Open Access Article
Open Access ArticleA new approach for authentication of four ginseng herbs and their related products based on the simultaneous quantification of 19 ginseng saponins by UHPLC-TOF/MS coupled with OPLS-DA†
Bao-ming Huangab,
Ting-bo Chenab,
Sheng-yuan Xiaocd,
Qing-lin Zhae,
Pei Luob,
Ying-ping Wangd,
Xiu-ming Cuif,
Liang Liuab and
Hua Zhou *ab
*ab
aFaculty of Chinese Medicine, Macau University of Science and Technology, H837a, Block H, Taipa, Macau, P. R. China. E-mail: hzhou@must.edu.mo; huazhou2009@gmail.com; Fax: +853 28825886
bState Key Laboratory of Quality Research in Chinese Medicine, Macau University of Science and Technology, Taipa, Macau, P. R. China
cSchool of Life Science, Beijing Institute of Technology, Beijing, P. R. China
dInstitute of Special Wild Economic Animal and Plant Science, Chinese Academy of Agricultural Science, Changchun, P. R. China
eJiangxi University of Traditional Chinese Medicine, Nanchang, P. R. China
fKunming University of Science and Technology, Kunming, P. R. China
First published on 4th October 2017
Abstract
White ginseng, red ginseng, notoginseng and American ginseng are commonly called ginseng herbs and are frequently used as traditional Chinese medicine (TCM) and dietary supplements in China. Although their functions are evidently different, the discrimination of the four varieties still remains a challenge, especially for the deeply processed products which have lost their botanical morphological features and share similar chemical profiles. The chemometric approach is now the frequently used pattern recognition method for botanical origin discrimination. In the present study, OPLS-DA was employed to discriminate the four varieties using 19 major ginsenosides as variables, which were accurately measured by using UHPLC-TOF/MS. A clear separation of the four varieties was observed in the OPLS-DA score plot. The constructed model proved to be able to discriminate 10% of adulteration. Furthermore, this model showed high accuracy in authenticating the botanical origin of the deeply processed ginseng products. The results also showed that adulteration could be identified for some ginseng herb products in the market.
1. Introduction
Ginseng herbs generally refer to three species from the genus Panax, which are processed into four varieties for Chinese medicine, i.e., white ginseng (Ginseng Radix et Rhizoma), red ginseng (Ginseng Radix et Rhizoma Rubra), notoginseng (Notoginseng Radix et Rhizoma), and American ginseng (Panacis Quinquefolii Radix). These four varieties have been used as Traditional Chinese Medicine (TCM) and dietary supplements in China for centuries. The total production amount of the four varieties is over 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tons per year and estimated to be worth more than 2 billion USD (including the ginseng roots and their related products).1
000 tons per year and estimated to be worth more than 2 billion USD (including the ginseng roots and their related products).1
Though the four varieties originate from the same genus, their usages and properties are evidently different. For example, it has been reported that the white ginseng had no influence on the action of warfarin,2,3 which is an anticoagulant with a narrow safety margin usually used in the prevention of thrombosis and thromboembolism. However, American ginseng could significantly influence warfarin's anticoagulant activity.4 Therefore, the discrimination of the four varieties is crucial for the safety of the consumers. Nevertheless, the discrimination of the four varieties remains a challenge due to the similar morphological appearance and the chemical fingerprint. The discrimination is even harder when the raw herb is deeply processed into products. The DNA barcoding approach is an ideal method for discrimination of different species, however, the white ginseng and red ginseng are different processed forms of the same species, therefore, there is no gene marker that could be employed for the discrimination of these two varieties. Ginseng saponins (collectively called ginsenosides in this research) are the major active components in the four varieties, but the frequently used QC marker including the ginsenoside Rg1, Rb1, Re, and Rd are abundant in all the four varieties.5–8 Though American ginseng has one unique marker named pseudoginsenoside F11 (ref. 9) for inspection, the other three varieties are hardly to discover their corresponding unique markers. Therefore, the discrimination of the four varieties was still dependent on the direct comparison of the content of multiple markers.
Recently, the chemometric approach is frequently used to distinguish the biology origin of the herbal substance. Orthogonal projection to latent structures discriminant analysis (OPLS-DA) is a newly developed supervised method specific for the dataset with clear pre-defined class membership. The key point to apply the chemometric approach is to find the significant variables to establish a discriminant model. Conventionally, the data array for chemometric analysis is the untargeted chemical profiling measured by the hyphenated techniques such as liquid chromatography coupled with diode array detector (LC-DAD), and LC coupled with mass spectrometry (MS),10 or the direct profiling method such as nuclear magnetic resonance (NMR).11 The LC-MS based approach has been reported effective in the discrimination of the geographic origin of ginseng,10 and the NMR based method has been applied in an even more complicated issue, the discrimination of the age.11 However, the variables used in these reported methods included too many unidentified compounds, which significantly increased the uncertainty for the QC and would significantly reduce the reproductivity of the method. Furthermore, the profiling method was dependent on many compounds e.g. sucrose, maltose, glucose, malate, and glutarate,11 which are not the certified active constituent in ginsengs. Technically, it has certain advantage to use the reported active and commercial available compound to establish a QC method for the commercial products. As we all known, no official standard yet adopted the untargeted profiling method for the quality control of ginseng. Recently, it has been reported that, the discrimination of American ginseng and its related products from the other three varieties was carried out based on four identified variables coupled with the PCA model,12 i.e., the ratios of Rb1/Rg1 and Rg1/Re, the contents of pseudoginsenoside F11 and Rf. However, the separation of the four varieties was not complete in the PCA score plot, where individual clusters located closely to at least one of the other clusters and showed certain degree of overlap. The major reason of the unsatisfied discrimination of this reported method is the improper selection of variable.
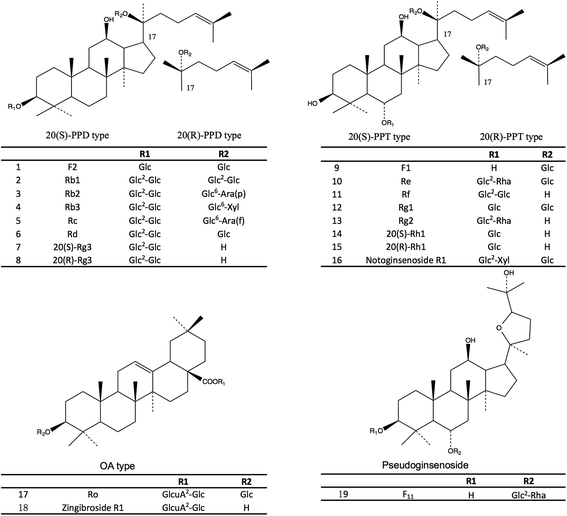
To select the suitable variables for the discrimination of the four varieties, the literature study was carried out to find the characteristic marker for individual varieties in this research. Each marker out of the 19 selected markers in this study could represent a certain aspect of the chemical feature of one variety. Therefore, the combinational use of these 19 ginsenosides as markers could provide sufficient systematic variation in the multivariate analysis for the discrimination of the four varieties. In the present study, 19 major ginsenosides (Fig. 1) in the four varieties were selected as the variables for the chemometric analysis and accurately measured by the ultra-high performance liquid chromatography coupled with time-of-flight mass spectroscopy (UHPLC-TOF/MS). The results showed that, a clear separation of the four varieties was observed in the OPLS-DA score plot by using the selected 19 ginsenosides as variables. The authentication result had been cross validated with the DNA sequencing method and this result has been published in our previous study.13
2. Experimental
2.1 Chemicals and reagents
Acetonitrile (Mass grade) and formic acid (Mass grade) were purchased from Sigma-Aldrich (Steinheim, Germany). The ultra pure water was prepared with the Milli-Q water purification system (Millipore, Bedford, MA, USA). The chemical reference substances (CRSs, purity >98%, HPLC-DAD) of ginsenoside F2, Rb1, Rb2, Rb3, Rc, Rd, 20(S)-Rg3, 20(R)-Rg3, F1, Re, Rf, Rg1, Rg2, 20(S)-Rh1, 20(R)-Rh1, notoginsenoside R1, ginsenoside Ro, and pseudoginsenoside F11 were obtained from Shanghai Winherb Medical Technology Co., Ltd (Shanghai, China). Zingibroside R1 (purity >98%, HPLC-DAD) was isolated, purified, and identified by Prof. Sheng-yuan Xiao from the Beijing Institute of Technology.2.2 Plant materials
The white ginseng samples were collected from four cultivation sites in Jilin and Liaoning Province of the northeast area of P. R. China. The white ginseng samples included garden ginsengs from 3 to 6 year old and wild ginseng in 6, 10 and 15 year old. The notoginseng samples were collected from 26 cultivation sites in Yunnan Province of the southwest area of P. R. China. The quality class of the notoginseng samples is 80 Tou (approximately 80 pieces of dried roots per 500 grams). The red ginseng samples were collected from 14 ginseng companies in P. R. China, South Korea and North Korea. The red ginseng samples included three quality classes, i.e., heaven (Tian), earth (Di), and good (Liang) corresponding to the superior, good, and regular quality. The American ginseng samples were collected from P. R. China, Wisconsin of the United State and Ontario of Canada. The samples of the four varieties were verified by Dr Wen-ru Wu from Guangzhou University of Chinese Medicine, who is an expert in herb authentication. Voucher specimens of the samples are available in the herbarium of the State Key Laboratory of Quality Research in Chinese Medicine (Macau University of Science and Technology). The detailed information of the samples could be found in ESI data, Table S-1.†The water contents of the ginseng samples were measured by a moisture balance system (Sartorius MA100, Gottingen, Germany) and accordance with the requirements in Chinese Pharmacopeia (Volume 1, Edition 2015), i.e., white ginseng <12.0%, red ginseng <12.0%, notoginseng <14.0%, and American ginseng <13.0%.
2.3 Ginseng herb products
The ginseng herb products are referred to the products containing the four ginseng herbs as raw materials and were collected at the pharmacy stores in Guangzhou City, including three batches of American ginseng tea (AGT1–4), three batches of red ginseng tea (RGT1–3), and four batches of white ginseng bolus and tea (WGB1–4).2.4 LC-MS analysis
The sample determination was performed on an Agilent 1290 UHPLC coupled with Agilent 6230 Accurate Mass TOF/MS system (Agilent technologies, Santa Clara, CA, USA) and the separation was based on an ACQUITY UPLC BEH® shield RP18 column (2.1 mm × 100 mm, 1.7 μm, Waters corporation, Santa Clara, CA, USA). The data of the UHPLC-TOF/MS was acquired by Agilent MassHunter Qualitative Analysis B.06.00.The autosampler was set at 10 °C and the column temperature was maintained at 40 °C. The separation was carried out with acetonitrile (A) and water (B) (both phases contain 0.1% formic acid) at a flow rate of 0.35 mL min−1 with a gradient program as follow: 25–43% A (0–14 min), 43–65% A (14–18 min), 65–80% A (18–20 min), 80–25% A (20–22 min), 25–25% A (22–25 min). The mass spectrometer was operated under the negative ionization mode with ESI (electronic spray ionization) iron source and the following settings: drying gas, N2; gas temperature, 325 °C; gas flow, 8.0 L min−1; nebulizer, 35 psi; VCap, 3500 V; nozzle voltage, 1000 V; fragmentor, 175 V. The data was acquired for each sample from m/z 100 to 1700. The molecule mass accuracy and reproducibility was maintained by using the independent reference lock-mass ions (API-TOF Reference Mass Solution Kit, G1969-85001, Agilent Technologies).
The following ions were extracted for the quantitative analysis of different compounds: m/z 845.49 ([M + HCOO]−, ginsenoside Rg1, Rf and pseudoginsenoside F11); m/z 991.55 ([M + HCOO]−, ginsenoside Re and Rd); m/z 829.49 ([M + HCOO]−, ginsenoside Rg2, F2, 20(S) and 20(R)-Rg3); m/z 683.43 ([M + HCOO]−, ginsenoside F1, 20(S) and 20(R)-Rh1); m/z 584.29 ([M + 2HCOO]2−, ginsenoside Rc, Rb2 and Rb3); m/z 977.53 ([M + HCOO]−, notoginsenoside R1); m/z 599.29 ([M + 2HCOO]2−, ginsenoside Rb1); m/z 955.48 ([M − H]−, ginsenoside Ro); and m/z 793.43 ([M − H]−, zingibroside R1).
2.5 Preparation of standard solutions
The 19 ginsenoside CRSs were accurately weighted approximately 1–2 mg and dissolved in 1 mL methanol as the stock solution individually whereas the 20(R)-Rg3 was dissolved in 25 mL methanol. Stock solutions of ginsenosides Re, Rb1, Rc, Rd and notoginsenoside R1 were diluted to a series of standard solutions in the concentration of approximately 0.15, 1.5, 7.5, 15, 40, 80, and 120 μg mL−1; ginsenosides Rg1, Rg2, Rf, Ro, and zingibroside R1 were diluted into a series of standard solutions in the concentration of approximately 0.08, 0.8, 4, 8, 20, 40, and 60 μg mL−1; ginsenosides 20(S)-Rh1, 20(R)-Rh1, F1, F2, Rb2, Rb3, 20(S)-Rg3, 20(R)-Rg3, and pseudoginsenoside F11 were diluted into a series of standard solutions in the concentration of 0.15, 0.75, 1.5, 4, 8, and 12 μg mL−1. All solutions were stored at −4 °C before use.2.6 Preparation of sample solutions from ginseng herbs
The dried roots were powdered to homogeneous size in a mill, sieved through a 40-mesh sieve. The sample powder (approximately 0.5 g) was accurately weighted in a centrifugal tube (50 mL, PP-single use; Thermo Fisher Scientific, Waltham, MA, USA) and shaken vigorously after the addition of 12.5 mL of 70% ethanol (water–ethanol, 30/70, v/v). Extraction was performed in an ultrasonic cleaner (Model 2800 HT; Crest Ultrasonics, Trenton, NJ, USA) for 1 hour. After ultrasonic extraction, centrifugal separation was performed for 10 minutes at 4000 rpm (Beckman Coulter, Brea, CA, USA). One fifth mL of the resulting supernatant solution was accurately diluted into 1.00 mL using methanol and mixed by vortex for 15 seconds. All samples were filtered through a 0.22 μm syringe filter (PTFE; Grace Davison Discovery Science, Columbia, MD, USA) before injected into the UHPLC-MS system.2.7 Preparation of sample solutions from ginseng tea
The content of the ginseng herbal tea bag was ground into fine powder, accurately weighted (approximately 0.5 g) and then extracted as the standard procedure described in the previous paragraph.2.8 Preparation of sample solutions from ginseng bolus
The ginseng bolus was accurately weighted (approximately 0.5 g) in a centrifugal tube and shaken vigorously after the addition of 12.5 mL of 70% ethanol. Extraction was performed in an ultrasonic cleaner for 1 hour. After ultrasonic extraction, centrifugal separation was performed for 10 minutes at 4000 rpm. The resulting supernatant solution was evaporated under 60 °C to remove the ethanol. The residual substance was dissolved in water and then loaded to the pre-treated macroporous resin (D101, column chromatography, volume 1.3 × 15 cm) according to previous method.14 The resin was eluted with water until the elution was colorless at a flow rate of 1.0 mL min−1, and then eluted with ethanol (80 mL) at the same flow rate until the elution was colorless. The ethanol elute was then evaporated under 60 °C to remove the solvent. The residual substance was accurately dissolved in 2 mL methanol, and then 0.1 mL solution was diluted with methanol to 1 mL and mixed by vortex for 15 seconds. All samples were filtered through a 0.22 μm syringe filter before injected into the UHPLC-MS system.2.9 Method validation
The method validation was performed in accordance with International Conference on Harmonization guidance documents.15,16 The intra-day precision of the method was analyzed using a mixture of ginseng saponin reference (approximate 5 μg/500 μL for each). The analyses ran 6 times. The inter-day precision was carried out over 3 consecutive days. Each analysis ran 6 times. The variations were expressed by the relative standard deviations (RSD%). The repeatability was evaluated by carrying out six replicate ginseng samples. The variation was expressed by the RSD (%).The recovery test was carried out by spiking the accurate amounts of the reference substance to the ginseng sample, the mixture was then processed as the description under Section 2.6. Three different levels of spiking amounts were carried out for the test. The original amount of ginsenosides in the ginseng sample was defined as the medium spiking level, whereas 50 and 150% of the medium level were defined as the low and high level to spike, respectively. Each level of test was carried out in triplicate. The recovery rate was calculated by eqn (1) below:
| Recovery (%) = 100 × (amount determined − original amount)/amount spiked | (1) |
The calibration curves were constructed by plotting the peak area against the concentration of each ginseng saponin. Nineteen ginsenosides were divided into three groups based on the concentration variation in the ginseng samples as described under Section 2.5 to prepare the calibration curves. Each concentration was measured in triplicate. The linearity was verified by correlation coefficients (R2).
The lower limit of quantification (LLOQ) and the lower limit of detection (LLOD) were defined as the concentration with the basic response of the signal-to-noise (S/N) ratio of 10 and 3, respectively.
The matrix effect of the ginseng herb extract was evaluated by spiking the known amount of the 19 CRSs into the ginseng herb extract and served as the mixed sample. In this experiment, the ginseng herb extract served as the matrix. The same amount of the CRSs spiked into the ginseng herb extract was dissolved in methanol as the reference solution to compare the signal deviation to the mixed sample. The matrix effect was calculated using the eqn (2) below:
| Matrix effect (%) = (target signal in mixed sample − target signal in matrix)/target signal in reference × 100% | (2) |
2.10 Data analysis
The OPLS-DA analysis was performed with the SIMCA software version 13.0 (Umetrics, Umeå, Sweden). The data was mean-centered and scaled to unit variance (UV) by the SIMCA software. The variable importance of projection (VIP) is the vector to summarize the total importance of the variable in explaining the model. The 19 ginsenosides were assessed as the independent variable and the OPLS-DA model was auto-fitted.The resolution of the OPLS-DA model was evaluated by the capability in separating a series of designated samples mixed with white ginseng and American ginseng. The mixed samples were produced in the following manner:
| Mixed sample = white ginseng × α + American ginseng × (1 − α) | (3) |
Data was expressed as the mean ± standard deviation (SD). Statistical analysis was performed using the SPSS 11.5 software. Significance was determined by one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test. Data were considered statistically significant if the p value was less than 0.05.
3. Results and discussion
3.1 The extraction of the 19 ginsenosides
The methods for quantitative measurement of the four ginsenosides (Rg1, Rb1, Re, and notoginsenoside R1) in the four varieties have been adopted in the Chinese Pharmacopeia (2015). However, the four varieties are extracted by different solvents systems i.e., white ginseng and red ginseng are extracted by chloroform (to remove the non-polar matrix) and then by butanol (to extract the semi-polar ginsenosides), notoginseng is extracted by methanol (to extract the semi-polar and polar ginsenosides), and the American ginseng is extracted by the butanol (to extract the semi-polar ginsenosides). Therefore, the extraction methods adopted in Chinese Pharmacopeia (2015) may be not suitable for the simultaneous extraction of the 19 ginsenosides in the four varieties. In the present study, the extraction method had been thus optimized to maximize the extraction efficiency of the target ginsenosides. The 70% ethanol has been reported to extract the multiple ginsenosides in the ginseng herbs.17–19 Because the 70% ethanol is ideal for the extraction of the semi-polar components such as the ginsenosides. Based on our experiment, using the 70% ethanol to extract the raw material two times could maximize the extraction efficiency, whereas approximately 80% of the extraction efficiency remained when it was extracted one time. It seemed more reasonable to extract two times, however, it was found that the fine powder of the ginseng roots would make the filtration very difficult to carry out, leading to time consuming. Due to the sample quantity used to establish the current model and the extension of the training set in the future, it would make the whole protocol not practical to execute when all samples are extracted two times. Furthermore, this study is aiming at discriminating the four varieties, therefore, the most important task is to make the data comparable among the four herbs but not the absolutely quantitative measurement of the ginsenosides. Therefore, the extraction method used in this study is a compromise of the extraction efficiency and the data comparability. Note that the reflux extraction could have make part of the primary ginsenosides decompose to secondary ginsenosides, and then change the chemical features of the samples. Therefore, the reflux extraction is not applied in this study.3.2 Chromatographic separation of 19 ginsenosides
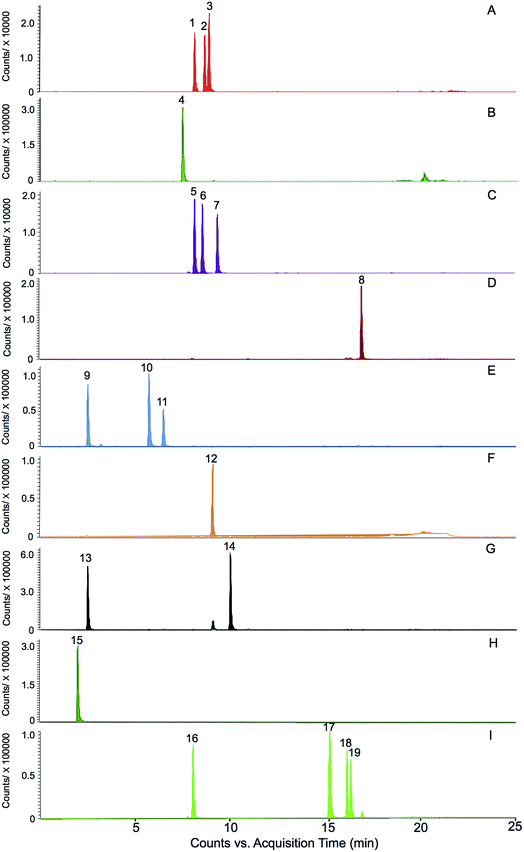
The selection of the 19 ginsenosides in this study was majorly according to their characteristic distribution in the four varieties reported in the literatures. The ginsenosides Rg1, Re, Rb1, Rd, and Rc were the five major markers used for quality assessment of the four varieties and adopted in several official quality standards, such as Chinese Pharmacopeia (2015 Edition), US Pharmacopeia (USP 35), and Japanese Pharmacopeia (JP XVI). The F11 and ginsenoside Rf were the certified markers for the discrimination of American ginseng from other species, and had been adopted in the Hong Kong Chinese Materia Medica Standards (Volume 5). The ginsenosides F1 and F2, notoginsenoside R1 were reported abundant in notoginseng.12 Furthermore, the steamed process of the red ginseng made some of the ginsenosides (such as the Rb1 and Re, termed primary ginsenosides) partly decompose to sub-grade ginsenosides (termed secondary ginsenoside) such as ginsenosides Rg2, Rg3, and Rh1.20 These compounds were supposed to be effective for discrimination. In addition, a rare ginsenoside named zingibroside R1 (one glycosyl less than the ginsenoside Ro) was purified in our lab and used as marker of ginseng herbs. The results of this study showed that, the zingibroside R1 is important for the discrimination of notoginseng from the other three varieties because of its extremely low abundance in notoginseng. The present study is also the first report to apply zingibroside R1 for the discrimination of ginseng herbs. Moreover, two pairs of rare epimers, i.e., 20(S/R)-Rh1 and 20(S/R)-Rg3 were also selected because the steam process of red ginseng can cause the transformation between the 20(S)- and 20(R)-epimers in ginseng herbs.20The challenge of simultaneous determination of the nineteen saponins is to separate the several groups of isomers i.e., (1) ginsenosides Rc, Rb2, and Rb3, (2) ginsenosides Rg1, Rf, and pseudoginsenoside F11, (3) ginsenosides Re and Rd, (4) ginsenosides F1 and Rh1, (5) ginsenosides F2, Rg2, and Rg3, especially the two groups of epimers i.e., 20(S/R) ginsenoside Rh1 and 20(S/R) ginsenoside Rg3. The methods previously reported were time consuming (approximately 60 minutes).21,22 In this study, the nineteen ginsenosides could be separated within 20 minutes (Fig. 2) due to the use of a 1.7 μm particle column and the gentle gradient (<1.5% organic phase increasing per min) optimized for epimers separation.
3.3 Method validation
Relative standard deviation (RSD) of the peak areas of a ginsenoside was used to evaluate the precision of the quantification. The intra-day precisions of the peak areas of all 19 ginsenosides ranged from 0.05 to 3.40% and their inter-day precisions ranged from 1.52 to 5.21%. The precision of the low-abundance ginsenosides, e.g. Rg3, is much better than previously described methods (RSD ≥ 20%).21,23 This phenomenon could be due to the higher sensitivity of a MS detector, which has better reproducibility in the low analyte concentration. The LLOQ of the 19 ginsenosides ranged from 0.0373 to 0.1220 ng (Table 1), which is approximately 30 to 150 folds better than an UV detector,23 and 1000 folds better than an ELSD detector.21,22 The LLOQ of the established method fulfilled the need of the determination of the lowest concentration in the real sample solutions.| Compound name | Precision | Matrix effect (inhibition, %) | LLOQ and LLOD | Repeatability (%) | Recovery (average ± SD, %) | ||
|---|---|---|---|---|---|---|---|
| Intra-day (%) | Inter-day (%) | LLOQ (ng μL−1) | LLOD (ng μL−1) | ||||
| Rg1 | 1.42 | 3.80 | 3.10 | 0.08 | 0.04 | 1.67 | 101.94 ± 5.01 |
| Re | 1.2 | 3.43 | 4.06 | 0.07 | 0.04 | 1.25 | 100.46 ± 4.83 |
| F11 | 1.06 | 3.41 | 3.25 | 0.04 | 0.02 | 2.52 | 95.91 ± 4.01 |
| Rf | 1.62 | 2.46 | 3.88 | 0.04 | 0.02 | 2.24 | 97.43 ± 4.68 |
| 20(S)-Rh1 | 0.95 | 2.57 | 2.56 | 0.08 | 0.04 | 0.71 | 96.82 ± 4.46 |
| Rg2 | 1.88 | 2.31 | 4.14 | 0.08 | 0.04 | 1.51 | 98.17 ± 4.84 |
| Rb1 | 0.80 | 2.25 | 5.03 | 0.06 | 0.03 | 1.01 | 97.64 ± 4.61 |
| 20(R)-Rh1 | 1.25 | 1.72 | 2.80 | 0.09 | 0.04 | 1.28 | 96.82 ± 4.46 |
| Rc | 1.56 | 4.7 | 4.13 | 0.08 | 0.04 | 1.61 | 96.92 ± 3.28 |
| F1 | 0.79 | 2.03 | 2.61 | 0.08 | 0.03 | 0.35 | 102.91 ± 4.41 |
| Ro | 1.09 | 2.89 | 5.09 | 0.07 | 0.04 | 1.86 | 95.36 ± 4.65 |
| Rb2 | 2.48 | 5.21 | 2.50 | 0.09 | 0.04 | 3.16 | 103.18 ± 2.00 |
| Rb3 | 2.21 | 4.63 | 3.51 | 0.08 | 0.03 | 3.40 | 104.27 ± 4.72 |
| Rd | 1.19 | 1.94 | 5.08 | 0.09 | 0.05 | 0.92 | 95.72 ± 5.66 |
| F2 | 0.73 | 1.67 | 2.25 | 0.10 | 0.05 | 1.73 | 97.52 ± 3.67 |
| 20(S)-Rg3 | 2.16 | 2.35 | 1.55 | 0.08 | 0.04 | 2.13 | 103.53 ± 4.65 |
| Zingibroside R1 | 1.02 | 2.22 | 2.35 | 0.04 | 0.02 | 3.67 | 97.28 ± 6.01 |
| 20(R)-Rg3 | 0.95 | 2.60 | 2.11 | 0.12 | 0.04 | 1.05 | 99.43 ± 8.64 |
| Notoginsenoside R1 | 3.11 | 4.81 | 1.18 | 0.03 | 0.02 | 3.07 | 101.29 ± 4.00 |
The linearity of each ginsenoside was established within a range covering its content in the real samples. The method displayed good linearity (R2 ≥ 0.9989) for all the ginsenosides analyzed over the concentration range selected for quantification (Table 2). The effect of the matrix on the ionization was tested by comparing the peak intensities of a mixed sample solution to that of reference solution of ginsenosides. The matrix of ginseng extract caused slight decrease of the detector responses of the 19 ginsenosides (ranging from 1.20% to 3.96%, Table 1). The quantities of the ginsenosides in real samples were calculated based on the calibration curves established using pure solution of the CRSs. The RSD of the repeatability of the method was less than 4.0% (n = 6), which indicated good stability for sample determination. The recoveries of all the ginsenosides ranged from 95.36 to 104.27% (Table 1), indicating the acceptable accuracy of the constructed method.
| Compound name | Linear regression | R2 | Linear range (ng) |
|---|---|---|---|
| Rg1 | y = 180![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649.7194x − 126 649.7194x − 126![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 941.3534 941.3534 |
0.9990 | 0.21–53.19 |
| Re | y = 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 310.4922x + 77 310.4922x + 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 039.3910 039.3910 |
0.9990 | 0.14–111.50 |
| F11 | y = 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 972.8423x − 3037.1768 972.8423x − 3037.1768 |
0.9993 | 0.08–6.11 |
| Rf | y = 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 503.5377x + 30 503.5377x + 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 060.3449 060.3449 |
0.9996 | 0.08–60.42 |
| 20(S)-Rh1 | y = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 587 587![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 310.8890x − 50 310.8890x − 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 168.0092 168.0092 |
0.9996 | 0.15–11.40 |
| Rg2 | y = 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 951.3935x − 45 951.3935x − 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 846.5962 846.5962 |
0.9991 | 0.15–77.15 |
| Rb1 | y = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 622.0538x + 99 622.0538x + 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790.5649 790.5649 |
0.9996 | 1.29–118.64 |
| 20(R)-Rh1 | y = 151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 470.4706x − 43 470.4706x − 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 872.4194 872.4194 |
0.9997 | 0.17–12.90 |
| Rc | y = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 726.5619x + 68 726.5619x + 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 754.4890 754.4890 |
0.9990 | 1.20–90.24 |
| F1 | y = 139![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 811.2723x − 43 811.2723x − 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780.2867 780.2867 |
0.9998 | 0.21–16.26 |
| Ro | y = 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 296.2455x + 112 296.2455x + 112![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 452.8555 452.8555 |
0.9995 | 0.89–89.06 |
| Rb2 | y = 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 885.2749x + 12 885.2749x + 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 115.1330 115.1330 |
0.9990 | 0.72–57.80 |
| Rb3 | y = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 134.8515x + 6344.1261 134.8515x + 6344.1261 |
0.9998 | 0.79–11.91 |
| Rd | y = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 793.5520x − 28 793.5520x − 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 486.9530 486.9530 |
0.9993 | 0.18–45.44 |
| F2 | y = 62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 615.4804x − 33 615.4804x − 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 527.6046 527.6046 |
0.9993 | 0.20–15.61 |
| 20(S)-Rg3 | y = 59![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 486.9699x − 29 486.9699x − 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 294.8091 294.8091 |
0.9998 | 0.16–12.55 |
| Zingibroside R1 | y = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312.1603x + 99 312.1603x + 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 239.1211 239.1211 |
0.9989 | 0.74–37.26 |
| 20(R)-Rg3 | y = 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 635.9206x − 41 635.9206x − 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191.4388 191.4388 |
0.9996 | 0.24–18.29 |
| Notoginsenoside R1 | y = 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 491.4288x + 32 491.4288x + 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 769.0024 769.0024 |
0.9990 | 1–100 |
Over all, a quick and accurate method for simultaneously quantification of the 19 ginsenosides was developed in the present research; the precision, accuracy, linearity, and sensitivity of the method meet the requirement in ICH guidance documents.15
3.4 Ginsenoside content of the root of four varieties
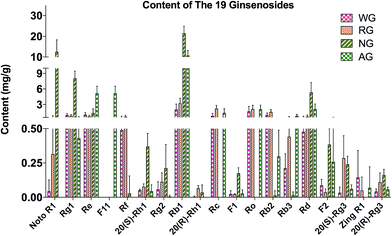
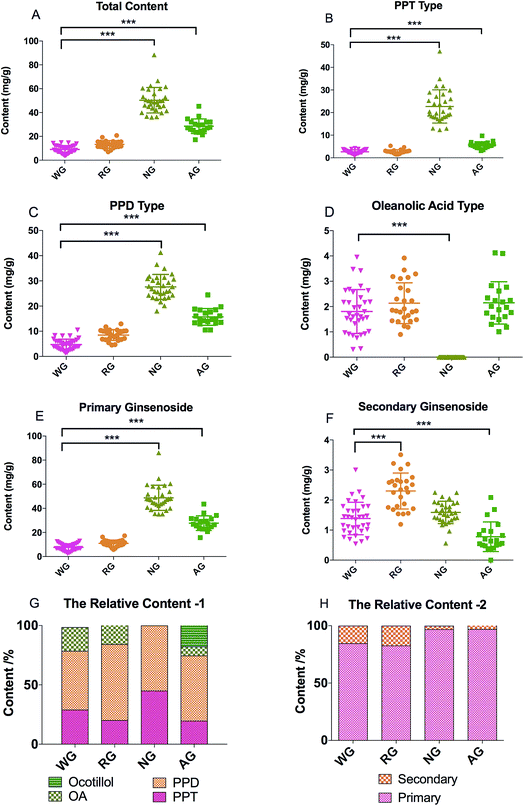
The total contents of the 19 ginsenosides ranked as follow: notoginseng > American ginseng > red ginseng > white ginseng (50.31 ± 10.53, 28.51 ± 5.98, 13.28 ± 3.10, and 9.03 ± 3.10 mg g−1, Fig. 4A). Notoginseng showed the highest content of protopanaxatriol (PPT, Fig. 1) type ginsenosides (22.71 ± 7.24 mg g−1), protopanaxadiol (PPD, Fig. 1) type ginsenosides (27.60 ± 4.86 mg g−1) and primary ginsenosides (48.72 ± 10.28 mg g−1, Rb1, Rc, Rd, Re, Rf, Rg1, Ro, Rb3, and notoginsenoside R1, Fig. 4B, C and E); American ginseng showed the highest content of oleanolic acid type (OA, Fig. 1) ginsenosides (2.14 ± 0.83 mg g−1, Fig. 4D) and a unique marker, pseudoginsenoside F11 (5.13 ± 1.40 mg g−1, ocotillol type); red ginseng showed the highest content of the secondary ginsenosides (2.30 ± 0.59 mg g−1, ginsenosides Rg2, 20(R/S) Rg3, 20(R/S) Rh1, F1, and F2, Fig. 4F).F11 was a unique ginsenoside in American ginseng while ginsenoside Rf was absent (Fig. 3), which was the critical chemical feature for American ginseng.9 Ginsenosides Re and Rg1 showed a characteristic ratio approximate 10 (Re/Rg1) in American ginseng.24 This ratio was less than or approximate to 1 in the other three herbs.21,22 The ratio of Rb3/Rb2 was larger than 1 in American ginseng, while the ratio was less than 1 in the other three herbs.5,21,23 The 20(R)-Rh1 was not detected in American ginseng, and was detected in one white ginseng sample, 9 notoginseng samples (0.03 ± 0.05 mg g−1), and in all red ginseng samples (0.06 ± 0.02 mg g−1). Therefore, the 20(R)-Rh1 was also a potential marker for the discrimination of American ginseng from other three ginseng herbs. In previous reports, the combination of 20(S/R)-Rh1 (ref. 21 and 25) was used for the quality assessment of American ginseng. But the present study showed that the distributions of the 20(S) and 20(R)-Rh1 in the four varieties were different. The 20(S)-Rh1 could not be detected in American ginseng as well as the 20(R)-Rh1,8 but notoginseng showed the highest content of 20(R)-Rh1 (0.18 ± 0.05 mg g−1) even higher than that in the red ginseng (0.04 ± 0.01 mg g−1). Therefore, the 20(S) and 20(R)-Rh1 were also potential markers for the discrimination of the four varieties.
Though the content of notoginsenoside R1 in the red ginseng and white ginseng was 50 to 100 folds lower than that in notoginseng (12.51 ± 5.82 mg g−1), the notoginsenoside R1 was not unique in the notoginseng. The signal intensity of OA type ginsenosides in notoginseng including Ro and zingibroside R1 was approximately 30 folds lower than the lower limit of the linear range, which could not cover the trace amount of the OA type ginsenosides in notoginseng by using a practical injection volume. The OA type ginsenosides in notoginseng were therefore excluded in the further discussion. The content of zingibroside R1 in the other three varieties ranged from 0.01 to 0.02 mg g−1, but not detected in notoginseng. This was the first report of accurately quantification of the zingibroside R1 in the ginseng herbs and then applied for the discrimination of notoginseng from other three varieties. The result showed that, the zingibroside R1 could be useful for the discrimination of notoginseng from the other three varieties due to the significant different content. The content of the three PPD type isomers Rc, Rb2, and Rb3 was merely approximately 0.05% of the total amount of ginseng saponins in notoginseng, which was much lower than that in the American ginseng (9.85%), white ginseng (21.66%) and red ginseng (32.24%). Therefore, the low content of the three isomers (Rc, Rb2, and Rb3) could also be one feature of notoginseng.
The direct comparison of the ginsenosides among the four varieties showed that, the F11 was the only unique marker for the discrimination of American ginseng from the other three. Though several markers such as the 20(S/R)-Rh1, notoginsenoside R1, and zingibroside R1 showed special distribution in the four varieties but not unique in any of the four.
Furthermore, due to the limited quantity of samples collected in present study, it was too risky to define a certain content of a marker as a threshold to exclude or include a sample for one variety. Therefore, the similarity among the four varieties made it difficult to discriminate by the direct comparison of the markers, particularly for the white ginseng and red ginseng. The difference was only from the relative content of the secondary ginsenosides, which occupied 17.32% of the total content of the ginsenosides in red ginseng, whereby 15.34, 3.16, and 2.86% in white ginseng, notoginseng, and American ginseng (Fig. 4H). The relative content of the secondary ginsenoside was also approximate between the white ginseng and red ginseng. It was too difficult to discriminate these two varieties by the direct comparison of the markers. Current studies have revealed that pattern recognition of multiple markers could be a feasible way for discrimination, and therefore the multivariate analysis is then used to discriminate the four ginseng herbs.
3.5 Discrimination of four ginseng herbs by OPLS-DA
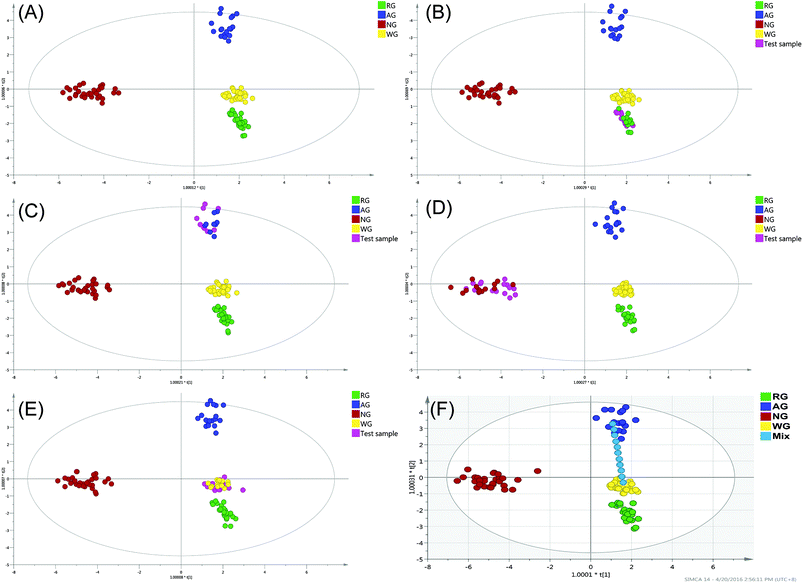
The OPLS-DA was employed to establish a model allowing the discrimination of the four varieties. The OPLS-DA extends a regression of principal component analysis (PCA)12 and involves an orthogonal signal correction (OSC) filter to divide the systematic variation into two parts: one is correlated (predict) to the y variable while the other part is uncorrelated (orthogonal) to the y variable (categorical y variables in OPLS-DA). The OPLS-DA uses the class membership to maximize the variation among groups and therefore leads to better interpretability for the classification compared to the unsupervised methods.26 In the past decade, the unsupervised method such as the hierarchical cluster analysis (HCA) and PCA were frequently applied for the classification of the natural products. The HCA can exhibit the natural grouping of the samples based on the distance calculation (such as k-mean and Euclidean distance), but the natural grouping is not always in the direction of the separation. PCA is a different model based on the projection to provide a path for the data observation in the most informatics viewpoint of the data, but similar to the HCA, the grouping in PCA might not be in the desirable direction. Correspondingly, the supervised method uses the class membership to guide the grouping to the designated direction and therefore is more convenient to find the markers responsible for the designated effect. Furthermore, the supervised method, such as the OPLS-DA could separately manage the variation that correlated and uncorrelated to the result variables and therefore also has better capacity to handle the within-class variation,27 which is usually large in the data from the natural products.The model was conducted using 7-fold cross-validation in this research, with the cross-validation parameters R2 and Q2 describing the goodness of fit and the predictive ability of the model, respectively. The R2 and Q2 close to 1 indicated the OPLS-DA model is excellent fitted. The constructed model was performed by considering four classes (107 samples), resulting in three predictive and six orthogonal components (overall goodness of fit: R2X = 0.865, R2Y = 0.944, Q2 = 0.929). The model explained 86.5% of the total variance of the data array, R2Y > 0.9 indicated an excellent fitted model, and Q2 > 0.9 indicated the excellent predictive ability. The outlier is the plot out of the ellipse, which is defined as the Hotelling's T2 range 95% confidence. No serious outlier was observed. To validate the constructed model, 50% of the samples from each class were excluded randomly and then served as the test samples (Fig. 5B–E). The results showed that even up to 50% of the samples of each class were excluded from the training set, the constructed model could identify these test samples accurately. The results indicated that, the constructed model was greatly fitted and it was strong enough for the further application to identify the deeply processed products.
A clear separation of the four classes was observed in Fig. 5A, the notoginseng and the American ginseng located in the end of the t[1] and t[2] direction, respectively. Both the white and red ginseng located near the center of the model plane and stayed close to each other but had no sample overlapping. As can be seen, the white ginseng and red ginseng could be separated only in the t[1] direction but completely overlapped in the t[2] direction. In the model plane, the samples stayed close indicating the positive correlated (similar). It can be concluded that even the red ginseng was processed in high temperature for the decomposition of the primary ginsenoside, the decomposition of the primary ginsenoside was not sufficient to create complete new chemical species. The chemical stability within the P. ginseng led to the similarity of the white ginseng and red ginseng. The OPLS-DA score plot showed that, the white ginseng was different from red ginseng, but not significant as the difference observed in the other two species.
Nine ginsenosides mainly responsible (VIP > 1.0) for the separation of the four varieties in the OPLS-DA including the ginsenoside Rc, pseudoginsenoside F11, ginsenoside Re, notoginsenoside R1, ginsenoside Rd, 20(R)-Rh1, Rb1, F1, and 20(S)-Rh1 (VIP ranged from 1.38 to 1.01). The clustering in the score plot was majorly caused by the significant different content of the VIP compounds in the four varieties. For notoginseng, the ginsenoside Rc was extremely lower while the notoginsenoside R1 was extremely higher than in the other three varieties, in addition to the significant higher content of the 20(S)-Rh1 and Rb1. For American ginseng, it had the unique maker pseudoginsenoside F11, the significant higher content of Re and no 20(R)-Rh1. But for the white ginseng and red ginseng, there was no unique marker or significantly different marker between these two varieties. Therefore, the pattern recognition method was necessary for the discrimination of the red and white ginseng due to their high similarity.
3.6 The resolution of the constructed model
The resolution of the model was evaluated by the separating of one set of designated mixture samples, which were mixed with white ginseng and American ginseng in different proportions (Fig. 5F). The mixture samples scattered from negative to positive direction of t[2], which was the major principal component for clear separation of white ginseng and American ginseng. As can be seen, the pure white ginseng (no adulteration) and pure American ginseng were classified into the corresponding group correctly, indicating the accuracy of the constructed model. The mixture samples were placed between these two pure samples and showed clear separation to each other, where the plots scattered according to the mixture proportion. Therefore, the resolution of the constructed model was at least at the level of 10% adulteration. However, when the adulteration proportion was 10 or 20%, the mixture samples were still classified into the white ginseng or American ginseng cluster, suggesting that the lower limitation of discrimination (LLOD) of the constructed model was 20% adulteration. The suboptimal LLOQ was caused by the relative large within-class variation of individual classes. Since the samples in present study were collected from different suppliers around the world and included different cultivars, the within-class variation was unavoidable. Therefore, based on the collected samples, the constructed method could merely to identify the mixture sample with at least 20% adulteration.3.7 Discrimination of ginseng herbal products by OPLS-DA
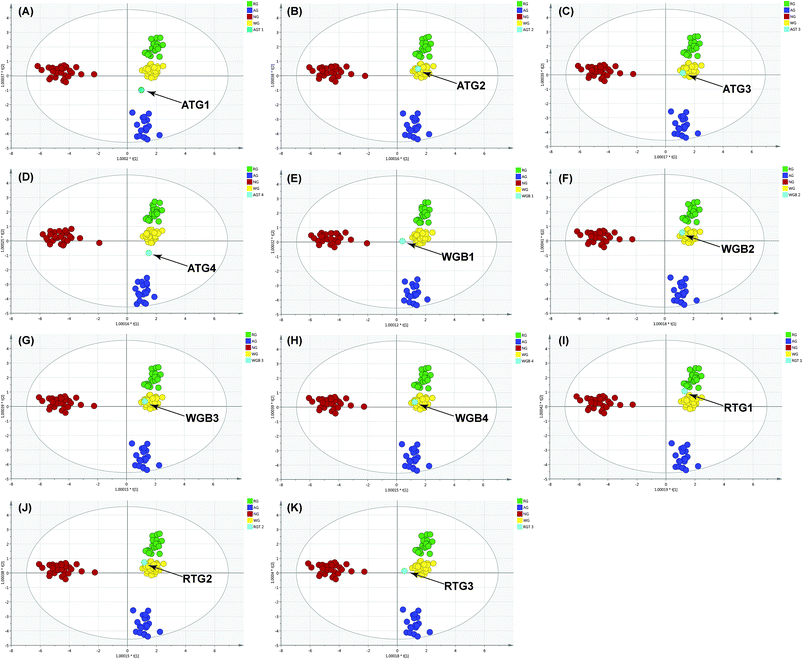
The results in previous section showed that the constructed model discriminated the herb origin correctly and efficiently. Therefore, the model was further employed to discriminate the herb origin in the deeply processed products. To maintain the scattering pattern of the constructed model, the ginseng product sample was assessed by the model individually.The result showed that AGT-1 and 3 (Fig. 6A and C) were mixture of American ginseng and P. ginseng, whereas AGT-2 (Fig. 6B) was a pure white ginseng sample. The white ginseng and red ginseng products showed no adulteration by other species. These results were highly consistent to the DNA sequencing approach13 (ESI data, Table S-2†). In the adulterated samples, the chemical features of different varieties could be observed simultaneously. In AGT-1 and 3, the presence of F11, Rb3/Rb2 >1 are the chemical feature of the American ginseng, whereas the chemical features of P. ginseng, the ginsenoside Rf, was also observed. But the AGT-3 located at the edge of the white ginseng cluster indicated the low abundance of the American ginseng in the product (Fig. 6C). Except the RGT-1 (Fig. 6I), the RGT-2 and RGT-3 (Fig. 6J and K) located in the white ginseng cluster indicated the adulteration of white ginseng in the red ginseng products or the red ginseng used in RGT-2 and RGT-3 were not processed enough to obtain enough secondary ginsenosides. However, AGT-4 (Fig. 6D) showed different discrimination results between DNA sequencing and OPLS-DA. AGT-4 contained only P. ginseng based on DNA sequencing, however, a mixture in OPLS-DA. In this sample, both the chemical markers of American ginseng and P. ginseng could be observed: the simultaneously presence of F11 and Rf. According to the literature, the AGT-4 might be a hybrid of American ginseng and P. ginseng and thus contained the F11 and Rf in the same plant.28 The results showed that the OPLS-DA could provide sufficient discrimination accuracy for real ginseng products by the cross validation with the DNA sequencing. Furthermore, the discrimination using OPLS-DA could be used for a rough estimation of the proportion of the foreign materials in the product.
The big difference of the price among the four varieties is the major reason of the intentional adulteration. In the international trade market, the price of the white ginseng from China is approximate 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 USD per ton, the red ginseng from Korea is 250
000 USD per ton, the red ginseng from Korea is 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 USD per ton, the American ginseng from Canada is 70
000 USD per ton, the American ginseng from Canada is 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 USD per ton, and the American ginseng from U.S. is 200
000 USD per ton, and the American ginseng from U.S. is 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 USD per ton.1,29–31 Therefore, the adulteration using the low-price varieties could bring huge illegal profit. Therefore, an efficient discrimination method for the four ginseng varieties is necessary. Compared to the DNA sequencing approach, the OPLS-DA approach is easy to operate and more effective for high-throughput analysis. Furthermore, it is also an optimal complement to the DNA sequencing approach for the discrimination of the samples with the same botanical origin, such as the white ginseng and red ginseng.
000 USD per ton.1,29–31 Therefore, the adulteration using the low-price varieties could bring huge illegal profit. Therefore, an efficient discrimination method for the four ginseng varieties is necessary. Compared to the DNA sequencing approach, the OPLS-DA approach is easy to operate and more effective for high-throughput analysis. Furthermore, it is also an optimal complement to the DNA sequencing approach for the discrimination of the samples with the same botanical origin, such as the white ginseng and red ginseng.
4. Conclusion
In summary, the present study established a method based on the 19 ginsenosides and the OPLS-DA model for the discrimination of the four widely used ginseng herbs and their related products. The method validation of the UHPLC-TOF/MS quantitative analysis and the validation of the OPLS-DA model indicated the established method was precise, accurate, and sensitive. Furthermore, the cross-method validation with the DNA sequencing method provided the solid evidence for the discrimination accuracy of the established method. The established method can be the complement for the QC methods adopted in the official standards and particularly for the DNA sequencing method when the within-species discrimination is performed. In addition, it is more practical than the conventional fingerprint profiling methods due to exclude the numerous unidentified markers. Analogously, the utilization of many other TCMs is in various processed forms or even different medical parts (such as root, leaf and stem), which share similar chemical features. Therefore, the constructed method in present study can be promising not only for the ginseng QC but also for many other TCMs. It would be valuable for other researchers of TCM and be benefit for millions of customers.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was financially supported by the Macao Science and Technology Development Fund (Project No. 037/2014/A1 and 071/2011/A3).References
- I. H. Baeg and S. H. So, J. Ginseng Res., 2013, 37, 1–7 CrossRef PubMed.
- S. H. Lee, Y. M. Ahn, Y. M. Ahn, H. K. Doo and B. C. Lee, J. Altern. Complement. Med., 2008, 14, 715–721 CrossRef PubMed.
- Y. H. Lee, B. K. Lee, Y. J. Choi, I. K. Yoon, B. C. Chang and H. S. Gwak, Int. J. Cardiol., 2010, 145, 275–276 CrossRef PubMed.
- C. S. Yuan, G. Wei, L. Dey, T. Karrison, L. Nahlik, S. Maleckar, K. Kasza, M. Ang-Lee and J. Moss, Ann. Intern. Med., 2004, 141, 23–27 CrossRef CAS PubMed.
- J. R. Wang, L. F. Yau, W. N. Gao, Y. Liu, P. W. Yick, L. Liu and Z. H. Jiang, J. Agric. Food Chem., 2014, 62, 9024–9034 CrossRef CAS PubMed.
- S. P. Li, C. F. Qiao, Y. W. Chen, J. Zhao, X. M. Cui, Q. W. Zhang, X. M. Liu and D. J. Hu, J. Chromatogr. A, 2013, 1313, 302–307 CrossRef CAS PubMed.
- W. Shi, Y. T. Wang, J. Li, H. Q. Zhang and L. Ding, Food Chem., 2007, 102, 664–668 CrossRef CAS.
- C. L. Qu, Y. P. Bai, X. Q. Jin, Y. T. Wang, K. Zhang, J. Y. You and H. Q. Zhang, Food Chem., 2009, 115, 340–346 CrossRef CAS.
- L. Li, G. A. Luo, Q. L. Liang, P. Hu and Y. M. Wang, J. Pharm. Biomed. Anal., 2010, 52, 66–72 CrossRef CAS PubMed.
- H. H. Song, D. Y. Kim, S. Woo, H. K. Lee and S. R. Oh, J. Ginseng Res., 2013, 37, 341–348 CrossRef PubMed.
- S. O. Yang, Y. S. Shin, S. H. Hyun, S. Cho, K. H. Bang, D. Lee, S. P. Choi and H. K. Choi, J. Pharm. Biomed. Anal., 2012, 58, 19–26 CrossRef CAS PubMed.
- Y. Shi, C. J. Sun, B. Zheng, B. Gao and A. M. Sun, Food Anal. Methods, 2013, 6, 112–122 CrossRef.
- C. Chun-song, T. A. N. Tian-qi, L. Ze, L. I. U. Zhi-zu, W. U. Wen-ru, W. Ying-ping, L. I. U. Liang and Z. Hua, Chin. Tradit. Herb. Drugs, 2015, 46, 2549–2555 Search PubMed.
- Y. N. Zhao, Z. L. Wang, J. G. Dai, L. Chen and Y. F. Huang, Chin. J. Nat. Med., 2014, 12, 382–392 CAS.
- I. Taverniers, M. De Loose and E. Van Bockstaele, TrAC, Trends Anal. Chem., 2004, 23, 535–552 CrossRef CAS.
- J. Ermer, J. Pharm. Biomed. Anal., 2001, 24, 755–767 CrossRef CAS PubMed.
- L. Li, G. A. Luo, Q. L. Liang, P. Hu and Y. M. Wang, J. Pharm. Biomed. Anal., 2010, 52, 66–72 CrossRef CAS PubMed.
- Y. Wang, J. You, Y. Yu, C. Qu, H. Zhang, L. Ding, H. Zhang and X. Li, Food Chem., 2008, 110, 161–167 CrossRef CAS PubMed.
- J. Liu, Y. Liu, L. Zhao, Z. H. Zhang and Z. H. Tang, SpringerPlus, 2016, 5, 1770 CrossRef PubMed.
- X. G. Li, L. Fu, Q. Lu and X. Li, J. Jilin Agric. Univ., 2000, 22, 1–9 Search PubMed.
- S. N. Kim, Y. W. Ha, H. Shin, S. H. Son, S. J. Wu and Y. S. Kim, J. Pharm. Biomed. Anal., 2007, 45, 164–170 CrossRef CAS PubMed.
- B. S. Sun, L. J. Gu, Z. M. Fang, C. Y. Wang, Z. Wang, M. R. Lee, Z. Li, J. J. Li and C. K. Sung, J. Pharm. Biomed. Anal., 2009, 50, 15–22 CrossRef CAS PubMed.
- H. W. Park, G. In, S. T. Han, M. W. Lee, S. Y. Kim, K. T. Kim, B. G. Cho, G. H. Han and I. M. Chang, J. Ginseng Res., 2013, 37, 457–467 CrossRef CAS PubMed.
- L. P. Christensen, M. Jensen and U. Kidmose, J. Agric. Food Chem., 2006, 54, 8995–9003 CrossRef CAS PubMed.
- L. Li, J. L. Zhang, Y. X. Sheng, D. A. Guo, Q. Wang and H. Z. Guo, J. Pharm. Biomed. Anal., 2005, 38, 45–51 CrossRef CAS PubMed.
- H. M. Zhang, S. L. Li, H. Zhang, Y. Wang, Z. L. Zhao, S. L. Chen and H. X. Xu, J. Pharm. Biomed. Anal., 2012, 62, 258–273 CrossRef CAS PubMed.
- M. Bylesjö, M. Rantalainen, O. Cloarec, J. K. Nicholson, E. Holmes and J. Trygg, J. Chemom., 2006, 20, 341–351 CrossRef.
- J. Song, Master, Chinese Academy of Agricultural Sciences, 2014.
- Askci Corporation, Analysis on the Price Trends of Notoginseng in China, ASKCI.com, http://www.askci.com/news/201309/25/2513582833836.shtml (accessed September 25, 2013).
- Askci Corporation, Analysis on the Production and Sales of Global American Ginseng, ASKCI.com, http://www.askci.com/news/201309/26/261632833853.shtml (accessed September 25, 2013).
- Askci Corporation, Analysis on the Current Situation of the Global Ginseng Industry Market, ASKCI.com, http://www.askci.com/news/201309/25/2513575233834.shtml (accessed September 25, 2013).
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06812c |
| This journal is © The Royal Society of Chemistry 2017 |