Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Combining paired analytical metabolomics and common garden trial to study the metabolism and gene variation of Ginkgo biloba L. cultivated varieties†
Xin-Guang Liu‡
,
Xu Lu‡ ,
Ji-Xin Wang,
Bin Wu,
Lin Lin,
Hui-Ying Wang,
Ru-Zhou Guo,
Ping Li
,
Ji-Xin Wang,
Bin Wu,
Lin Lin,
Hui-Ying Wang,
Ru-Zhou Guo,
Ping Li * and
Hua Yang*
* and
Hua Yang*
State Key Laboratory of Natural Medicines, China Pharmaceutical University, No. 24 Tongjia Lane, Nanjing 210009, People's Republic of China. E-mail: liping2004@126.com; 104yang104@163.com
First published on 5th December 2017
Abstract
Secondary metabolites play a pivotal role in plant physiology and medicinal function. Much work has been carried out to uncover the genetic basis of plant secondary metabolite variation, but direct screening of gene variation in the whole genome is extremely time- and labor-consuming. The prediction of a candidate locus of a single nucleotide polymorphism (SNP) will save much time and resource. In this work, we combined a paired analytical metabolomics and a common garden trial to bridge the association of plant metabolism and related gene variation of Ginkgo cultivated varieties. Firstly, the leaves of 30 cultivated varieties of Ginkgo biloba L. grown in the same garden since 1990 were analyzed by UHPLC-QQQ MS/MS. Thirty-six metabolites in the flavonoid biosynthetic pathway were quantified. The biosynthetic rate of flavonoids in different cultivated varieties could reflect the related enzyme gene variation, since the environmental influence was minimized. Thus, the role of SNPs in possible varied genes was further associated with flavonoid synthesis. Results showed that after long term environment and artificial influence, different accessions of G. biloba showed a varied ability in flavonoid aglycone synthesis due to some gene polymorphism; this difference may be heritable but not obvious. Compared with previous methods, this strategy is advantageous in accuracy, low sample requirement and easier operation, providing effective information in phenotype–genotype association, and it can also be used in the heritability study of artificial breeding before large-area introduction.
1. Introduction
Ginkgo biloba is the oldest gymnosperm. The leaves and standard extracts are consumed in both eastern and western countries as the best-selling functional food.1 G. biloba is widely distributed in China and long-term effects of natural conditions as well as artificial selections resulted in many cultivated varieties or strains. Generally, G. biloba was cultivated to show certain morphological traits, such as the size or shape of a kernel and leaf, as well as the texture of the wood.2 However, its commercial value is dependent on the content of the functional flavonoids, which play key roles in plant physiology as attractants that enhance pollination and seed dispersal, and as a part of the plant defense mechanism and also possess excellent health promotion abilities.3–6 In China, many efforts were put into breeding the strain of G. biloba that accumulates a higher concentration of total flavonoids. In G. biloba, most flavonoids belong to the glycosides of quercetin, kaempferol and isorhamnetin, and structural modifications of the flavonoid skeleton have produced different flavonoids. The types and concentrations of flavonoids may be varied among different cultivated varieties. Thus, it is crucial to know whether these cultivated varieties differed in their abilities to form target flavonoid metabolites, and whether this trait is decided by genetic variation or environmental conditions, because only inheritable traits can be introduced by other cultivars.4,5Direct screening of gene variation in the whole genome is extremely time- and labor-consuming. If evidence is available to predict the candidate locus of single nucleotide polymorphism (SNP), then we can use simpler methods like direct sequencing, Taqman makers or restriction fragment length polymorphism PCR to save time and resources.8,9 Metabolomics can reveal differently expressed metabolites which are reflective of plants grown in different environments or diseased.10,11 However, untargeted metabolomics may not provide detailed information for a concerned pathway due to non-individual optimized conditions. Metabolic flux analysis is an interesting tool to facilitate a better understanding of physiological processes. Isotopic enrichments of the metabolites are necessary to obtain metabolic flux, and this is an accurate way to map the overall cellular functions;12 however, this experiment is still hardly operative. Considering the matrix effect, few analyses were conducted for the quantification of plant metabolites, while in most cases, the matrix effect of a plant extract is much lower than in bio-samples. In addition, the matrix effect can also be minimized by using structurally similar internal standards, and with a paired analytical metabolomics that calculates and compares the ratio of product/substrate of the enzymes in a pathway, a mapping flux might also be conducted in an accurate and easy way.
In the present work, we proposed a reliable strategy to refine the application of metabolomics in a plant genome study; an overview of our scheme is shown in Fig. 1. This study carried out a common garden trial where target plants were grafted from different origins to minimize environmental influence. A “paired analysis” of targeted metabolomics, which comprehensively considers the conversion rate of each biosynthetic reaction in a pathway, was used to analyze the samples. The different cultivate varieties of G. biloba L. grafted from different origins since 1990 were selected as a case study of our experimental design. Thirty-six metabolites in the flavonoid biosynthesis pathway were profiled in each variety. Combining the proposed paired analytical strategy, metabolic flux and possible varied genes were predicted. Then the relationship between SNPs and flavonoid synthesis was further associated by partial least squares regression analysis and homology analysis. The results demonstrated that Ginkgo cultivate varieties could be divided into 3 clusters based on flavonoid synthesis ability and possible varied genes were predicted. Compared to a genome sequencing study, this method was accurate in method validation and has a low sample requirement, which proves that targeted metabolomics could also be applied as a convenient strategy in studies of plant physiology.
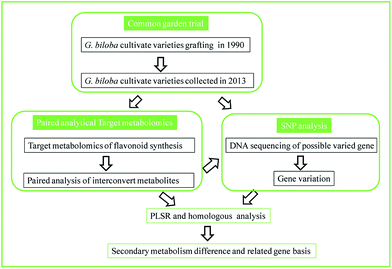 | ||
| Fig. 1 Scheme of the paired analytical metabolomics to bridge the association of metabolism and gene variation. | ||
2. Materials and methods
2.1 Plant materials
The tested materials were collected from National Ginkgo seed base in Pizhou (34°10′∼34°40′ N, 117°42′∼118°10′ E), Jiangsu, China. Tree branches with superior economic characters were introduced from the main Ginkgo producing areas since 1990, including Jiangsu, Shandong, Guangxi, Guizhou provinces, 2–3 meters high, 5 year-old seedlings were used as rootstocks, grafting site starting between 1.8–2 meters, each rootstock grafted 3–4 rootstock scions, each clone grafted 3–6 strains. The origin of the grafted strains is shown in Table S1.† Fresh leaves were collected from three branches from 30 accessions in the same time and the specimens were stored in the State Key Laboratory of Nature Medicines, China Pharmaceutical University.Kaempferol-3-O-rutinoside (Kaem-3-RU), isorhamnetin 3-O-rutinoside (Isor-3-RU), quercetin (Quer), kaempferol (Kaem) and isorhamnetin (Isor) were purchased from Zelang Medical Technology Co. Ltd. (Nanjing, China). Dihydromyricetin (Dmyri), dihydrokaempferol (DKaem), naringenin (Nari), protocatechuic acid, chlorogenic acid, catechin, caffeic acid (CA), procyanidin B2, epicatechin, p-coumaric acid (p-Coum), ferulic acid, clitorin, (−)-epigallocatechin (EGC), (−)-gallocatechin gallate (GCG), (−)-epicatechin gallate (ECG), epigallocatechin-3-gallate (EGCG), isorhamnetin-3-O-glucoside (Isor-3-G), quercetin-3-O-glucoside (Quer-3-G), apigenin-7-O-glucoside (Apig-3-G), quercetin-3-O-rhamnoside (Quer-3-R), kaempferol-7-O-β-D-glucoside, myricetin (Myri), luteolin (Lute), apigenin (Apig), quercetin-3-O-β-D-glucopyranosyl-(1–2)-α-L-rhamnoside (QGR), quercetin 3-O-2′′-(6′′-p-coumaroyl)-glucosyl-rhamnoside (QRCG), kaempferol 3-O-2′′-(6′′-p-coumaroyl)-glucosyl-rhamnoside (KRCG) and hesperidin (IS) were purchased from MUST Bio-technology Co. Ltd. (Chengdu, China). Ginkgetin, isoginkgetin, bilobetin, amentoflavone, sciadopitysin, hydroxybenzoic acid (HA) were purchased from Biopurify Co. Ltd. (Chengdu, China). Rutin and andrographolide were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). Syringetin was purchased from Extrasynthese (Genay Cedex, France). Dihydroquercetin (Dquer) was purchased from Phystandard bio-Tech Co., Ltd. (Shenzhen, China).
2.2 Metabolomics analysis of flavonoid biosynthetic pathway by UHPLC-QQQ-MS/MS
The powder of dried Ginkgo leaves was extract with 70% aqueous methanol, and diluted before MS analysis. The metabolomics analysis was performed by a Shimadzu LC-30AD UHPLC tandem Shimadzu 8050 (Shimadzu, Japan) QQQ-MS/MS. The mass spectrometer was operated in negative ion mode. The ion source parameters were set as: nebulizing gas flow, 3 L min−1; heating gas flow, 10 L min−1; interface temperature, 300 °C; DL temperature, 250 °C; heat block temperature, 500 °C; drying gas flow, 10 L min−1. Shimadzu Labsolution provided instrument control, data acquisition, and data processing. Analytes were quantified in multiple reaction monitoring mode – the selection of precursor ion, product ion, collision energy, Q1/Q3 pre bias was automatically optimized by Labsolution. The parameters are listed in Table 1. The sample vials were maintained at 4 °C in a thermostatic autosampler. The chromatographic separation was achieved on an Agilent Zorbax Extend C18, Narrow-bore column (2.1 × 150 mm, 5micron, 80 A). The mobile phase consisted of solvent A (H2O containing 1‰ formic acid) and solvent B, acetonitrile.| Metabolite | Retention time (min) | Precursor m/z | Product m/z | Q1 pre bias (V) | CE | Q3 pre bias (V) | |
|---|---|---|---|---|---|---|---|
| 1 | (−)-Epigallocatechin | 1.52 | 305.1 | 125.0 | 14 | 23 | 23 |
| 2 | p-Hydroxybenzoic acid | 3.22 | 137.1 | 93.0 | 28 | 17 | 17 |
| 3 | Chlorogenic acid | 3.40 | 353.2 | 191.1 | 25 | 14 | 19 |
| 4 | Catechin | 3.49 | 289.2 | 245.0 | 20 | 14 | 26 |
| 5 | Caffeic acid | 4.53 | 179.1 | 134.1 | 12 | 25 | 25 |
| 6 | Procyanidin B2 | 5.03 | 577.1 | 407.1 | 20 | 24 | 28 |
| 7 | Epicatechin | 6.20 | 289.1 | 245.1 | 20 | 15 | 26 |
| 8 | Dihydromyricetin | 6.31 | 319.1 | 193.1 | 22 | 12 | 20 |
| 9 | p-Coumaric acid | 7.53 | 163.1 | 119.1 | 11 | 16 | 22 |
| 10 | Clitorin | 9.31 | 739.2 | 284.1 | 20 | 45 | 29 |
| 11 | Quercetin-3-O-rutinoside | 9.75 | 609.2 | 300.1 | 20 | 38 | 30 |
| 12 | Dihydroquercetin | 9.85 | 303.1 | 285.0 | 21 | 13 | 30 |
| 13 | Quercetin-3-O-β-D-glucoside | 10.12 | 463.1 | 300.1 | 10 | 27 | 20 |
| 14 | Quercetin-3-O-β-D-glucopyranosyl-(1–2)-α-L-rhamnoside | 10.73 | 609.2 | 300.1 | 20 | 37 | 30 |
| 15 | Kaempferol-3-O-rutinoside | 10.83 | 593.2 | 285.1 | 20 | 32 | 30 |
| 16 | Isorhamnetin-3-O-rutinoside | 11.04 | 623.2 | 315.1 | 20 | 31 | 21 |
| 17 | Quercetin-3-O-α-L-rhamnoside | 11.23 | 447.1 | 300.0 | 10 | 26 | 20 |
| 18 | Isorhamnetin-3-O-glucoside | 11.41 | 477.1 | 314.0 | 10 | 28 | 20 |
| 19 | Kaempferol-7-O-β-D-glucoside | 11.44 | 447.1 | 285.0 | 12 | 24 | 30 |
| 20 | Apigenin-7-O-D-glucoside | 11.47 | 431.1 | 268.0 | 10 | 32 | 28 |
| 21 | Dihydrokaempferol | 11.84 | 287.1 | 259.1 | 20 | 14 | 28 |
| 22 | Myricetin | 11.98 | 317.1 | 151.1 | 11 | 24 | 30 |
| 23 | Quercetin-3-O-α-L-rhamnopyranosyl-2′′-(6′′′-p-coumaroyl)-β-D-glucoside | 12.62 | 755.0 | 300.1 | 20 | 47 | 30 |
| 24 | Kaempferol-3-O-α-L-rhamnopyranosyl-2′′-(6′′′-p-coumaroyl)-β-D-glucoside | 13.47 | 739.2 | 284.1 | 20 | 47 | 30 |
| 25 | Luteolin | 14.31 | 285.1 | 133.1 | 10 | 33 | 24 |
| 26 | Quercetin | 14.38 | 301.1 | 151.0 | 10 | 21 | 28 |
| 27 | Apigenin | 16.20 | 269.0 | 117.1 | 12 | 34 | 22 |
| 28 | Naringenin | 16.26 | 271.1 | 151.1 | 19 | 18 | 29 |
| 29 | Kaempferol | 16.64 | 285.0 | 93.0 | 10 | 35 | 17 |
| 30 | Syringetin | 16.66 | 345.1 | 315.0 | 12 | 26 | 21 |
| 31 | Isorhamnetin | 16.87 | 315.1 | 300.0 | 10 | 20 | 13 |
| 32 | Amentoflavone | 18.18 | 537.1 | 375.1 | 20 | 32 | 25 |
| 33 | Bilobetin | 19.34 | 553.1 | 521.0 | 40 | 31 | 36 |
| 34 | Isoginkgetin | 21.51 | 565.1 | 533.1 | 20 | 28 | 38 |
| 35 | Ginkgetin | 21.87 | 567.2 | 535.1 | 20 | 29 | 38 |
| 36 | Sciadopitysin | 25.51 | 579.2 | 547.1 | 28 | 27 | 26 |
| IS | Hesperidin | 11.68 | 609.1 | 301.1 | 22 | 25 | 20 |
The mobile phases were eluted at 0.5 mL min−1 with the gradient as follows: 0–5 min, 8–10% B; 5–15 min, 10–30% B; 15–20 min, 0–49% B; 20–23 min, 49% B; 23–25 min, 49–60% B; 25–26 min, 60–80% B; 26–28 min, 80% B; 28–30 min, 80–8% B; 30–32 min, 8% B. The injection volume was 2 μL. 36 metabolites in the flavonoid biosynthesis were quantified in this work. The analytical methods for all the compounds were validated for linearity, accuracy, precision, repeatability, stability and recovery. The data of regression equation and method validation were listed in Tables S2 and S3.†
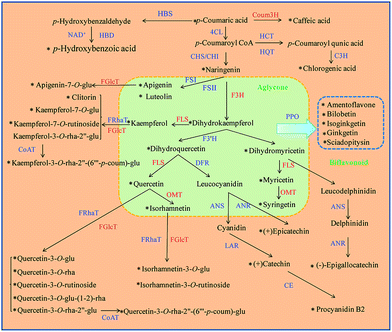
2.3 Paired analysis of flavonoid/modified flavonoid compared with individual compound evaluation
In order to analyze the conversion rate of each flavonoid biosynthetic reaction, the ratio of both flavonoid/modified flavonoid needed quantitation. Thus, we conducted an analysis of a nearly whole panel of flavonoid biosynthesis (Fig. 2). Conversion rates of eleven reactions were monitored, including: flavonoid reduction reaction at position 2–3 catalyzed by FLS (including the reactions between dihydrokaempferol/kaempferol, dihydroquercetin/quercetin and dihydromyricetin/myricetin); methylation of phenolic hydroxyl catalyzed by OMT (for quercetin/isorhamnetin, myricetin/syringetin), hydroxylation at position 3 catalyzed by F3H (the reaction between naringenin/dihydrokaempferol), hydroxylation at the benzene ring catalyzed by 4-coumarate 3-hydroxylase, Coum3H, (the reaction between p-coumaric acid/caffeic acid) and glycosidation catalyzed by FGlcT including the form of mono glycosides belonging to isorhamnetin, quercetin, kaempferol and apigenin.2.4 Single nucleotide polymorphism (SNP) analyses
The total DNA was isolated from Ginkgo leaves described above using the Plant Genomic DNA Kit (Tiangen Biotech Co., Ltd, Beijing, China). The assay principle is described online.13 Complete CDS of FLS were derived from the published data (GenBank: GQ994432.1).6,7 The primers used were purchased from GENEWIZ, Inc. (Beijing, China) as:FLS-ex1-for: 5′GGCAACTATCTTTGGAGTCGAG3′
FLS-ex1-rev: 5′GTCCATAGATCTGAAACAA3′
FLS-ex2-for: 5′AAATAAGCTACTGTCGGCGC3′
FLS-ex2-rev 5′TGTGCAGTGATCCATTTGTCA3′
FLS-ex3-for 5′CGTTCTGCACCGGAGTTTAG3′
FLS-ex3-rev 5′GTCTTGGCATTGTATAAGGGAGG3′
Amplification was carried out by two respective PCR programs (Bio-Rad, Hercules, CA, USA): FLS ex1 using 10 cycles of 95 °C for 30 s, 61 °C for 30 s, 72 °C for 30 s, and 32 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s with a final extension at 72 °C for 5 min. FLS ex2, ex3, using 10 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, and 32 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s with a final extension at 72 °C for 5 min. The PCR products were purified and sequenced by ABI 3730 (Majorbio, Shanghai, China) at GENEWIZ, Inc. (Beijing, China). The sequences were analyzed by SepManPro 7.1.
2.5 Data analysis
The deduced amino acids of homologous sequences of all exons in FLS from various varieties were aligned using MEGA 5.1 and clustalx.14 The figures were constructed by Graphpad prism 6.01. Multiple alignments and DNA translation were performed by DNAMAN 6.03. The concentration of flavonoid metabolites was illustrated by a heatmap using HemI 1.0.3.15To reveal the relationship between SNPs and the GbFLS activity, a partial least squares regression (PLSR) method was conducted to calculate the coefficients between 14 SNPs and the relative ratios of dihydrokaempferol/kaempferol, dihydroquercetin/quercetin and dihydromyricetin/myricetin in 30 Ginkgo cultivated varieties (Minitab 17.1.0, Minitab Inc., USA). PLSR finds a linear regression model by projecting the predicted variables and the observable variables. In this model, the SNP locus with one amino acid was set as “0” in the model, whereas the SNP locus with two heterozygous amino acids was identified as “1”. The ratios of dihydrokaempferol/kaempferol, dihydroquercetin/quercetin and dihydromericetin/myricetin were chosen as the “response”.
3. Results and discussion
3.1 Targeted metabolomics profiling of Ginkgo flavonoid biosynthesis pathway
Based on the scheme of flavonoid biosynthesis, a target metabolomics analysis of 36 flavonoids was achieved by UHPLC-QQQ MS, covering the most vital positions in the flavonoid as well as anthocyanidin and hydroxycinnamate biosynthesis. As we know, many factors will influence the accuracy of metabolomics. Among them, experimental design, controlled quenching of the metabolism, extraction from plant tissue and subsequent preparation remain the most crucial steps in carrying out a meaningful metabolomics study.16 In order to standardize the varied techniques and experimental designs for such studies, the metabolomics standards initiative has provided guidelines for metabolomics experiments.17 However, no analytical method can provide an absolute unbiased result because uncontrollable errors may happen in some necessary procedures. For the experimental design, individual differences must be minimized by a large amount of samples; usually hundreds to thousands of samples are needed. If the number of samples is limited, the outcome could be a false result. The metabolite extraction and sample preparation procedure can also easily result in errors.18Untargeted metabolomics usually provides a comprehensive identification of all the metabolites. It is limited in the sense that it is practically impossible to optimize the conditions for each metabolite. These include extraction solvents, solvents for redissolution and MS or NMR parameters. For instance, polar and non-polar compounds may not be extracted from tissues with the same solvent.
Thus, a target metabolomics method was chosen for the flavonoid biosynthesis study, and a total run of 32 min was carried out for quantitative analysis all 36 analytes. The method was well validated for each compound. As shown in Table S2,† each calibration curve was linear over the studied concentration ranges with satisfactory correlation coefficients (R2 > 0.99). The proposed method was sensitive with LOQs ranging from 0.001 to 16 ng mL−1 (Table S2†). Precision was evaluated using quality control solutions at three concentrations. The RSD values across the various concentrations were less than 13.97% for intra-day precision analysis while the inter-day precisions were less than 14.5%. The repeatability RSD values of all the compounds were less than 8.78%, indicating that the method was reliable and repeatable. The overall recoveries fell within the range of 85.15–113.38% after spiking high, middle and low amounts of analytes before extraction, with RSDs less than 6.69%.
Our strategy arouses several considerations to minimize the errors in metabolomics. Firstly, when a pathway is known to be significant for a plant physical or commercial value, targeted metabolomics is superior in accuracy to comprehensive metabolomics. In this work, 70% aqueous methanol was used to extract flavonoids of high polarity. MS data were acquired in the negative ion mode because flavonoids are easier to dehydrogenate, leading to higher responses in this mode. All the methods were strictly validated to ensure the quantitation for each metabolite was reliable, stable, repeatable and precise. Secondly, a self-controlled trial can provide a great improvement in credibility and reduce the necessary sample number. In disease or diagnostic metabolomics, a paired experimental design is more recommended than an unpaired comparison to reduce the sample amount required.19
The chromatograms of 36 flavonoids in G. biloba leaves are shown in Fig. 3A. In Fig. 3B, the results showed that flavonol glycosides and biflavonoids were the major compounds in all leaves, while few aglycones and organic acids were present. The individual level of flavonoids among Ginkgo accessions were also different, such as TM-2, TC231 and SNFS showed a higher content of flavonol glycosides such as rutin, while YZYX and GL-9 proved to have more biflavonoids.
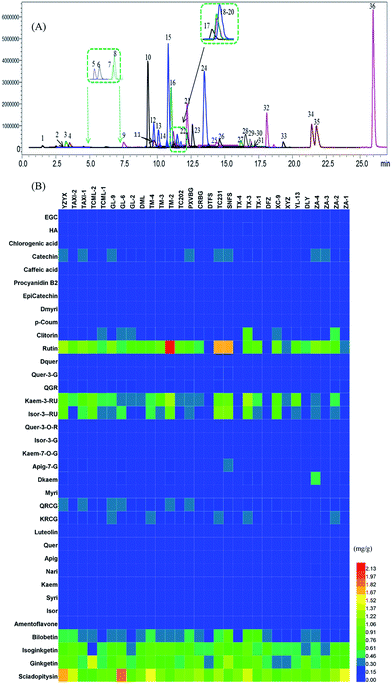 | ||
| Fig. 3 (A) The chromatograms of 36 flavonoids of Ginkgo leaf samples in a multiple reactions monitoring mode. The numbers of the peaks are according to Table 1. (B) The concentrations of 36 flavonoid metabolites in 30 Ginkgo cultivated varieties. | ||
3.2 Differences in conversion rate of flavonoid biosynthesis among Ginkgo cultivated varieties
In a plant physiology study, the environmental influence or genetic change needs considerable time, so a self-controlled trial is difficult to conduct. In this work, we evaluated the conversion rate of flavonoid biosynthesis by calculating the relative ratio of flavonoid/modified flavonoid in many flavonoid biosynthetic reactions, and all the data compared were from the same plant. This is also similar to a paired design to minimize individual errors.Fig. 4 shows individual levels and relative ratios of the flavonoids involved in the reactions catalyzed by FLS, OMT and FGlcT. It shows that the absolute amount of flavonoids is extremely varied among different samples (Fig. 4A–C), so it is difficult to find their internal relationship. However, as each flavonoid metabolite was formed by the catalysis of a certain enzyme, the conversion rate in a reaction may indirectly reflect the activity of a target enzyme. Fig. 4A shows the individual level of aglycones and their glycosides, and their relative ratios are presented in Fig. 4B. We can see inconsistencies with the two results, for example in Fig. 4A and B, a sample with a higher glycoside level may not always have a higher glycoside/aglycone ratio, accession 9 had a middle level of Kaem-7-G, but the ratio versus kaempferol was the highest, which indicated this accession may have a high ability to synthesize glycosides. In addition, some reactions, such as that catalyzed by FGlcT (Fig. 4A and B), FLS (Fig. 4C and D) and OMT (Fig. 4E and F), have more than one substitute; the paired analysis also indicated that when the ratio of different substitutes changed consistently, the activity or expression level of that enzyme may simply increase or decrease. If this change was different in a sample, we might predict that the selectivity of the substitute varied. The phenomenon may be similar to the turnover of neurotransmitters in some human diseases.20 Fig. 4E and F indicate that the selectivity on quercetin and myricetin of methyltransferase (OMT) was not changed in most samples, but the selectivity of FLS and FGlcT differed in many cases. Conventional metabolomics evaluate the compound level just based on the absolute concentration or response. However, a higher response of metabolite cannot result from the different enzyme catalytic abilities or further association with gene variation due to the individual plant differences. A systematic analysis of the conversion rate is necessary to find the relationship between metabolomics result and gene variation.
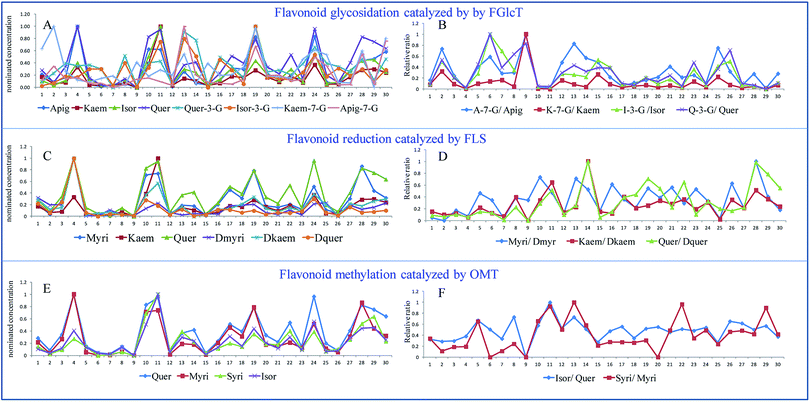 | ||
| Fig. 4 The nominated concentrations (A, C and E) and their relative ratios (B, D and F) of the flavonoids involved in the reactions catalyzed by FLS, OMT and FGlcT of 30 Ginkgo cultivated varieties. | ||
3.3 Clustering of Ginkgo cultivated varieties based on the flavonoid forming rate
As discussed earlier, the conversion rate may give more reliable information than the single compound level. In order to show the metabolic flux change in the flavonoid synthesis, Ginkgo cultivated varieties were clustered based on these 11 monitored reactions (K-means clustering). Three clusters were assumed in this work, the final cluster membership and centers are listed in Tables 2 and 3. The accessions in cluster 1 were characterized by lower levels of flavonoid glycosides, with the ratio of glycosides/aglycone ranging from 1.29 to 5.73. Correspondingly, these accessions showed high levels of aglycone due to lower glycosylation. Accessions in cluster 2 contained moderate levels of most flavonoid synthetic ability. Only one accession in cluster 3, because trace amounts of aglycone was detected, was associated with an extremely high level of glycoside/aglycone ranging from 14.47 to 27.77. (The ratio of Isor-3-G/Isor cannot be calculated because Isor was not detected.) Clustering showed a metabolic flux preference to glycoside production and also showed a lower reverse rate of aglycone/dihydroflavonoid, such as the samples in cluster 3, which indicated a high expression or enzyme ability of FGlcT and FRhaT. This difference probably resulted from their gene variations, since they were grown in the same garden. Ginkgo is a highly conserved species, but it has been planted for thousands of years in extremely different environments, and long-term environmental influence may result in some gene variations.21| Strain | Cluster | Distance | Strain | Cluster | Distance |
|---|---|---|---|---|---|
| ZA-1 | 1 | 2.82 | CRBG | 2 | 14.31 |
| ZA-2 | 2 | 5.64 | PXVBG | 1 | 2.19 |
| ZA-3 | 1 | 3.62 | TC202 | 1 | 2.19 |
| ZA-4 | 1 | 4.71 | TM-2 | 1 | 3.13 |
| DLY | 2 | 4.86 | TM-3 | 1 | 1.32 |
| YL-13 | 2 | 17.24 | TM-4 | 1 | 5.40 |
| XYZ | 2 | 6.71 | DML | 1 | 2.69 |
| XC-9 | 2 | 5.51 | GL-2 | 2 | 3.99 |
| DFZ | 3 | 0.00 | GL-6 | 1 | 1.91 |
| TX-1 | 1 | 3.59 | GL-9 | 2 | 6.80 |
| TX-3 | 1 | 4.23 | TCML-1 | 2 | 6.72 |
| TX-4 | 1 | 7.80 | TCML-2 | 1 | 3.09 |
| SNFS | 2 | 6.99 | TAXI-1 | 1 | 3.61 |
| TC231 | 1 | 8.55 | TAXI-2 | 1 | 4.94 |
| DTFS | 2 | 5.41 | YZYX | 1 | 2.42 |
| Cluster | |||
|---|---|---|---|
| 1 | 2 | 3 | |
| CA/p-Coum | 1.62 | 2.42 | 0.37 |
| Dkaem/Nari | 3.77 | 3.91 | 2.54 |
| Myri/Dmyri | 1.97 | 1.92 | 1.88 |
| Kaem/Dkaem | 0.07 | 0.05 | 0.02 |
| Quer/Dquer | 0.89 | 0.37 | 0.14 |
| Isor/Quer | 0.77 | 0.74 | 0.00 |
| Syri/Myri | 0.14 | 0.10 | 0.00 |
| Apig-7-G/Apig | 3.50 | 7.27 | 14.47 |
| Kaem-7-G/Kaem | 1.29 | 3.66 | 23.29 |
| Isor-3-G/Isor | 2.39 | 8.39 | 0.00 |
| Quer-3-G/Quer | 5.73 | 18.21 | 27.77 |
3.4 Single nucleotide polymorphism (SNP) analyses of possible varied gene
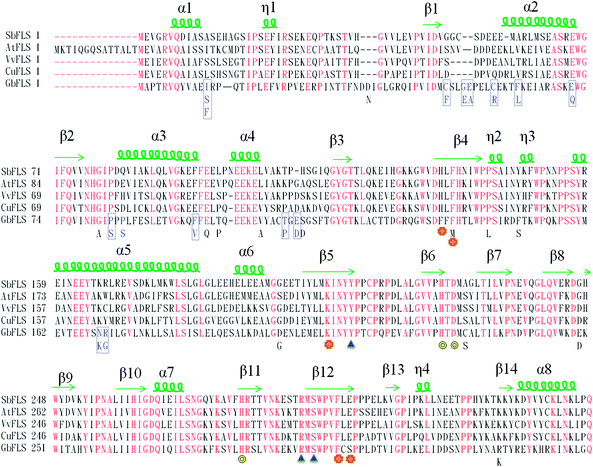
We intended to investigate whether these Ginkgo cultivated varieties had inheritable variations in flavonoid accumulation. Since the influence of the environment was minimized by using a common garden trial, we assumed that there may be possible genetic variations in corresponding enzymes. Based on this assumption, GbFLS was used as a case study because FLS plays a central function at the branch point of the flavonol derivative metabolism.22,23 The GbFLS gene in different cultivated varieties was sequenced and the coding SNP (cSNP) was analyzed. As in our prediction, 39, 9 and 5 SNPs were found, respectively, in three exons of GbFLS. Compared with the reported sequences, most of them were consistent in the test samples and turned out to be heterozygous mutations. These results were consistent with the report that FLS was encoded by a multi-copy gene in plants.24 Since most of these SNPs were heterozygous mutations, the sequence was only partly reported.25 After alignment, 34, 4 and 2 mutations were proven to be “truly” SNPs in these cultivated varieties. Among these SNPs, 12 and 2 SNPs were found as non-synonymous mutations in exons 1 and 2 of the GbFLS gene. There was no SNP in amino acid difference in the exon 3.To define the central role of these amino acid substitutions, in Fig. 5, the GbFLS cDNA sequence was translated into an amino acid sequence and then aligned with Scutellaria Baicalensis FLS (SbFLS), Vitis Vinifera FLS (VvFLS), Citrus Unshiu FLS (CuFLS) and Arabidopsis thaliana FLS (AtFLS). GbFLS is highly homologous with described proteins. The structure of FLS is composed of a β jelly-roll fold surrounded by ten different length α-helix. The iron metal may ligate with a bidentate group of cosubstrate 2-oxoglutarate on 3 side-chain residues, H228, D230 and H282, which conserved all FLS proteins. Hydrogen bonds may form between the other side of 2OG with 3 conserved residues, Y211, R292 and S294. The substrate might locate near an iron metal ion, in the possible residues of H137, F139, K207, F298 and E300.26 These substrate and catalyzed residues may play a vital role in the function of GbFLS. From Fig. 5 we can see no SNPs occurred in these positions or neighboring alleles, which indicates that these gene variations might not heavily influence the enzyme function.
Actually, 12 of 14 SNPs were found in exon 1, which is far from the substrate and catalyzed residues of GbFLS in the exon 2 and exon 3 (this region is still conserved). However, some of those SNPs located in the α-helices of exon 1 might affect the GbFLS structure and activity, including F62/L62, E71/Q71, P85/S85, F97/V97, N169/K169 and R170/G170, so this indicated the probability of the enhancement or inhibition of enzyme activity in the formation of quercetin, kaempferol and isorhamnetin. The plot of the PLSR coefficient against predictors (14 SNPs, predictors 1–14, represent the 14 SNPs according to the order in Fig. 5) is shown in Fig. 6. No SNP showed extremely higher coefficients than others, which may be due to the absence of SNPs in the catalyzing or substrate binding allele.
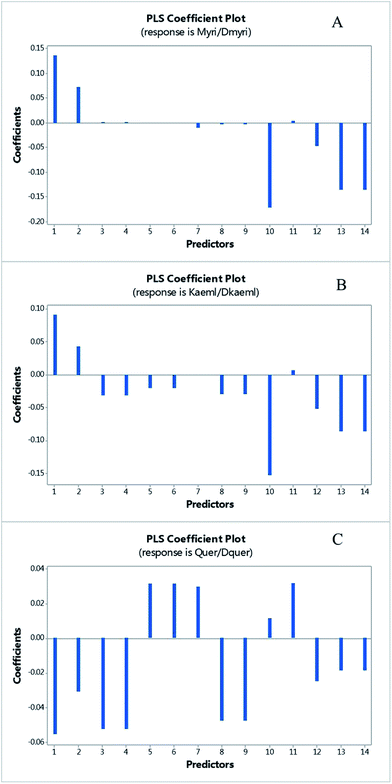 | ||
| Fig. 6 The plot of PLS coefficient versus predictors of myricetin/dihydromyricetin (A), kaempferol/dihydrokaempferol (B) and quercetin/dihydroquercetion (C). The predictors 1–14 represent the SNPs among these cultivated varieties in order with Fig. 5. | ||
Among them, predictor 1, 10, 13 and 14 showed a relatively significant influence on the synthesis of myricetin and kaempferol, which represented the SNPs of I12/F12/S12, F97/V97, N169/K169 and R170/G170, respectively. Three of these SNPs located in an allele of the protein form α-helices, which may have a higher possibility to influence the structure and function of GbFLS. The results indicated the probability of an enhancement or inhibition of enzymes in the formation of quercetin, kaempferol and isorhamnetin. This may contribute to FLS displaying variable substrate preferences and loose catalytic activities. For instance, in Fig. 6, predictors 1 and 2, which correspond to the mutation from I12 to F12/S12 and C50 to F50, showed a positive influence at converting dihydrokaempferol to kaempferol and dihydromyricetin to myricetin, but negatively regulated the reaction of converting dihydroquercetin to quercetin. This mechanism may be similar to a number of mutations of C. unshiu FLS reported in in vitro studies.27 This FLS had a higher affinity for dihydrokaempferol than for dihydroquercetin. A functional FLS cloned from Z. mays also showed a different Km for converting dihydrokaempferol and dihydroquercetin.28
The FLS protein in the substrate binding residue is evolutionarily conserved, including the residue ligate with iron metal, 2-oxoglutarate and flavonoid. Mutations occurring in these alleles may significantly affect the function of GbFLS. A SNP in these residues can be used as a general marker in Ginkgo breeding for improving nutritional quality and in the formation of target flavonoids.29,30 In addition, technology, like genome-editing, can be performed based on these potential functional SNPs as a novel control for the precise alteration of native genomes.31,32 Genome-editing technology may be used to specifically edit a single nucleotide, resulting in the change of the evolutionarily conserved GbFLS proteins in Ginkgo cultivated varieties. This could then trigger functional flavonoid biosynthesis without any visible deleterious effect on plant performance.
4. Conclusions
In summary, compared with untargeted metabolomics, a paired analytical targeted metabolomics study can provide a higher accuracy by individual method optimization and self-controlled calibration. Comprehensive analysis of the conversion rate of biochemical synthesis can provide more information of plant physiology similar to a metabolic flux analysis. In addition, combining a common garden trial with accurate quantitation of a complete pathway panel meant it was easier to associate the gene variation with the metabolite level. In this work, we proved that after long term environmental and artificial influence, different accessions of G. biloba showed a varied ability in flavonoid aglycone synthesis. The proposed strategy can be used as a pre-experiment or a complementary study to genome screening in plant physiology which was useful in the heritability study of other artificial breeding before large-area introduction. Based on the potential functional SNPs found by this method, genome-editing can be performed to precisely control the alteration of artificial varieties for different purposes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported in part by the Natural Science Foundation of Jiangsu Province (Grant No. BK20140673), the National Natural Science Foundation of China (Grant No. 81403043), the Qing Lan project and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The authors thank Prof. Fu-Liang Cao and Dr Wan-Wen Yu from Nanjing Agriculture University for providing plant materials.Notes and references
- T. A. Van Beek and P. Montoro, J. Chromatogr. A, 2009, 1216, 2002–2032 CrossRef CAS PubMed
.
- F. L. Cao, An Illustrated Monograph of Ginkgo Biloba L. Cultivars in China, Science Press, Bejing, China, 2011 Search PubMed
.
- P. Mukhopadhyay and A. K. Prajapati, RSC Adv., 2015, 5, 97547–97562 RSC
.
- S. A. Mohamed, M. A. Awad, E. F. A. El-Dengawy, H. M. Abdel-Mageed, M. O. El-Badry, H. A. Salah, A. M. Abdel-Aty and A. S. Fahmy, RSC Adv., 2016, 6, 44814–44819 RSC
.
- H. T. Maria, et al., New Phytol., 2013, 198, 1085–1095 CrossRef PubMed
.
- Y. Z. Pang, Molecular cloning and characterization of important genes involved in the biosynthetic pathways of flavonoids and terpenoids from Ginkgo biloba L., PhD thesis, Fudan University, Shanghai, China, 2005
.
- C. Zhang, et al., Acta Hortic. Sin., 2012, 39, 355–362 CAS
.
- J. H. Li, et al., New Phytol., 2015, 208, 137–148 CrossRef CAS PubMed
.
- L. Mehli, T. D. Kjellsen, F. M. Dewey and A. M. Hietala, New Phytol., 2005, 168, 465–474 CrossRef CAS PubMed
.
- Y. Wang, S. Y. Liu, Y. J. Hu, P. Li and J. B. Wan, RSC Adv., 2015, 5, 78728–78737 RSC
.
- Q. Liang, H. Liu, L. X. Xie, X. Li and A. H. Zhang, RSC Adv., 2017, 7, 2587–2593 RSC
.
- S. Acket, A. Degournay, F. Merlier and B. Thomasset, Anal. Biochem., 2017, 527, 45–48 CrossRef CAS PubMed
.
- http://www.tiangen.com/?productShow/t1/1/id/15.html.
- K. Tamura, K. Peterson, N. Peterson, G. Stecher, M. Nei and S. Kumar, Mol. Biol. Evol., 2011, 28, 2731–2739 CrossRef CAS PubMed
.
- W. K. Deng, Y. B. Wang, Z. X. Liu, H. Cheng and Y. Xue, PLoS One, 2014, 9, e111988 Search PubMed
.
- K. Saito and F. Matsuda, Annu. Rev. Plant Biol., 2010, 61, 463–489 CrossRef CAS PubMed
.
- N. Morrison, D. Bearden, J. G. Bundy, T. Collette, F. Currie, M. P. Davey, N. S. Haigh, D. Hancock, O. A. H. Jones, S. Rochfort, S. A. Sansone, D. Stys, Q. Teng, D. Field and M. R. Viant, Metabolomics, 2007, 3, 203–210 CrossRef CAS
.
- V. Ossipov, S. Ossipova, V. Bykov, E. Oksanen, J. Koricheva and E. Haukioja, Metabolomics, 2008, 4, 39–51 CrossRef CAS
.
- X. E. P. Rhee, C. B. Clish, K. A. Pierce, A. Saad, L. O. Lerman and S. C. Textor, J. Hypertens., 2015, 33, 836–842 CrossRef PubMed
.
- X. Zheng, et al., Anal. Chem., 2012, 84, 10044–10051 CrossRef CAS PubMed
.
- N. Morrison, D. Bearden and J. G. Bundy, et al., Metabolomics, 2007, 3, 203–210 CrossRef CAS
.
- C. L. Li, et al., Cloning, J. Agric. Food Chem., 2012, 60, 5161–5168 CrossRef CAS PubMed
.
- T. T. Vu, et al., J. Agric. Food Chem., 2015, 63, 7819–7829 CrossRef CAS PubMed
.
- D. K. Owens, et al., Plant Physiol., 2008, 147, 1046–1061 CrossRef CAS PubMed
.
- F. Xu, et al., Mol. Biol. Rep., 2012, 39, 2285–2296 CrossRef CAS PubMed
.
- B. G. Kim, E. J. Joe and J. H. Ahn, Biotechnol. Lett., 2010, 32, 579–584 CrossRef CAS PubMed
.
- F. Wellmann, R. Lukačin, T. Moriguchi, L. Britsch, E. Schiltz and U. Matern, Eur. J. Biochem., 2002, 269, 4134–4142 CrossRef CAS PubMed
.
- M. L. F. Ferreyra, et al., Plant J., 2010, 62, 77–91 CrossRef CAS PubMed
.
- C. S. Chua, D. Biermann, K. S. Goo and T. S. Sim, Phytochemistry, 2008, 69, 66–75 CrossRef CAS PubMed
.
- G. Tzuri, X. J. Zhou, N. Chayut, H. Yuan, V. Portnoy, A. Meir, U. Sa'ar, F. Baumkoler, M. Mazourek, E. Lewinsohn, Z. J. Fei, A. A. Schaffer, L. Li, J. Burger, N. Katzir and Y. Tadmor, Plant J., 2015, 82, 267–279 CrossRef CAS PubMed
.
- J. Miao, D. S. Guo, J. Z. Zhang, Q. P. Huang, G. J. Qin, X. Zhang, J. M. Wan, H. Y. Gu and L. J. Qu, Cell Res., 2013, 23, 1233–1236 CrossRef CAS PubMed
.
- Q. W. Shan, Y. P. Wang, J. Li, Y. Zhang, K. L. Chen, Z. Liang, K. Zhang, J. X. Liu, J. Z. Jeff Xi, J. L. Qiu and C. X. Gao, Nat. Biotechnol., 2013, 31, 686–688 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06229j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |