Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
One-step trapping of droplets and surface functionalization of sensors using gold-patterned structures for multiplexing in biochips†
T. M. Dias *abc,
E. Fernandesd,
S. Cardosocd,
G. Monteirobc and
P. P. Freitasad
*abc,
E. Fernandesd,
S. Cardosocd,
G. Monteirobc and
P. P. Freitasad
aINESC Microsystems and Nanotechnologies, Instituto de Nanociencias e Nanotecnologias, Rua Alves Redol, 1000-029 Lisbon, Portugal. E-mail: tdias@inesc-mn.pt
bIBB–Institute for Bioengineering and Biosciences, Av. Rovisco Pais 1, 1049-001 Lisboa, Portugal
cInstituto Superior Tecnico, Universidade de Lisboa, Av. Rovisco Pais 1, 1049-001 Lisbon, Portugal
dInternational Iberian Nanotechnology Laboratory, Av. Mestre José Veiga, 4715-330, Braga, Portugal
First published on 7th September 2017
Abstract
Multiplexing in Point-of-Care (POC) diagnostics is crucial for a comprehensive readout of multifactorial diseases such as cancer or fast-acting complications such as heart attacks. In multiplexed POC (MPOC) testing, groups of sensors are functionalized with different biological probes, enabling the simultaneous capture and detection of various analytes. Current literature demonstrates MPOC systems using microfluidics, capable of compartmentalizing only a few sensing regions. In contrast, miniaturized robotic spotting (microspotting) theoretically enables the independent functionalization of every sensor in a microarray. However, its use is still challenging in biosensors which require the combination of different materials at the surface because microspotted droplets tend to move to areas of higher surface energy, thus affecting the spotting precision. To counteract this phenomenon, we have combined shapes of gold-patterned films for the functionalization of magnetoresistive (MR) sensors and simultaneous trapping of droplets over the sensing regions, previously passivated with silicon nitride (Si3N4). Due to the higher hydrophobicity of gold when compared to Si3N4, the droplets remain immobilized inside the gold frames and functionalization of the sensors is accurately achieved. With this strategy, we demonstrate the microspotting of different DNA probes and antibodies and the specific hybridization and binding of complementary DNA targets and proteins, attached to magnetic beads. This combined trapping and functionalization system shows promise in the implementation of MR-based systems in multiplexing applications. This work can also be of interest to the wider community working in the development of other surface-based assays, also developed through microfabrication techniques, as the same trapping methodology is conceivable for adaption to other biochip formats.
1. Introduction
The implementation of microsystems capable of multiple analyte detection from a single sample is gaining increased importance at the Point-of-Care (POC). A recent review published by Gerald A. Urban and co-workers broadly highlights the importance of such systems in POC applications.1 In comparison to tests performed in centralized laboratories, the use of miniaturized POC systems enables on-the-spot testing where the patient needs, rapid analysis, the use of a smaller volume of reagents/samples which is, in general, associated with lower costs.2,3Because of these advantages, miniaturized POC systems are gaining increased interest in applications where time is critical, for instance, in the emergent triage of imminent diseases (e.g. heart attacks);4 regions where laboratory facilities are nonexistent and resources are low (e.g., developing countries);5 or as a methodology for analysis of multifactorial diseases such as cancer for rapid treatment decision.6 In most of complex diseases, the diagnosis of a single biomarker does not provide a definitive diagnostic and multiplexing approaches are required.7
Several bio-sensing technologies can be adapted for this purpose and many formats are available. Although some exceptions exist, most of the sensor methodologies are broadly classified into label-based and label-free. In our group, we research the potential implementation of magnetoresistive (MR) sensors in bio-sensing applications. MR sensors detect target entities when attached to magnetic nanoparticles (MNPs) and are included in the classification of label-based systems.8 When compared to traditional label-based technologies, usually employing optical labels (e.g. colorimetric, fluorescent or chemiluminescent), MR sensors offer advantages in terms of sensitivity because biological entities and bio-fluids (blood, urine, serum) naturally lack any detectable magnetic content and thus, magnetic signals are obtained solely from magnetically-labeled targets.9 In contrast, the excitation and emission spectra of fluorescent or chemiluminescent labels may overlap with different biomolecules or optical properties of the media, generating a background signal, diminishing its sensitivity, usually problematic in the analysis of low abundant species from highly complex samples.10 Label-free methods, such as Surface Plasmon Resonance (SPR), Quartz Crystal Microbalances (QCM) or electrochemical sensors, present outstanding limits-of-detection (LOD) and provide real-time readings.11 However, as duly noted by J. L. Arlett ad co-workers, the detection of a given analyte mixed in a pool of other molecules, i.e. a complex sample, is not straightforward and the biological noise floor (signal created from the sum of all the noise sources) largely arises from the non-specific binding (NSB) of interfering molecules to the surface, possibly leading to false-positives.12
Thus, apart from surface recognition, target labeling is still desirable as it grants a second degree of specificity, making the use of MR biochip-based platforms appealing in bio-sensing. MR sensors can easily be integrated in cost-effective and portable systems, providing real-time measurements with high sensitivity and specificity, similarly to the aforementioned techniques.13,14
Because MR sensors are fabricated using typical integrated circuits (IC) technologies, and in the scope of this work, hundreds or thousands of biochips can be fabricated simultaneously and each biochip can contain as well hundreds to theoretically thousands of sensors,15,16 opening the possibility of truly developing multiplexed POC (MPOC) systems for diagnostic purposes. In this case, each sensor or group of sensors in an array, must be functionalized with different DNA capture probes, aptamers or antibodies, enabling the simultaneous capture and detection of various analytes.
During the last years, various MR biochip formats were reported for the detection of different molecules such as nucleic acids, proteins, chemical analytes, or other biological entities such as bacteria, microvesicles or human cells.17–21 However, although arrays of dozens to hundreds of MR sensors have been fabricated and reported, the simultaneous multiplexed detection of analytes has not been yet deeply explored. Current literature demonstrates MPOC MR biochip systems suitable for a combinatorial analysis of few samples, aided by the integration of microfluidics as compartmentalization modules.17,22,23 Extensive examples comprising other sensing technologies have also been described, including the already mentioned optical, SPR and electrochemical sensors.24–26 Nonetheless, the focus of this work centers on expanding the use of MR sensors for multiplexing purposes due to their large-scale fabrication ease.
Contrarily to the use of microfluidics, assisted miniaturized robotic spotting (microspotting), enables the independent biomolecular functionalization of every sensor in an array since it surpasses the space limitations inherent to the integration of microfluidics with micropatterned sensors.
Compared to traditional liquid dispensing systems, microarray spotters allow one to dispense extremely small volumes (50 pL to 950 nL) of biological samples, in microtiter plates, on glass slides or on polymer-based chips. Microspotters have proven to be successful and well adapted for the preparation of conventional DNA microarrays and cell-based matrixes, allowing to save resources and maximize the number of assays per experiment.27,28 However, the integration of microspotting with most of the emergent microsystem technologies present a few challenges. All integrated sensing devices, either MR sensors or other microfabricated technologies, require the deposition of different thin films and/or micropatterns at the surface for functionalization and passivation, amongst other functions.29–32 The presence of different materials gives rise to different wettability properties along the surface area of a chip. Moreover, at the microscale, droplets are spherical and surface tension forces are predominant, making the droplets to move for areas of higher surface energy (where attractive forces are predominant), thus affecting greatly the spotting precision.33–36
To design a stable microspotting methodology, we have combined the deposition of gold pads over the sensors, commonly reported by our group as the element of choice for bio-functionalization,13,20,21 to design as well, patterned gold traps to enclose the dispensed droplets on top of the sensing sites, achieving both functions in one step. In various sensing methodologies (e.g. SPR, electrochemistry, Bio-FETs), gold surfaces are commonly used as the material of choice for functionalization. As an example, microspotting of molecules on gold surfaces of SPR sensors was previously performed.37 We want to demonstrate, nonetheless, as a step further in the field, the effect of gold traps in terms of efficient droplet separation and fixation, compared to similar structures, free of barriers, and using the same methodology. For this purpose, samples of DNA probes and antibodies are used and, after microspotting, the specific hybridization/binding to complementary target DNA or proteins, previously attached to magnetic beads, is assessed. We start our study with manual spotting using a DNA probe-target model previously reported by our group, for validation and translation to our microspotting technique. Our study is then expanded for the validation of antibody-protein interactions, using in overall two different robotic systems and two types of magnetic nanoparticles (MNPs), namely streptavidin-coated dextran-based 250 nm and streptavidin-coated bionized-nanoferrite Texas-Red Fluorescent 100 nm MNPs.
2. Materials and methods
2.1 Gold patterned substrates and MR sensors microfabrication
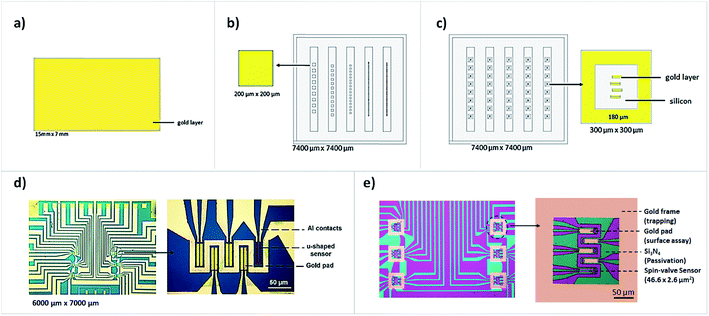
For spotting optimization and validation, three different substrates made of a bilayer of Ti 5 nm/Au 40 nm were fabricated. One consisted of a monolayer of bare gold, diced in 7 × 15 mm2 individual pieces (Disco, DAD-321) (Fig. 1a), and two were previously passivated with silicon nitride (Si3N4) 300 nm, defined by lift-off and diced in 7.4 × 7.4 mm2 individual pieces (Fig. 1b and c). The micropatterned substrates consisted of (b) individual gold squares with various dimensions (200 × 200 μm2; 100 × 100 μm2; 50 × 50 μm2; 20 × 20 μm2 and 10 × 10 μm2) and (c) several structures of four 10.9 × 40 μm2 patterns of gold, mimicking the gold pads deposited on top of MR sensors, enclosed by gold frames of 300 × 300 μm, 60 μm2 wide. All substrates were meticulously rinsed with isopropanol (IPA), ultra-pure milli-Q grade water and blown-dried with compressed air. For further cleaning from residual organic contaminants, ultraviolet light/ozone plasma was applied for 15 min at 28 mW cm−2 (UVO cleaner, Jelight™, USA). Two biochip sensor designs were fabricated, with 30 MR sensors each, arranged in arrays of six groups of five sensors with the following stack deposited by ion beam deposition in a Nordiko 3000 tool: Ta 1.5/NiFe 3.6/CoFe 2.5/Cu 2.1/CoFe 3.3/MnIr 11/Ta 10 (thicknesses in nm) as described by Dias et al. 2016.21 Each group of MR sensors are passivated with a Si3N4 layer of 300 nm and both biochip configurations include four “bioactive” sensors, with the described gold layer (40 nm) deposited on top, plus a reference sensor, without gold and inert for our functionalization chemistry (based on thiol–gold interaction) (Fig. 1d). Biochips were also fabricated with a patterned gold frame of 300 × 300 μm2, 60 μm2 wide enclosing MR sensors in its interior (Fig. 1e).2.2 Robotic arraying systems
Two microspotting systems were used: (1) the Nanoplotter NpC2.1 (GeSim™) and (2) the SciFlexArrayer S3 (Scienion™). Although each system comprises its own user interface specifications, both are composed by: (1) a liquid handling station; (2) the automated robotic apparatus and (3) humidity and temperature controllers. Their mechanism of operation relies on a piezoelectric dispensing non-contact system capable of handling ultra-low volumes of samples (nano-to picoliter range). The liquid handling station includes system liquid reservoirs, tubing and pumps connected to the piezo-dispensing pipettes used for the automatic washing of the system and pipette tips, controlled aspiration of the required volume of samples and spotting onto pre-designated sites at a substrate's surface. Both automated robotic apparatus are enclosed by a lid, for environment control, which accommodates different modules: a target tray for handling and positioning of substrates for spotting; a washing and a drying station for maintenance of the piezoelectric pipette tips; a robotic xyz-stage arm where the tips are installed, a cooled microplate holder where 96- or 384-well plates are positioned to store the samples used for spotting in different wells. Additionally, each system comprises a stroboscope and water sensitive paper to monitor or optimize the drop size, drop velocity and drop trajectory and a microscope connected to a CCD camera for image visualization and acquisition of the formed spots. In the Nanoplotter NpC2.1, a z-sensor tip may be temporally connected to measure the height distance before placing the piezoelectric pipette in the xyz stage. The z distance was adjusted to a separation height of 5297 μm in the SciFlexArrayer S3 and of 42.56 μm in the Nanoplotter NpC2.1. A piezoelectric pipette capable of dispensing 50 pL up to 600 pL per droplets was used in the Nanoplotter NpC2.1 (Pico-Tip J, GeSim) while in the sciFlexArrayer a pipette capable of dispensing 50 pL up to 800 pL per droplets was used (sciDrop Pico, SciFlex). Both microspotters are controlled by software products developed by each of the providers. Spotting plans are created and read by the robotic apparatus which control all aspects related to the motion of the xyz-stage arms and liquid handling stations for each microspotting procedure.2.3 Microspotting process
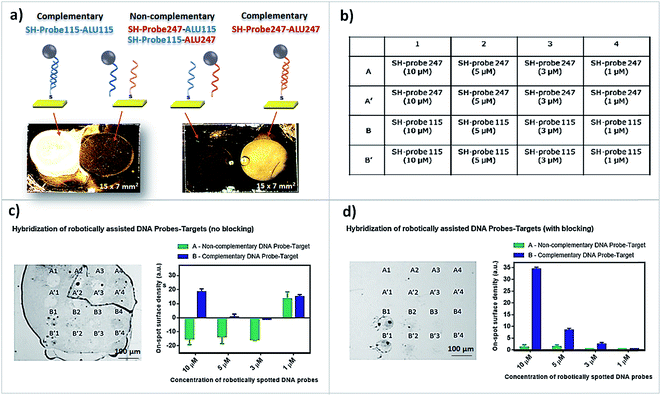
Although picoliter volumes of samples are dispensed onto each of the substrates, for a precise aspiration of the samples into the pipettes, it is recommended to have a substantial volume of samples available. Therefore, prior to the microspotting procedure, 100 μL of the DNA probe solutions or antibodies, at the different concentrations used, were dispensed into different wells of the microtiter plates. TRIS-10 mM-EDTA 1 mM (TE buffer) complemented with 5% glycerol was the buffer used for oligonucleotide surface immobilization while for the immobilization of antibodies, phosphate buffer 10 mM pH 7.4 (PB buffer), supplemented with 5% glycerol was used. Glycerol is important to avoid the evaporation of the droplets after spotting.In this work, two oligonucleotide strands, serving as DNA probes, were synthesized and purchased from StabVida Portugal, and designated here as SH-Probe115:
(5′SH-TTTTTTTTTTTTTTTGTAGGTACCCTGAGGTCAGG-3′) and SH-Probe247: (5′SH-TTTTTTTTTTTTTTTGATCTGCAGTGGCTCACGCCTGT-3′). They are complementary, respectively, to DNA fragments ALU115 and ALU247, as previously validated by manual spotting (T. Dias et al., 2016).21 The oligonucleotide DNA probe samples were diluted sequentially to 10 μM, 5 μM, 3 μM and 1 μM in TE-buffer and used for the microspotting, immobilization and hybridization assays on gold-patterned substrates and biochips. Purified anti-human fibronectin antibody was microspotted over prepared biochips for the capture of cellular fibronectin (c-Fn) and later detection using a biotinylated anti-human fibronectin antibody attached to magnetic beads in a magnetic sandwich-like assay. A non-related human fibronectin antibody anti-human platelet derived growth factor-CC (PDGF-CC) was also microspotted and used as a negative control (all purchased from Immunostep™). The antibodies were diluted to a concentration of 250 μg mL−1 in PB buffer.
Both systems were optimized for the spotting of 10 droplets of 56 pL each, making a total volume of 560 pL per spot. For these conditions, the nozzle piezoelectric parameters were adjusted to a voltage, pulse width and frequency of 44 V, 17 μs and 100 Hz for the Nanoplotter NpC2.1 and 87 V, 56 μs and 500 Hz for the SciFlexArrayer, respectively in the piezoelectric pipette to dispense the droplets.
The temperature and humidity were controlled by a chilling and a humidifying system connected to the Nanoplotter chamber and maintained along the experiments. It was found that a temperature of 17 °C and a relative humidity of 60% was able to prevent the condensation of water molecules inside the target chamber and prevent the evaporation of spotted samples, achieving therefore an equilibrium state important for surface functionalization during an incubation period of 2 h. The surface was blocked with bovine serum albumin (BSA) 1% to avoid unspecific adsorption outside the bioactive sensing regions (the gold pads) before the microspotting of the antibodies. DNA does not adsorb to silicon nitride (Si3N4), thus BSA was only used, at 5%, in the experiments performed on bare gold substrates. Between the spotting of different probes or antibodies, the tip of the pipettes was cleaned by series of automatic immersions in the washing units and dried after. Before the spotting of each sample, functionality tests were also performed during the procedure, using the stroboscope and the water sensitive paper test, to optimize the droplet dispensing. The stroboscope analyzes the droplets shape and dispensing efficiency (flow of the fluids and if any time delay is observed which could be explained by the formation of air bubbles). The water sensitive paper test evaluates if (after passing the stroboscope test) the alignment of the dispensed droplets was correct.
All structures were symmetrically spaced from each other. The center-to-center spot spacing was 300 μm for the gold patterned structures depicted in Fig. 1b and 500 μm for the gold patterned structures in Fig. 1c and sensors shown in Fig. 1d and e.
2.4 Biomolecule target conjugation with 250 nm and 100 nm MNPs
By asymmetric PCR, single-stranded ALU115 and ALU247 biotinylated targets were prepared. The reaction mixture, amplification program and purification method are provided in the ESI.† Streptavidin-coated 250 nm (Nanomag-D 250 nm) and 100 nm (BNF-Dextran-redF) MNPs (supplied by Micromod™) were washed from stocking preservatives with the aid of a magnetic concentrator (DynaMagTM-2 supplied by Invitrogen) following a standard procedure and using PB-buffer during the process. The MNPs were diluted 10× from the stock concentration in PB-Tween-20 0.05% buffer. From the prepared suspension of beads, 10 μL were placed in contact with 1 μL of the solution containing the amplified DNA fragments at a concentration of 50 ng μL−1 (measured using the NanoValue Plus Spectrophotometer, GE Healthcare). To maintain consistency within the experiments, the biotinylated anti-human fibronectin antibody was diluted to 50 ng μL−1 and all the samples were incubated with MNPs for 1 h at RT and under agitation. After functionalization of the MNPs with antibodies, a step for blocking with BSA 5% was performed during an incubation period of 40 min. At last, a final step for the capture of the magnetically labeled DNA targets and antibodies was performed by placing them on the magnetic concentrator and discharge of the reminiscent supernatant before the resuspension of the conjugates in 10 μL of PB-buffer.2.5 DNA hybridization and protein binding with surface microspotted probes
The complexes of MNPs attached to DNA molecules were dispensed over previously prepared gold substrates and biochips and were let to settle down and react for 45 min in a humid chamber. For the antibody-protein assays, 1 μL fibronectin (10 μg mL−1) was manually spotted over the surface functionalized sensors and incubation occurred for 45 min before dispensing the MNPs conjugated to biotinylated anti-fibronectin antibody. After incubation, in both cases, washing of the unbound or weekly bound complexes at the reactionary surfaces was performed. The density of beads over the gold substrates or biochips was analyzed by optical or fluorescent microscopy.2.6 Optical microscopy and imageJ analysis
The gold substrates and biochips were analyzed by optical microscopy. The accumulation of MNPs due to the specific hybridization of DNA targets or specific sandwich binding of antibodies to fibronectin can be visualized by the density of beads in the formed spots. To further and more accurately analyze the MNPs density on the gold surfaces, ImageJ was used. Images from outside the microspotted areas were analyzed and used as the background reference values for comparison with the spotting densities on the microspotted areas. A relative surface coverage density was determined. The percentage values obtained with ImageJ are not absolute values but represent a relative comparison among different spots (equally analyzed).2.7 Fluorescent microscopy analysis
Fluorescent microscopy was employed for discrimination between MNPs and residual salts at the surface using the streptavidin-coated bionized-nanoferrite Texas-Red Fluorescent 100 nm MNPs. Wide-field Fluorescence Microscopy was employed, by excitation of Texas-Red using an upright microscope for excitation and emission at appropriate wavelengths for wide red spectrum analysis (Nikon™). After computer digitalization, fluorescent images were recorded and analyzed.2.8 Contact angle measurements
The contact angle (θ) of TE and PB droplets on gold and Si3N4 substrates was measured in a Drop Shape Analyzer DSA100 (Kruss) using the sessile drop method. Constant volumes of 2 μL were manually spotted over the surfaces after cleaning with isopropanol, Mili-Q water and UV-ozone exposure for 5 min at 55 °C. Droplet profiles were automatically detectable, and the analysis was recorded using the DSA3® software. All measurements were performed at room temperature and the droplets were fitted to the Young–Laplace equation.3. Results and discussion
3.1 Microspotting enables surface functionalization and biomolecule capture
Initially, the hybridization of conjugates SH-Probe115-ALU115 and SH-Probe247-ALU247 and the non-specific interaction of SH-Probe115-ALU247 and SH-Probe247-ALU115 was assessed by manual spotting on bare gold substrates (Fig. 1a and 2a). The fragments ALU115 and ALU247 were previously attached to 250 nm MNPs (as explained in the Materials and Methods Section 2.3). As expected, complementary conjugates of manually spotted DNA probe-target fragments hybridized as seen through the accumulation of magnetic beads on the spotting areas (brighter spots) while the conjugation of probes with non-complementary targets led to an insignificant accumulation of beads on the spotting areas (darker spots). The experiment was further extended for microspotting to assess if the hybridization behavior of the conjugates were comparable at the microscale. A spotting plan was created (Fig. 2b) and employed directly on bare gold substrates for hybridization with target ALU115 without blocking (Fig. 2c) or after blocking the gold free spaces at the surface with BSA 5% (Fig. 2d). In both cases, SH-Probe247 was microspotted in rows identified as A and A′ and SH-Probe115 in rows identified as B and B′ (duplicates in both cases). Four different concentrations of DNA probes were dispensed according to the spotting plan, 10 μM, 5 μM, 3 μM and 1 μM, respectively identified by 1, 2, 3 or 4. DNA spontaneously adsorbs to gold if free spaces are available at the surface. Because no blocking agents were utilized in the experiment correspondent to Fig. 2c, target ALU115 adsorbed outside the microspotted areas, accumulating as well in those regions the conjugated magnetic beads. In Fig. 2d, no adsorption of DNA outside the microspotted areas have occurred due to the blocking with BSA. In the microspotted areas, a higher blocking efficiency for the unspecific cross-reaction or adsorption of ALU115 was achieved by increasing the concentration of non-complementary probe SH-Probe247 whereas by increasing the concentration of the complementary probe SH-probe115 an increased capture of specific target fragments occurred. This experiment corroborates our previous findings on the role of DNA probes as simultaneous blocking agents of unspecific fragments and capture of specific targets if used at optimal conditions, without the need of blocking agents.21 Moreover, we proved that microspotted DNA probes retain their functionality in terms of surface binding and specific target recognition (Fig. 2c and d). In cases where blocking agents are required, microspotting is proved to be equally compatible.3.2 Spotting precision is affected by the combination of different materials at the surface
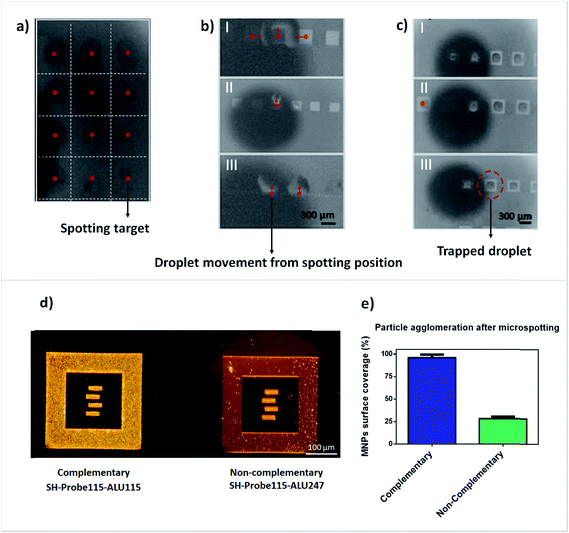
After analyzing the suitability of the microspotted probes for surface immobilization and efficient target capture, we analyzed the effect of the surface materials present in our biochips for the precise spotting of samples as an initial approach for multiplexing. At first, and to show that when microspotting is performed on a monolayer of a single material (the bare gold substrates), the surface energy is homogenous throughout the substrate and the dispensed droplets remain immobilized accurately on the spotted sites (Fig. 3aI). However, when two or more materials are used and micropatterns are designed (Fig. 1b and c), droplets show a different behavior. The images presented in Fig. 3aII, show the exact same amount of volume dispensed per spotting site (560 pL total) as in Fig. 3aI, however, these structures combine gold and Si3N4, at the surface, to simulate the surface materials found in our MR biochips. In all images shown, the Nanoplotter NpC2.1 was programmed to spot directly on top of the gold structures. However, as can be observed (Fig. 3aI–III), the droplets moved out of the spotting areas towards the Si3N4 and, at times, joined together, proving how standard microspotting in biochips is rather difficult to implement in MPOC testing for analysis of different analytes.Taking advantage of the seemingly higher hydrophobicity of gold in comparison to Si3N4, we fabricated trapping structures made of gold as an attempt to immobilize the dispensed droplets inside these traps and to promote the contact of the samples with the gold pads deposited on top of the MR sensors, used for surface functionalization. As can be seen in Fig. 3c(I–III), the droplets containing DNA probes remained trapped inside the gold microfabricated structures and are also functional for hybridization with complementary target DNA (ALU115) while they block the non-complementary target (ALU247) (Fig. 3d and e).
3.3 Surface-liquid contact angle measurements
To support our results and to prove that gold films can be used as a trapping system to enclose droplets of samples when Si3N4 is used as a passivation layer, contact angle measurements of the buffers used in our experiments for DNA immobilization (TE-buffer) and antibody immobilization (PB-buffer) in contact with bare gold substrates and bare Si3N4 substrates were performed (Fig. 4). A contact angle (θ) of 27.3 ± 0.6° was obtained when PB buffer was placed in contact with gold substrates (Fig. 4a and e); 25.6 ± 0.7° when TE buffer was also placed in contact with gold substrates (Fig. 4c and e); 14.3 ± 0.3° when PB buffer was placed in contact with Si3N4 substrates (Fig. 4b and e) and, finally, 19.3 ± 0.5° when TE buffer was placed in contact with Si3N4 substrates (Fig. 4d and e). Despite of the relatively different contact angle of TE buffer and PB buffer in both surfaces, they vary accordingly when placed over the gold or Si3N4 substrates.The higher contact angle measured for the interfaces with the gold substrates when compared to the interfaces with Si3N4 suggests that the droplets would likely remain trapped inside the gold traps, in contact with the Si3N4 as the higher the contact angle is, less are the chances for the liquid droplets to spread out onto the surface, justifying why the gold traps effectively prevent the microspotted droplets from moving in contrast to when they are absent.
Additionally, despite of the higher contact angle between the droplets and the gold substrates, the measured values are small, meaning that the molecules in the liquids will preferentially interact with the surface, justifying why gold can be combined simultaneously for trapping of molecules and be used successfully for surface functionalization.
3.4 Patterned gold traps opens a new multiplexing approach in biochips
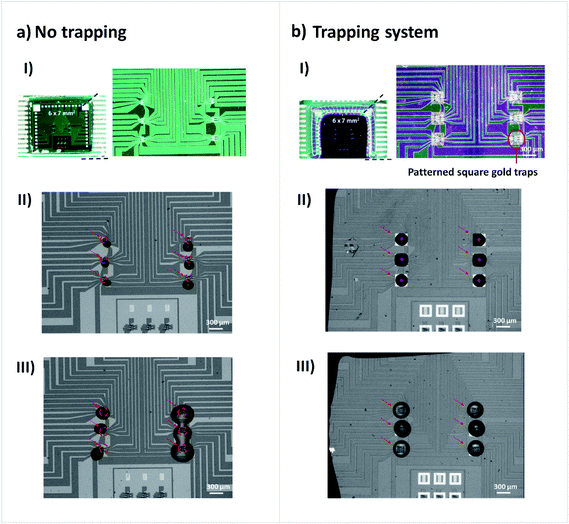
As mentioned so far, our MR biochips have gold films deposited for surface functionalization and Si3N4 for passivation of the surrounding areas. However, our MR biochips are more complex than the substrates mentioned so far, as they encompass as well a combination of ferromagnetic and non-magnetic materials for sensor fabrication along with aluminum contacts which altogether may exert additional effects on the dispensed droplets. To validate our previous results, we have performed the microspotting of DNA probes and antibodies on biochips designed with and without the gold traps (Fig. 5aI and bI).The same amount of liquid was dispensed on all assays. Fig. 5aII and bII represent the spotting of DNA probes in TE-buffer. Without the gold traps, all droplets ended up moving from the spotting position, losing contact with the sensors for functionalization. The effect on droplet stability over the dispensed sites in the presence of the traps is clearly shown. Moreover, in Fig. 5aIII apart from the spotting of DNA probe samples in the left column, antibodies diluted in PB buffer were microspotted and all droplets, undesirably, mixed together. In Fig. 5bIII the use of gold traps prevented the droplets to spread out and mix.
3.5 Specific target binding to microspotted probes
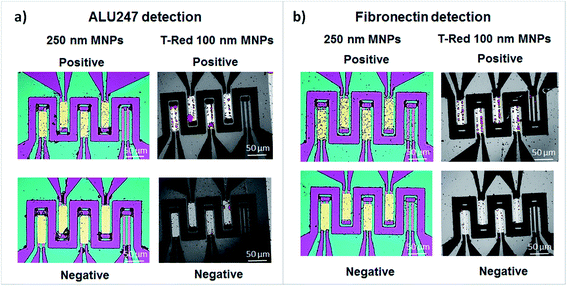
Finally, the microspotted DNA probes and antibodies were reacted with MNPs conjugated to complementary DNA fragments and fibronectin plus the conjugates of beads attached to biotinylated anti-human fibronectin antibody. Analysis of the results show that positive signal was obtained, as expected, after incubation of complementary probe-target DNA and negative after incubation of non-complementary probe-target (Fig. 6a). The same occurred for the binding of fibronectin at the surface. After incubation, fibronectin successfully attached to the microspotted anti-human fibronectin antibody while it did not interact with the non-related antibody anti-human platelet derived growth factor-CC (Fig. 6b). Both for DNA and protein detection, efficient droplet fixation and surface functionalization was thus achieved showing that this methodology may overcome the challenges associated to the microspotting and multiplexing over complex microfabricated structures.4. Conclusions and future work
Our initial attempts to perform microspotting of different droplets on top of the sensors, were inconsistent or failed, as the droplets moved out of the sensors surface region and/or different droplets ended up mixing together. By realizing that the droplets were moving preferentially towards the Si3N4, we took use of our established method for gold deposition over the sensors surface for the fabrication of traps that could increase, in a restricted space, the contact of the droplets with the Si3N4 but, at the same time, promoting as well the contact with the sensors for functionalization. With this work, we prove that simply by combining gold structures as traps and pads for functionalization (in a single step) with the aid of robotic spotting, various sensors can be functionalized with different probes for the capture of various molecules, as much as needed, for the demanding applications in MPOC of multifactorial diseases as it will allow to perform the correlation of a panel of different biomarkers in a single biochip. With the prepared gold traps, there are no additional costs or fabrication steps involved. As for the future, we are using this methodology for the analysis of a relevant panel of six different proteins for rapid response to stroke attacks. If successful, it will prove to be an advance in the field of MPOC systems and a step towards their implementation in pre-clinical and clinical test studies.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The author Tomás Dias thanks FCT for his PhD Grant SFRH/BD/81537/2011. INESC-MN acknowledges FCT funding through the IN Associated Laboratory Pest-OE/CTM/LA0024/2011 and project EXCL/CTM-NAN/0441/2012.References
- C. Dincer, et al., Multiplexed Point-of-Care Testing – xPOCT, Trends Biotechnol., 2017, 35(8), 728–742 CrossRef CAS PubMed.
- P. Yager, et al., Microfluidic diagnostic technologies for global public health, Nature, 2006, 442(7101), 412–418 CrossRef CAS PubMed.
- C. D. Chin, V. Linder and S. K. Sia, Lab-on-a-chip devices for global health: Past studies and future opportunities, Lab Chip, 2007, 7(1), 41–57 RSC.
- A. B. Storrow, et al., A Systematic Review of Emergency Department Point-of-Care Cardiac Markers and Efficiency Measures, Point of Care, 2009, 8(3), 121–125 Search PubMed.
- P. J. Garcia, et al., Point-of-care diagnostic tests for low-resource settings, Lancet Glob. Health, 2015, 3(5), e257–e258 CrossRef PubMed.
- S. H. Bossmann and D. L. Troyer, Point-of-care routine rapid screening: the future of cancer diagnosis?, Expert Rev. Mol. Diagn., 2013, 13(2), 107–109 CrossRef CAS PubMed.
- P. B. Luppa, et al., Clinically relevant analytical techniques, organizational concepts for application and future perspectives of point-of-care testing, Biotechnol. Adv., 2016, 34(3), 139–160 CrossRef PubMed.
- D. L. Graham, H. A. Ferreira and P. P. Freitas, Magnetoresistive-based biosensors and biochips, Trends Biotechnol., 2004, 22(9), 455–462 CrossRef CAS PubMed.
- D. A. Hall, et al., GMR Biosensor Arrays: A System Perspective, Biosens. Bioelectron., 2010, 25(9), 2051–2057 CrossRef CAS PubMed.
- E. Trinquet and G. Mathis, Fluorescence technologies for the investigation of chemical libraries, Mol. BioSyst., 2006, 2(8), 380–387 RSC.
- E. Helmerhorst, et al., Real-time and Label-free Bio-sensing of Molecular Interactions by Surface Plasmon Resonance: A Laboratory Medicine Perspective, Clin. Biochem. Rev., 2012, 33(4), 161–173 Search PubMed.
- J. L. Arlett, E. B. Myers and M. L. Roukes, Comparative advantages of mechanical biosensors, Nat. Nanotechnol., 2011, 6(4), 203–215 CrossRef CAS PubMed.
- V. C. Martins, et al., Femtomolar limit of detection with a magnetoresistive biochip, Biosens. Bioelectron., 2009, 24(8), 2690–2695 CrossRef CAS PubMed.
- J. Germano, et al., A Portable and Autonomous Magnetic Detection Platform for Biosensing, Sensors, 2009, 9(6), 4119–4137 CrossRef CAS PubMed.
- D. A. Hall, et al., A 256 pixel magnetoresistive biosensor microarray in 0.18 μm CMOS, IEEE J. Solid-State Circuits, 2013, 48(5), 1290–1301 CrossRef PubMed.
- L. Jogschies, et al., Recent Developments of Magnetoresistive Sensors for Industrial Applications, Sensors, 2015, 15(11), 28665–28689 CrossRef PubMed.
- G. Kokkinis, et al., Microfluidic platform with integrated GMR sensors for quantification of cancer cells, Sens. Actuators, B, 2017, 241, 438–445 CrossRef CAS.
- A. C. Mak, et al., Sensitive Giant Magnetoresistive-based Immunoassay for Multiplex Mycotoxin Detection, Biosens. Bioelectron., 2010, 25(7), 1635–1639 CrossRef CAS PubMed.
- R. S. Gaster, et al., Quantification of protein interactions and solution transport using high-density GMR sensor arrays, Nat. Nanotechnol., 2011, 6(5), 314–320 CrossRef CAS PubMed.
- S. Cherre, et al., Rapid and specific detection of cell-derived microvesicles using a magnetoresistive biochip, Analyst, 2017, 142(6), 979–986 RSC.
- T. M. Dias, et al., Implementing a strategy for on-chip detection of cell-free DNA fragments using GMR sensors: A translational application in cancer diagnostics using ALU elements, Anal. Methods, 2016, 8(1), 119–128 RSC.
- D. J. B. Bechstein, et al., Microfluidic multiplexed partitioning enables flexible and effective utilization of magnetic sensor arrays, Lab Chip, 2015, 15(22), 4273–4276 RSC.
- C. Duarte, et al., Semi-Quantitative Method for Streptococci Magnetic Detection in Raw Milk, Biosensors, 2016, 6(2), 19 CrossRef PubMed.
- D.-S. Wang and S.-K. Fan, Microfluidic Surface Plasmon Resonance Sensors: From Principles to Point-of-Care Applications, Sensors, 2016, 16(8), 1175 CrossRef PubMed.
- B. Zribi, et al., A microfluidic electrochemical biosensor based on multiwall carbon nanotube/ferrocene for genomic DNA detection of Mycobacterium tuberculosis in clinical isolates, Biomicrofluidics, 2016, 10(1), 014115 CrossRef CAS PubMed.
- J. Cao, et al., A microfluidic multiplex proteomic immunoassay device for translational research, Clin. Proteomics, 2015, 12, 28 CrossRef PubMed.
- J. Zhu, et al., Identification of Glycoprotein Markers for Pancreatic Cancer CD24+CD44+ Stem-like Cells Using Nano-LC–MS/MS and Tissue Microarray, J. Proteome Res., 2012, 11(4), 2272–2281 CrossRef CAS PubMed.
- R. P. Auburn, et al., Robotic spotting of cDNA and oligonucleotide microarrays, Trends Biotechnol., 2005, 23(7), 374–379 CrossRef CAS PubMed.
- M.-D. Cubells-Beltrán, et al., Integration of GMR Sensors with Different Technologies, Sensors, 2016, 16(6), 939 CrossRef PubMed.
- Z. Zhao and H. Jiang, Enzyme-based Electrochemical Biosensors, in Biosensors, ed. P. A. Serra, InTech, Rijeka, 2010, ch. 1 Search PubMed.
- B. Špačková, et al., Optical Biosensors Based on Plasmonic Nanostructures: A Review, Proc. IEEE, 2016, 104(12), 2380–2408 CrossRef.
- A. Gao, et al., Robust ultrasensitive tunneling-FET biosensor for point-of-care diagnostics, Sci. Rep., 2016, 6, 22554 CrossRef CAS PubMed.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, The present and future role of microfluidics in biomedical research, Nature, 2014, 507(7491), 181–189 CrossRef CAS PubMed.
- M. Kondo, S. Koshizuka and M. Takimoto, Surface tension model using inter-particle potential force in Moving Particle Semi-implicit method, Transactions of the Japan Society for Computational Engineering and Science, 2007, vol. 2007, p. 20070021 Search PubMed.
- Y. Yuan and T. R. Lee, Contact Angle and Wetting Properties, in Surface Science Techniques, ed. G. Bracco and B. Holst, Springer Berlin Heidelberg, Berlin, Heidelberg, 2013, pp. 3–34 Search PubMed.
- J. Wang, et al., Surface structure determines dynamic wetting, Sci. Rep., 2015, 5, 8474 CrossRef CAS PubMed.
- J. S. Shumaker-Parry, et al., Microspotting Streptavidin and Double-Stranded DNA Arrays on Gold for High-Throughput Studies of Protein–DNA Interactions by Surface Plasmon Resonance Microscopy, Anal. Chem., 2004, 76(4), 918–929 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra06085h |
| This journal is © The Royal Society of Chemistry 2017 |