A. Fokin and P. R. Schreiner, Band gap tuning in nanodiamonds: first principle computational studies, Mol. Phys., 2009, 107(8–12), 823–830 CrossRef CAS.
- M. Vörös, T. Demjén, T. Szilvási and A. Gali, Tuning the optical gap of nanometer-size diamond cages by sulfurization: a time-dependent density functional study, Phys. Rev. Lett., 2012, 108(26), 267401 CrossRef PubMed.
- G. Sivaraman and M. Fyta, Chemically modified diamondoids as biosensors for DNA, Nanoscale, 2014, 6, 4225 RSC.
- F. C. Maier, G. Sivaraman and M. Fyta, The role of a diamondoid as a hydrogen donor or acceptor in probing DNA nucleobases, Eur. Phys. J. E: Soft Matter Biol. Phys., 2014, 37(10), 1 Search PubMed.
- M. Orita, H. Iwahana, H. Kanazawa, K. Hayashi and T. Sekiya, Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms, Proc. Natl. Acad. Sci. U. S. A., 1989, 86(8), 2766–2770 CrossRef CAS.
- T. J. Chen, R. G. Boles and L. J. C. Wong, Detection of mitochondrial DNA mutations by temporal temperature gradient gel electrophoresis, Clin. Chem., 1999, 45(8), 1162–1167 CAS.
- F. Patolsky, A. Lichtenstein and I. Willner, Detection of single-base DNA mutations by enzyme-amplified electronic transduction, Nat. Biotechnol., 2001, 19(3), 253–257 CrossRef CAS PubMed.
- E. M. Boon, D. M. Ceres, T. G. Drummond, M. G. Hill and J. K. Barton, Mutation detection by electrocatalysis at DNA-modified electrodes, Nat. Biotechnol., 2000, 18(10), 1096–1100 CrossRef CAS PubMed.
- A. C. Rajan, M. R. Rezapour, J. Yun, Y. Cho, W. J. Cho and S. K. Min, et al., Two dimensional molecular electronics spectroscopy for molecular fingerprinting, DNA sequencing, and cancerous DNA recognition, ACS Nano, 2014, 8(2), 1827–1833 CrossRef CAS PubMed.
- S. Kanvah, J. Joseph, G. B. Schuster, R. N. Barnett, C. L. Cleveland and U. Landman, Oxidation of DNA: Damage to Nucleobases, Acc. Chem. Res., 2010, 43(2), 280 CrossRef CAS PubMed.
- P. W. Laird, The power and the promise of DNA methylation markers, Nat. Rev. Cancer, 2003, 3, 253 CrossRef CAS PubMed.
- P. Hohenberg and W. Kohn, Inhomogeneous Electron Gas, Phys. Rev., 1964, 136, B864 CrossRef.
- W. Kohn and L. J. Sham, Self-Consistent Equations Including Exchange and Correlation Effects, Phys. Rev., 1965, 140, A1133 CrossRef.
- J. M. Soler, E. Artacho, J. D. Gale, A. García, J. Junquera and P. Ordejón, et al., The SIESTA method for ab initio order-N materials simulation, J. Phys.: Condens. Matter, 2002, 14, 2745 CrossRef CAS.
- J. P. Perdew, K. Burke and M. Ernzerhof, Generalized Gradient Approximation Made Simple, Phys. Rev. Lett., 1996, 77, 3865 CrossRef CAS PubMed.
- N. Troullier and J. L. Martins, Efficient pseudopotentials for plane-wave calculations, Phys. Rev. B: Condens. Matter Mater. Phys., 1991, 43, 1993 CrossRef CAS.
- N. W. Ashcroft and N. D. Mermin, Solid State Physics, Holt, Rinehart and Winston, New York, 1976 Search PubMed.
- G. Sivaraman and M. Fyta, Chemically modified diamondoids as biosensors for DNA. Nanoscale. 2014;6(8):4225–4232 Search PubMed.
- M. Brandbyge, J. Mozos, P. Ordejón, J. Taylor and K. Stokbro, Density-functional method for nonequilibrium electron transport, Phys. Rev. B: Condens. Matter Mater. Phys., 2002, 65, 165401 CrossRef.
- M. Di Ventra, Electrical transport in nanoscale systems, Cambridge University Press Cambridge, 2008, vol. 14 Search PubMed.
- S. Datta, Quantum transport: atom to transistor, Cambridge University Press, 2005 Search PubMed.
- M. Paulsson and M. Brandbyge, Transmission eigenchannels from nonequilibrium Green’s functions, Phys. Rev. B: Condens. Matter Mater. Phys., 2007, 76(11), 115117 CrossRef.
- S. K. Min, W. Y. Kim, Y. Cho and K. S. Kim, Fast DNA sequencing with a graphene-based nanochannel device, Nat. Nanotechnol., 2011, 6(3), 162 CrossRef CAS PubMed.
- A. Fanget, F. Traversi, S. Khlybov, P. Granjon, A. Magrez and L. Forro, et al., Nanopore integrated nanogaps for DNA detection, Nano Lett., 2013, 14(1), 244–249 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2017 |