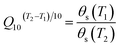Open Access Article
Open Access ArticleQualitative and quantitative analysis of fatty acid profiles of Chinese pecans (Carya cathayensis) during storage using an electronic nose combined with chemometric methods
Shui Jiang,
Jun Wang * and
Yubing Sun
* and
Yubing Sun
Department of Biosystems Engineering, Zhejiang University, 866 Yuhangtang Road, Hangzhou 310058, PR China. E-mail: jwang@zju.edu.cn; Fax: +86-571-88982192; Tel: +86-571-88982178
First published on 2nd October 2017
Abstract
Chinese pecans (Carya cathayensis) continuously deteriorate during storage because of their high fatty acid contents. In this study, an electronic nose (E-nose) was introduced to characterize Chinese pecans with different storage times. Chemometric methods (principal component analysis (PCA), partial least squares regression (PLSR), and back propagation neural networks (BPNNs)) were employed to analyze E-nose data. For qualitative analysis, PCA could visualize the discrimination between different pecans based on the E-nose data. For quantitative analysis, the results indicated that BPNN models performed better both in predicting storage times and fatty acid contents than the PLSR models. In addition, a multi-target BPNN regression model was built to simultaneously predict the contents of the six main fatty acids, and the results (R2 > 0.95 in calibration sets and R2 > 0.88 in validation sets) were satisfactory. This study provides a potentially viable method for determining the storage times and fatty acid profiles of nut products.
1. Introduction
Nuts, as the essence of plants, are generally nutrient dense since they have high content of fat, abundant protein, and mineral substances. The composition of nuts makes them beneficial food supplements for reducing the risks of malnutrition and some chronic diseases such as cardiovascular and cerebrovascular diseases.1 Thus, their high nutritional and medical value make the economic value of nut products increasingly prominent. However, during long storage, a high content of fat makes nuts prone to rancidity, and the economic value is significantly decreased. This is because the fatty acid profiles of most nuts include polyunsaturated fatty acids such as oleic acid and linolenic acid.2 Therefore, it is important to monitor the internal quality of nuts during storage.Traditionally, the rancidity of nuts is often evaluated by determining several indices such as the acid value, peroxide value, and fatty acid profile.3 Among these evaluation indices, fatty acid profile is the most important and direct indicator of the internal quality of nuts. Nkwonta stated that the fatty acid profile of nuts was significantly affected by postharvest storage and processing techniques, and it could be used to characterize the internal quality of nuts.4 Normally, fatty acid profiles can be detected via analytical techniques, such as gas chromatography-mass spectroscopy (GC-MS) and high performance liquid chromatography (HPLC), by determining the oil extracted from nut meat;4,5 although these two methods exhibit good objectivity and repeatability, they suffer from the disadvantages of complex sample preparation and long detection times. For example, in the study reported by Li, who determined the fatty acid profiles of heartnut and Persian walnut, the total run time of GC-MS was 86 min, which was very long.6 In summary, the oxidization procedure of nuts is rarely observed from the outside directly.
To resolve the aforementioned issues, the development of a new method to detect the internal quality of nuts is urgently needed. Recently, some scholars carried out research on the relationship between volatile compounds and the internal quality of nuts.7–9 Mexis10 and Abdallah11 have stated that the components, such as acids, alcohols, esters, and ketones, of sample gases will be changed as a result of changes in nut compositions. Normally, the sample gas escapes from the nuts through their microporous shells; considering that the unique odors of nuts are altered during rancidity, an electronic nose was introduced to detect the internal quality.12 An electronic nose (E-nose) is an instrument that analyzes samples by sensing the sample gases without complex preparation.13,14 A typical E-nose consists of several gas sensors that exhibit the characteristics of cross-sensitivity and a broad spectrum response. As a result, the electrical signal generated by an E-nose contains abundant and overlapping information about the test sample.15 Via multivariate statistical techniques and artificial neural networks, the electrical signal can be used to distinguish different samples or predict the indices of internal quality.16
To extract more useful information from the E-nose data, scholars have made some attempts to test the performances of different algorithms. Therein, artificial neural networks (ANNs) have shown great potential to build input–output relationships between complex nonlinear data obtained from the E-noses.17 Based on different architectures, transfer functions, and learning strategies, there are many types of ANNs. Among these, BPNN has received extensive attention because of its universal approximation capability.18 The work principle of BPNNs is to establish a model by learning and storing the mapping relations between the input and output.19 As a multilayer algorithm, a BPNN consists of an input layer, a hidden layer, and an output layer. During the modeling process, it is not necessary to define the mathematical equation between the input and output; this makes the operation of BPNNs very convenient. Thus, BPNNs have been widely applied in the fields of food safety and food detection. Many studies have been reported on the application of BPNN algorithms to deal with the E-nose data obtained from the detection of food products such as meat,20 fruit,21 rice,22 egg,23 and tea.24
Upon summarizing the abovementioned studies, a conclusion can be made that BPNN was just used to build the regression model to predict a single index. However, during the spoilage of food, changes in internal compositions will cause changes in many evaluation indices. Moreover, all these indices reflect the internal quality to a certain extent. Therefore, it becomes significantly important to determine these indices rapidly and precisely. Nowadays, most commonly used algorithms, such as PLSR, MLR, and SVM, can only be used to build single-variable prediction models.25 Compared to these algorithms, BPNN has a special characteristic: its architecture (the number of layers and nodes) can be designed; this means that the number of nodes (objective variables) in the output layers can be greater than one. Thus, multi-target prediction models can be established based on the BPNN algorithm. The applications of multi-target prediction models have been proved in other fields such as materials processing and welding processes.26,27
To verify the feasibility of multi-target models in the food detection field, a BPNN was applied to analyze the E-nose data of Chinese pecans. Detailed information about Chinese pecans has been provided in previous studies, and the literature proved that it is possible to discriminate between different pecan samples and predict their internal quality based on the E-nose signals.16 The main objectives of this study are as follows: (1) to characterize pecans with different storage times using an E-nose and to determine their fatty acid profiles by GC-MS; (2) to qualitatively and quantitatively analyze the E-nose data based on the PLSR and BPNN algorithms; and (3) to compare the performance of a multi-target BPNN model with that of currently popular methods according to the determination coefficient (R2), root mean square error (RMSE), and relative standard deviation (RSD).
2. Materials and methods
2.1. Sample preparation
The experimental materials used in this study were Chinese pecans (Carya cathayensis), which were supplied by Tuanyuanren Company. These pecans were harvested in Longgang Town, Linan City of Zhejiang province, China (119.72 E, 30.22 N).The internal quality of pecans, which were protected by shells, showed little change in a short period of time. To decrease the time required for this experiment, an accelerated storage simulation was applied. During the simulation, the in-shell pecans were placed in an incubator (STIK (Shanghai) CO., China) at a temperature of 35 °C and relative humidity (RH) of 30%. According to the reported studies, pecans stored in this environment for 10 d and 20 d could simulate those stored in a 4 °C storehouse for approximately 1 and 2 years, respectively.28,29 This accelerated storage simulation is based on the Q10 value, which is calculated as follows:
The supplied pecans were randomly divided into 5 groups (15 sample sets per group and 20 pecans (about 70 g) in each set). During the process, 4 groups were placed in an incubator for the artificial process, and one group was taken out every 5 days. The original samples were defined as day 0, and processed samples were defined as day 5, day 10, day 15, and day 20. The processed samples were exposed to clean air for enough time to cool them from 35 °C to room temperature, which was 20 °C ± 1 °C. Then, E-nose detection was performed. After the detection, all the samples were cracked carefully, and the pecan kernels were taken out. On the same day, pecan oil was extracted from these kernels and then analyzed by GC-MS.
2.2. Electronic nose detection
In this study, an electronic nose (E-nose, PEN2, Airsense Company, Germany) was used to detect sample gases. This E-nose system was equipped with 10 metal oxide semiconductor (MOS) sensors, which were sensitive to specific groups of volatile compounds. The 10 sensors constituted a sensor array, and the sensor arrays were located in a chamber in which the sample gases were exposed to the sensors. These MOS sensors are W1C (aromatic), W5S (broad range sensitivity), W3C (ammonia and aromatic), W6S (hydrogen), W5C (arom-aliph), W1S (broad-methane), W1W (sulph-chlor), W2S (broad-alcohol), W2W (sulph-chlor), and W3S (methane-aliph).32Before the detection of pecans, the working conditions (sample weight, temperature, beaker volume, and headspace generation time) of the E-nose system were optimized by a set of experiments to obtain the best performance. Detailed information is provided in previously reported studies.28 In this study, we have briefly listed some important parameters. The sample weight was about 70 g (20 pecans), the beaker volume was 500 mL, and the head space generated time was 45 min. The monitoring and cleaning processes took 80 and 70 s, respectively. The flow rates of clean air and sample gas through the gas path and sensor chamber were 600 mL min−1 and 200 mL min−1, respectively. During the detection, one signal per second for each sensor was obtained by the WinMuster software (version 1.6.2, Airsense Analytics, Germany). Finally, a data matrix (10 × 80) for each sample was obtained for the following analysis. The output signal of the E-nose was represented as G/G0. G0 and G represent the electronic conductivity of the sensor while detecting clean air and sample gas, respectively. During the monitoring process, 10 response curves of E-nose were obtained. Typical response curves for E-nose in different sample detections have been described in the previous literature.
2.3. Determination of the fatty acid profiles
As abovementioned, the internal quality of pecan kernels with different storage times varied because of the fatty acid rancidity. Therefore, the profile of fatty acids in pecans was regarded as the indicator of internal quality, and the fatty acid profiles were determined by GC-MS in this study. After detection via the E-nose, pecan samples from each group were broken carefully to remove the kernels. Afterwards, these pecan kernels were ground into powder by a squeezer, and these powders were randomly divided into three groups for the following oil extraction. Then, three oil samples were extracted by the Soxhlet extractor (SER148/6, VELP Company, Italy). By repeating the abovementioned steps, a total of 15 oil samples were obtained (3 repeats × 5 groups of pecans). To avoid breakage of the long chain structures of fatty acids in the high temperature environment, fatty acids were processed into stable fatty acid methyl esters (FAMEs). The FAMEs were prepared by methyl esterification, as described previously by Torres, with slight modifications.7 Subsequently, all the FAME solutions were detected by GC-MS. The identification of FAMEs was carried out by comparing the retention times of standards, and the contents of each FAME were calculated using the standard curves. In this experiment, the preparation and detection of FAMEs were conducted in triplicate. Detailed information about the preparation of sample oils and FAME solutions and the working conditions of the GC-MS instruments can be found in our previous study.34,352.4. Analysis of the E-nose data
With regard to the qualitative and quantitative analysis of GC-MS results, the identification of individual peaks was carried out by comparing the retention times of the standards (Aladdin-Jingchun [Shanghai] Biology and Technology Co., Ltd). The contents of fatty acids were calculated using standard curves, and the results were expressed as g kg−1 of pecan oils. In addition, all measurements were replicated three times. The results have been reported as mean ± standard deviation (SD).For the chemometric analysis, the 75th second values of E-nose response signals were extracted as features for subsequent analysis. PCA was applied to discriminate samples with different storage times. PLSR and BPNNs were used to predict the storage time and precise contents of fatty acids. The prediction models were evaluated by determination coefficient (R2), root mean standard error (RMSE), and relative standard deviation (RSD).
In this study, PCA was performed by Statistical Product and Service Solutions v18.0 (International Business Machines Corporation, USA), the PLSR method was performed by Minitab 14 (Minitab Inc., USA), and BPNN was run in MATLAB 2010b (The MathWorks Inc., USA).
3. Results and discussion
3.1. Results of the E-nose detection
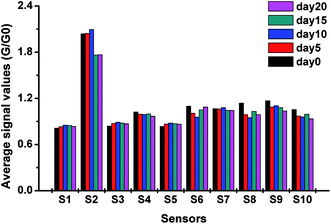
In this study, the response values of each sensor at the 75th second were selected as features for the following analysis. To explore the differences between signals among different groups, the average signal values at the 75th second were calculated, and the result is presented in Fig. 1. In this study, S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10, respectively, represent the W1C, W5S, W3C, W6S, W5C, W1S, W1W, W2S, W2W, and W3S sensors.As shown in Fig. 1, the average signal values of S2 were larger than those of other sensors in the experiment; this indicated that S2 was most sensitive to pecan samples. The response values of S1, S3, and S5 were minimum and almost remained constant. This might be because these three sensors were sensitive to aromatic compounds, and the changes in the main aromatic compounds in different pecan samples were not obvious. In contrast, the signals of S2, S6, S8, and S10 showed significant changes. This may be because new volatile compounds were produced or the concentration of some original compounds changed. These volatile compounds may be alcohols and alkenes, for which S2, S6, S8, and S10 are sensitive. It can be concluded from the average response values of the sensors that the E-nose has the potential to distinguish different pecans by detecting the sample gases.
The stability of each sensor value at the 75th second was analyzed by calculating the RSD, and the results of the RSD analysis are listed in Table 1. As shown in Table 1, all the RSD values are smaller than 3.52%, except the values for S2. The small RSD indicated a high stability for each sensor response.33 S2 has a broad range sensitivity and is easily influenced by small differences in sample gases. Thus, the RSD values of S2 are relatively higher than those of other sensors. According to a previous study, S2 plays an important role in distinguishing different Chinese pecan samples. Therefore, the response values of S2 were retained in the data matrix. Table 1 also presents the results of one-way ANOVA for the sensor array. The results indicated significant differences (P < 0.001) between different sample groups.
| No. | Day 0 (%) | Day 5 (%) | Day 10 (%) | Day 15 (%) | Day 20 (%) | One-way ANOVA | |
|---|---|---|---|---|---|---|---|
| F | P | ||||||
| a Fifteen samples of each group were tested, the RSD of sensor values demonstrated high stability of the test. Significance (p < 0.001) obtained from one-way ANOVA of sensor values measured for 75 samples in total. S1–S10, respectively, represent W1C, W5S, W3C, W6S, W5C, W1S, W1W, W2S, W2W, and W3S sensors. | |||||||
| S1 | 3.52 | 2.56 | 3.04 | 3.32 | 1.72 | 6.34 | <0.001 |
| S2 | 11.97 | 9.85 | 10.27 | 12.96 | 15.94 | 7.44 | <0.001 |
| S3 | 2.78 | 1.76 | 2.07 | 2.55 | 1.26 | 14.77 | <0.001 |
| S4 | 0.22 | 0.46 | 1.75 | 0.43 | 0.90 | 70.03 | <0.001 |
| S5 | 2.75 | 1.91 | 2.19 | 2.57 | 1.37 | 11.03 | <0.001 |
| S6 | 2.32 | 1.93 | 1.38 | 1.20 | 1.24 | 168.64 | <0.001 |
| S7 | 1.91 | 1.84 | 2.09 | 1.30 | 1.27 | 11.66 | <0.001 |
| S8 | 1.88 | 1.94 | 1.53 | 0.95 | 2.37 | 234.36 | <0.001 |
| S9 | 2.99 | 1.51 | 1.91 | 1.81 | 1.39 | 69.46 | <0.001 |
| S10 | 0.42 | 1.24 | 1.29 | 0.80 | 1.37 | 283.65 | <0.001 |
3.2. Results of the fatty acid profiles
In this study, the in-shell pecans were analyzed by E-nose during storage, and then, fatty acids of pecans were analyzed as FAMEs using GC-MS. FAME compositions of pecan oil solutions were obtained from the GC chromatogram, and six main peaks were identified; these were methyl esters of palmitic acid (C16:0), palmitoleic acid (C16:1, cis-9), stearic acid (C18:0), oleic acid (C18:1, cis-9), linoleic acid (C18:2, cis-9, 11), and linolenic acid (C18:3, cis-9, 12, 15) in the elution order. The GC chromatogram can be found in our previous study.34 Recently, many studies have been reported on the fatty acid compositions of different nuts such as hazelnut, peanut, and walnut. However, in these studies, the obtained fatty acid compositions were expressed as percentage of content. As an example, Moser36 determined the fatty acid profiles of hazelnut, peanut, and walnut oil by calculating the area values from their GC chromatograms. Mexis10 researched the effect of gamma irradiation on the fatty acid profiles of raw unpeeled almond kernels, and one of the conclusions was that the fatty acid profiles changed after processing with different doses of gamma irradiation. These studies explored the fatty acid profiles of nuts and their influencing factors, but no precise contents of fatty acids were obtained in these studies. To solve this problem, in this study, we applied GC-MS combined with standard curves to determine the precise contents of FAMEs. Then, the contents of FAMEs were translated to the final contents of fatty acids in the oil samples using the molecular weight ratio of fatty acids and FAMEs. The accurate fatty acid contents obtained from GC-MS are summarized in Table 2 and are expressed as mean ± SD. As shown in Table 2, the determined fatty acid profile of Chinese pecans mainly contained two saturated and four unsaturated fatty acids. For the fresh pecan samples, the content of unsaturated fatty acids was 86.08%, and oleic acid was most abundant with a percentage of 63.97%. Other fatty acid constituents of Chinese pecan were linoleic acid (18.73%), palmitic acid (9.62%), stearic acid (4.30%), linolenic acid (2.11%), and palmitoleic acid (1.26%). The obtained fatty acid compositions were similar to those reported in a previous study;37 however, in this article, only the fatty acid composition of Chinese pecans has been reported, and no further research on the influence of storage on the fatty acid profiles has been carried out.| Fatty acids | Day 0 | Day 5 | Day 10 | Day 15 | Day 20 |
|---|---|---|---|---|---|
| a Values are expressed as mean (n = 3) ± SD. Means in the same row followed by different inline letters (a, b, c, d, and e) are statistically different according to the Tukey's HSD test (P < 0.05). | |||||
| Oleic acid | 340.44 ± 9.93a | 325.10 ± 9.12a,b | 320.08 ± 13.89a,b | 301.60 ± 14.68b,c | 282.27 ± 9.48b,c |
| Linoleic acid | 99.68 ± 2.78a | 94.86 ± 3.99a,b | 90.77 ± 5.25a,b | 87.57 ± 4.49b | 85.44 ± 2.86b |
| Palmitoleic acid | 6.72 ± 0.19a | 6.01 ± 0.06b | 5.72 ± 0.14b | 5.28 ± 0.04c | 5.06 ± 0.07c |
| Linolenic acid | 11.25 ± 0.26a | 10.21 ± 0.33b | 9.81 ± 0.19b | 9.73 ± 0.16b | 9.02 ± 0.12c |
| Palmitic acid | 51.22 ± 0.27a | 48.65 ± 0.34b | 47.37 ± 0.56c | 44.91 ± 0.23d | 42.69 ± 0.20e |
| Stearic acid | 22.91 ± 0.22a | 21.18 ± 0.38b | 19.80 ± 0.43c | 19.47 ± 0.27c | 18.67 ± 0.27d |
In this study, the influence of storage time on fatty acid profiles was explored by determining the precise contents of fatty acids. As shown in Table 2, it could be concluded that all the six fatty acid contents exhibited a decreasing trend during storage. According to the results, oleic acid became most abundant (340.44–282.27 g kg−1) as the storage time increased, followed by, in decreasing order of abundance, linoleic acid (99.68–85.44 g kg−1), palmitic acid (51.22–42.69 g kg−1), stearic acid (22.91–18.67 g kg−1), linolenic acid (11.25–9.02 g kg−1), and palmitoleic acid (6.72–5.06 g kg−1). With regard to the values for day 0, storage under the simulated conditions caused a significant reduction in the content of individual fatty acids. Specifically, palmitoleic acid showed the highest decrease (24.7%) followed by, in the decreasing order, linolenic acid (19.8%), stearic acid (18.5%), oleic acid (17.1%), palmitic acid (16.7%), and linoleic acid (14.3%) after 20-d storage.
3.3. Analysis of PCA
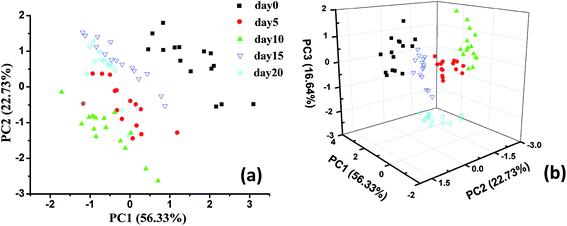
As described in the previous sections, the output of the E-nose is a type of non-representational data matrix. Thus, it is difficult to observe the data structure of the E-nose directly. Via appropriate pattern recognition, the E-nose data could be analyzed for qualitative discrimination and quantitative regression. In this study, principal component analysis (PCA) was applied to visualize the data structure and to qualitatively discriminate sample groups with different storage times.PCA two-dimensional and three-dimensional plots obtained from five groups of pecan samples with different storage times are shown in Fig. 2. As shown in Fig. 2(a), the sum of PC1 and PC2 was calculated and indicated that 79.06% information of the variance was contained in the score plot. In this plot, the sample groups day 0, day 5, day 10, and day 15 could be significantly distinguished from each other. However, some samples of day 20 were mixed with those of day 5, day 10, and day 15. This might be because the first two PCs explained only part of the total variance, and some important information from the sensors was lost during the PCA.
To solve this problem, we selected three components for qualitative discrimination, and the three-dimensional score plot of the PCA is shown in Fig. 2(b). As shown in this plot, the sum of the first three PCs was 95.7%, which meant that most of the information from the original data was considered during PCA. For qualitative discrimination, all the sample groups were separated by a clear boundary and rarely overlapped. It could be concluded from the dispersion of sample points that the PCA plots showed a satisfactory discrimination performance. The results of the qualitative analysis indicated that the E-nose data exhibited a good capability for discrimination of pecans with different storage times.
3.4. Prediction of storage time by E-nose
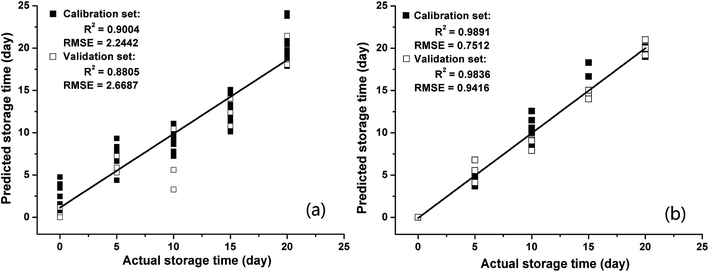
The storage time is the most important factor affecting the internal quality of Chinese pecans. In this study, PLSR and BPNNs were introduced to develop models for predicting storage times. The 75th second values were extracted from the response signals of the E-nose as the feature data. Before building the models, the feature data (15 samples × 5 categories) was divided randomly into calibration and validation subsets, with 60 samples (12 samples of each category) for the calibration set and 15 samples (3 samples of each category) for the validation set. The values in the calibration set were applied to build the PLSR and BPNN models as independent variables, and the simulated storage times were regarded as the dependent variables. The prediction performances of models were evaluated by calculating R2 and RMSE. A higher R2 and a lower RMSE represent a better prediction model. In addition, the R2 values and RMSEs of the calibration and validation sets were used to verify the performance of model overfitting.The regression results for PLSR and BPNN are shown in Fig. 3. Fig. 3(a) shows that the PLSR model does not perform well in predicting the storage time of pecans based on the E-nose response signals. The R2 between the actual storage times and predicted storage times in the calibration set (R2 = 0.9004) was slightly higher than that in the validation set (R2 = 0.8805); moreover, the RMSE in the calibration set (RMSE = 2.2442) was lower than that in the validation set (RMSE = 2.6687). The results of PLSR analysis were similar to those of Wei12 who applied the PLSR method to predict the storage time of peanuts based on the E-nose data. Although the R2 values of PLSR were acceptable, the RMSEs were quite high both in the calibration set and validation set. This indicated that the prediction precision of the PLSR models was not good. To more precisely predict the storage time of pecans, a BPNN was also applied to build a prediction model based on the E-nose data. As shown in Fig. 3(b), the R2 values were very high both in the calibration set (R2 = 0.9891) and validation set (R2 = 0.9836), and the RMSEs were very low both in the calibration set (RMSE = 0.7512) and the validation set (RMSE = 0.9416).
Regarding the quantitative analysis of storage time, the regression models PLSR and BPNN had satisfactory performances in prediction. Compared to the results of PLSR, the higher R2 values and the lower RMSEs led to the conclusion that the prediction model of storage time based on the BPNN was much better. In addition, the R2 values and RMSEs of the calibration and validation sets showed small differences; this indicated that the overfitting degrees of the PLSR and BPNN models are small. As abovementioned, the E-nose data had the capacity to accurately predict storage time via an appropriate regression algorithm.
3.5. Prediction of the fatty acid profiles by E-nose
In this study, an E-nose was introduced as a potential alternative method to GC-MS to analyze the fatty acid profiles of pecans. PLSR and BPNN were used to establish the correlation between the sensor signals of the E-nose and the fatty acid (i.e., oleic acid, linoleic acid, palmitoleic acid, linolenic acid, palmitic acid, and stearic acid) profiles of pecans.| Fatty acids | Calibration | Validation | ||||
|---|---|---|---|---|---|---|
| R2 | RMSE | RSD (%) | R2 | RMSE | RSD (%) | |
| Oleic acid | 0.8340 | 8.2927 | 5.70 | 0.8294 | 9.6127 | 6.29 |
| Linoleic acid | 0.7214 | 2.7063 | 4.77 | 0.7204 | 2.1965 | 4.51 |
| Palmitoleic acid | 0.7579 | 0.2887 | 8.92 | 0.7712 | 0.2331 | 8.34 |
| Linolenic acid | 0.8757 | 0.2599 | 6.92 | 0.8616 | 0.2994 | 6.70 |
| Palmitic acid | 0.7965 | 1.3419 | 5.54 | 0.7923 | 1.4833 | 5.87 |
| Stearic acid | 0.7635 | 0.7268 | 6.51 | 0.7574 | 0.7674 | 6.05 |
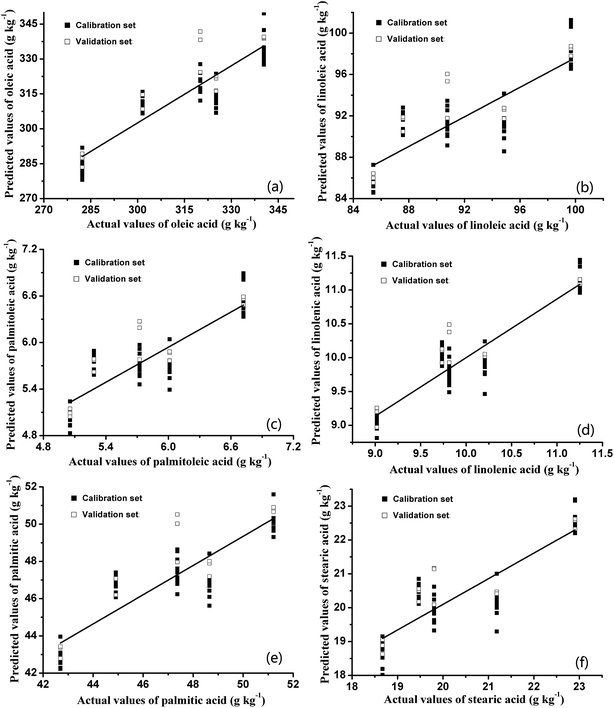
As shown in Table 3, there were small differences between the R2 values of the calibration and validation sets, which indicated that no overfitting occurred during the building procedure of prediction models based on the PLSR method. However, the RSD values in this table are smaller than 9%. The low R2 values (R2 < 0.88 in the calibration sets and R2 < 0.87 in the validation sets) led to a conclusion that the performances of the PLSR models were unsatisfactory. Furthermore, the RMSEs in both the calibration and validation sets were very high. As shown in Fig. 4, the data points were unorganized, and the data points for day 15 were far away from the fitted curves. Therefore, the fitting effects for fatty acids based on the PLSR method were unsatisfactory. This result might suggest that the latent correlation between the E-nose data and the experimental data of fatty acids cannot be completely based on the PLSR method. PLSR is a data analysis method based on the multivariate statistical analysis, which combines the properties of principal component analysis and multiple linear regression analysis.38 To a certain extent, PLSR can accept collinear data, separate out the sample noise, and make linear combinations in the dependent concentration matrix.39 However, the relationship between predicted indices and E-nose data was nonlinear, and the regression model based on PLSR could not perfectly characterize the mapping relation between the indices and E-nose data. To gain a better prediction performance, a BPNN was also applied to build the prediction model of fatty acid profiles based on the E-nose data.
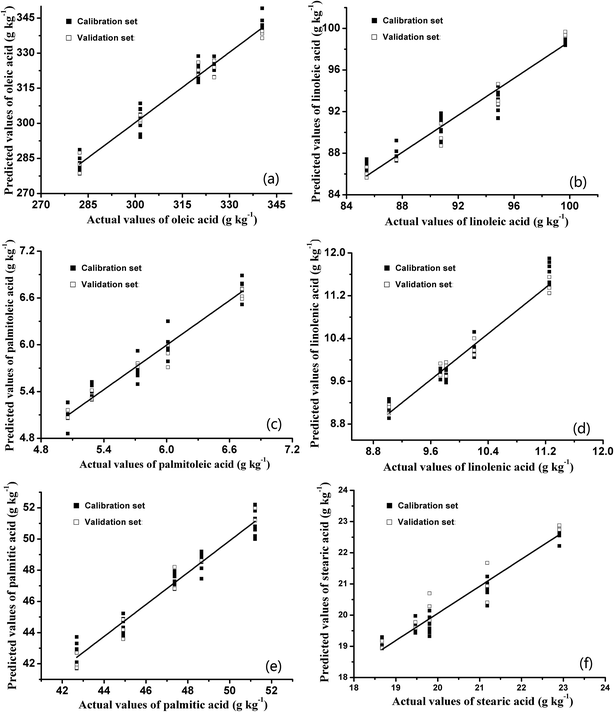
The distributions between predicted and experimental values of fatty acids based on the BPNN are shown in Fig. 5. The R2 values and RMSEs of the calibration and validation sets based on the BPNN are summarized in Table 4. Comparison of the regression results based on PLSR and BPNN led to the discovery that the R2 values of both the calibration sets (R2 > 0.96) and validation sets (R2 > 0.93) based on the BPNN were higher than those of the calibration sets (R2 < 0.88) and validation sets (R2 < 0.87) based on PLSR. Furthermore, the RMSEs of the calibration and validation sets based on the BPNN were much lower than those based on PLSR. The higher R2 values and the lower RMSEs indicated that there were close relationships between the fatty acid contents estimated by the BPNN models and those obtained via GC-MS. In addition, as another evaluation criterion, the RSDs in this table are smaller than 10%, which is acceptable in this study. It could be concluded that the BPNN algorithm could extract useful information from E-nose data more effectively than the PLSR method.
| Fatty acids | BPNN (single-target) | BPNN (multi-target) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calibration | Validation | Calibration | Validation | |||||||||
| R2 | RMSE | RSD (%) | R2 | RMSE | RSD (%) | R2 | RMSE | RSD (%) | R2 | RMSE | RSD (%) | |
| Oleic acid | 0.9777 | 3.1097 | 6.60 | 0.9765 | 3.1048 | 6.29 | 0.9685 | 4.4058 | 6.84 | 0.9620 | 3.9938 | 6.37 |
| Linoleic acid | 0.9643 | 1.2703 | 5.07 | 0.9678 | 1.0373 | 4.51 | 0.9779 | 0.7840 | 5.65 | 0.9528 | 1.1967 | 5.13 |
| Palmitoleic acid | 0.9695 | 0.1033 | 10.03 | 0.9751 | 0.1045 | 8.34 | 0.9759 | 0.0978 | 10.60 | 0.9569 | 0.1269 | 9.99 |
| Linolenic acid | 0.9673 | 0.1713 | 8.17 | 0.9771 | 0.1321 | 6.70 | 0.9648 | 0.1609 | 7.82 | 0.9486 | 0.1883 | 8.25 |
| Palmitic acid | 0.9678 | 0.5637 | 6.45 | 0.9803 | 0.6136 | 5.87 | 0.9734 | 0.5985 | 6.88 | 0.9459 | 0.6875 | 6.34 |
| Stearic acid | 0.9655 | 0.3153 | 6.51 | 0.9398 | 0.4090 | 6.05 | 0.9548 | 0.3243 | 7.11 | 0.8807 | 0.5274 | 6.97 |
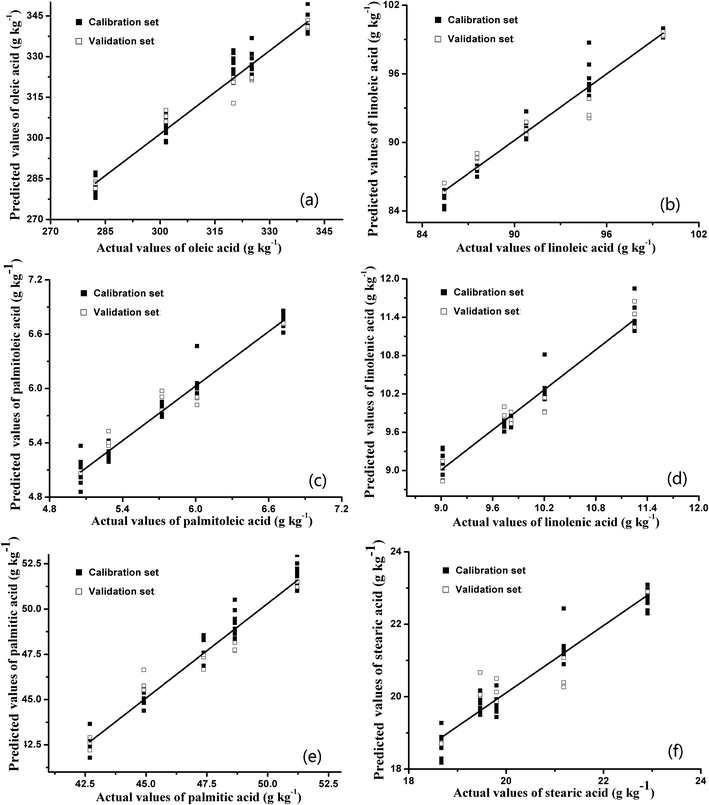
At present, most of the relevant reported studies mainly focus on building a regression model of single target. As abovementioned, the BPNN regression models performed well while singly predicting the content of each fatty acid. However, the internal quality of pecans could only be characterized by fatty acid profiles that include six main fatty acids. Therefore, single-target BPNN models of fatty acids were inconvenient for determining the fatty acid profiles of pecans in practical applications. In this study, a multi-target BPNN regression model was built to verify the possibility of predicting the six fatty acid contents simultaneously. Many reports have mentioned that multilayered networks are capable of handling a wider range of nonlinear functions than single-layer networks.40 However, the more complicated architecture of the network model will substantially increase the computational effort.27 For application in food quality detection, the BPNN model with one hidden layer is sufficient to deal with the E-nose data. Therefore, in this study, the developed model consisted of three layers: an input layer (10 nodes), a hidden layer (14 nodes), and an output layer (6 nodes). The 6 nodes in the output layer represented the predicted values of six fatty acids. The distributions between the predicted and experimental values of six fatty acid contents based on the multi-target BPNN prediction model are shown in Fig. 6, and the R2 values and RMSEs of the calibration and validation sets are summarized in Table 4.
As shown in Table 4, the performance of the multi-target BPNN model, which predicted the six fatty acid contents simultaneously, was acceptable. The R2 values in the calibration and validation sets were greater than 0.95 and 0.88, respectively. In addition, the RMSEs of the calibration and validation sets were almost the same as the results of single-target BPNN models, and the RSDs in this table were also acceptable. The satisfactory evaluation indexes (R2 and RMSE) indicated that the BPNN algorithm could provide the most useful information from the E-nose data to predict the six fatty acid contents simultaneously. The advantage of the obtained multi-target BPNN model was that the fatty acid profiles of pecans could be predicted directly by just inputting the E-nose data once. Moreover, the multi-target BPNN model makes the detection of fatty acid profiles more convenient and direct, avoiding complex experimental procedures and data processing.
To date, this is the first study on the assessment of the impact of postharvest storage on the fatty acid profile of Chinese pecans and prediction of the precise contents of each fatty acid based on the E-nose data. As abovementioned, the fatty acid profile of Chinese pecans contained mainly two saturated fatty acids (palmitic acid and stearic acid) and four unsaturated fatty acids (palmitoleic acid, oleic acid, linoleic acid and linolenic acid). The main types of fatty acids were similar to those of most walnuts.41,42 However, the precise fatty acid composition of Chinese pecans was distinctly different from that of others, which led to their unique economic value.7,37 Moreover, this study explored the feasibility of precisely predicting the fatty acid contents simultaneously based on the BPNN model. The established multi-target BPNN model could predict the fatty acid profile directly by inputting one data set. Compared with that of the common regression methods such as PLSR and the single-target BPNN, the prediction performance (high R2 and low RMSE) of the multi-target BPNN was satisfactory.
4. Conclusions
This study provided a nondestructive detection method to evaluate in-shell pecans using an E-nose. The main research and conclusions are as follows: (1) PCA has been applied to visualize the discrimination between different pecan samples, and the E-nose performs well in differentiating different samples with the first three PCs explaining 95.7% of the variation. (2) A total of six fatty acids (i.e., palmitic acid (C16:0), palmitoleic acid (C16:1), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), and linolenic acid (C18:3)) have been identified by GC-MS. During the storage, the measured values of the six fatty acids decrease by 16.7%, 24.7%, 18.5%, 17.1%, 14.3%, and 19.8%. (3) Single-target BPNN models perform better both in predicting storage time and fatty acid contents than PLSR models, and the results of the multi-target BPNN model, which has been built to simultaneously predict the contents of six fatty acids, are satisfactory (R2 > 0.95 in calibration sets and R2 > 0.88 in validation sets). To the best of our knowledge, no previous study has been performed to simultaneously predict the fatty acid profile of nuts based on a multi-target BPNN model. This study provided a potentially viable method for determining the storage times and fatty acid profiles of nuts. In the subsequent study, more effort will be focused on analyzing the aroma characteristics of nuts using a solid phase microextraction method (SPME) combined with GC-MS.Conflicts of interest
There are no conflicts to declare.Nomenclature
Detection instruments
| E-nose | Electronic nose |
| GC-MS | Gas chromatography-mass spectroscopy |
| SPME | Solid phase microextraction method |
| HPLC | High performance liquid chromatography |
Different types of sensors
Electronic nose: W1C, W5S, W3C, W6S, W5C, W1S, W1W, W2S, W2W, and W3SPattern recognition techniques
| PCA | Principal component analysis |
| PLSR | Partial least squares regression |
| BPNN | Back propagation neural network |
| MLR | Multivariable linear regression |
| SVM | Support vector machine |
Samples
Day 0, day 5, day 10, day 15, and day 20 represent Chinese pecan samples stored in an incubator at a temperature of 35 °C and relative humidity (RH) of 30% for 0 day, 5 days, 10 days, 15 days, and 20 days.| FAMEs | Fatty acid methyl esters |
Acknowledgements
The authors acknowledge the financial support received from the Chinese National Foundation of Nature and Science through Project 31370555.References
- C. Sarvamangala, M. V. C. Gowda and R. K. Varshney, Field Crop Res., 2011, 122, 49–59 CrossRef.
- E. Ros and J. Mataix, Br. J. Nutr., 2006, 96, S29–S35 CrossRef CAS PubMed.
- M. V. Christopoulos and E. Tsantili, Postharvest Biol. Technol., 2015, 104, 17–25 CrossRef CAS.
- C. G. Nkwonta, M. C. Alamar, S. Landahl and L. Terry, J. Food Compos. Anal., 2016, 45, 87–94 CrossRef CAS.
- X. Y. Mao, Y. F. Hua and G. G. Chen, Int. J. Mol. Sci., 2014, 15, 2003–2014 CrossRef PubMed.
- L. Li, R. Tsao, R. Yang, J. K. G. Kramer and M. Hernandez, J. Agric. Food Chem., 2007, 55, 1164–1169 CrossRef CAS PubMed.
- M. M. Torres, M. L. Martínez and D. M. Maestri, J. Am. Oil Chem. Soc., 2005, 82, 105–110 CrossRef CAS.
- S. Pastorelli, L. Torri, A. Rodriguez, S. Valzacchi, S. Limbo and C. Simoneau, Food Addit. Contam., 2007, 24, 1219–1225 CrossRef CAS PubMed.
- M. Cirlini, C. Dall'Asta, A. Silvanini, D. Beghe, A. Fabbri, G. Galaverna and T. Ganino, Food Chem., 2012, 134, 662–668 CrossRef CAS PubMed.
- S. F. Mexis, A. V. Badeka, K. A. Riganakos, K. X. Karakostas and M. G. Kontominas, Food Control, 2009, 20, 743–751 CrossRef CAS.
- I. B. Abdallah, N. Tlili, E. Martinez-Force, A. Rubio, M. C. Perez-Camino, A. Albouchi and S. Boukhchina, Food Chem., 2015, 173, 972–978 CrossRef CAS PubMed.
- Z. Wei, J. Wang and W. Zhang, Food Chem., 2015, 177, 89–96 CrossRef CAS PubMed.
- X. Z. Hong and J. Wang, J. Food Eng., 2014, 126, 89–97 CrossRef CAS.
- T. C. Pearce, S. S. Schiffman, H. T. Nagle and J. W. Gardner, in Handbook of Machine Olfaction: Electronic Nose Technology, Wiley-VCH, Germany, 2003, pp. 133–135 Search PubMed.
- L. Zhang, F. C. Tian, X. W. Peng and X. Yin, Sens. Actuators, A, 2014, 205, 170–176 CrossRef CAS.
- S. Jiang and J. Wang, Postharvest Biol. Technol., 2016, 118, 17–25 CrossRef CAS.
- M. F. Tahir, T. Hanssan and M. A. Saqib, Int. J. Electr. Power Energ. Syst., 2016, 83, 49–57 CrossRef.
- Y. Q. Huang, L. J. Kangas and B. A. Rasco, CRC Crit. Rev. Food Sci. Nutr., 2007, 47, 113–126 CrossRef CAS PubMed.
- Y. D. Liu, X. D. Sun and A. G. Ouyang, LWT--Food Sci. Technol., 2010, 43, 602–607 CrossRef CAS.
- X. Z. Hong, J. Wang and Z. Hai, Sens. Actuators, B, 2012, 161, 381–389 CrossRef CAS.
- H. M. Zhang, J. Wang and S. Ye, Food Bioprocess Technol., 2012, 5, 65–72 CrossRef.
- B. Zhou and J. Wang, Sens. Actuators, B, 2011, 160, 15–21 CrossRef CAS.
- Y. W. Wang, J. Wang and B. Zhou, Anal. Chim. Acta, 2009, 650, 183–188 CrossRef CAS PubMed.
- H. C. Yu, Y. W. Wang and J. Wang, Sensors, 2009, 9, 8073–8082 CrossRef PubMed.
- S. S. Qiu, J. Wang and L. P. Gao, J. Agric. Food Chem., 2014, 62, 6426–6434 CrossRef CAS PubMed.
- P. Saha, A. Singha, S. K. Pal and P. Saha, Int. J. Adv. Manuf. Tech., 2008, 39, 74–84 CrossRef.
- M. A. Moghaddam, R. Golmezergi and F. Kolahan, Measurement, 2016, 92, 279–287 CrossRef.
- P. S. Taoukis, T. P. Labuza and I. S. Saguy, in Handbook of food engineering practice, CRC Press, New York, 1997, pp. 369–394 Search PubMed.
- S. Wang, J. Tang, T. Sun, E. J. Mitcham, T. Koral and S. L. Birla, J. Food Eng., 2006, 77, 304–312 CrossRef.
- T. P. Labuza, J. Chem. Educ., 1984, 61, 348–358 CrossRef CAS.
- B. Ling, L. X. Hou and R. Li, LWT--Food Sci. Technol., 2014, 59, 786–793 CrossRef CAS.
- S. Benedetti, S. Buratti, A. Spinardi, S. Mannino and I. Mignani, Postharvest Biol. Technol., 2008, 47, 181–188 CrossRef CAS.
- N. Sadrieh, J. Brower, L. Yu, W. Doub, A. Straughn and S. Machado, Pharm. Res., 2005, 22, 1747–1756 CrossRef CAS PubMed.
- S. Jiang, J. Wang, Y. W. Wang and S. M. Cheng, Sens. Actuators, B, 2017, 242, 511–521 CrossRef CAS.
- S. Q. Cui, J. Wang and L. C. Yang, J. Pharm. Biomed. Anal., 2015, 102, 64–77 CrossRef CAS PubMed.
- B. R. Moser, Fuel, 2012, 91, 231–238 CrossRef.
- L. Ni and W. Y. Shi, Iran. J. Pharm. Res., 2014, 13, 221–226 CAS.
- S. Wold, A. Ruhe, H. Wold and W. J. Dunn, SIAM J. Sci. Stat. Comput., 1984, 5, 735–743 CrossRef.
- J. Lozano, J. P. Santos, T. Arroyo, M. Aznar, J. M. Cabellos, M. Gil and M. D. C. Horrillo, Sens. Actuators, B, 2007, 127, 267–276 CrossRef CAS.
- J. D. Cullen, N. Athi, M. Al-Jader, P. Johnson, A. I. Al-Shamma'a, A. Shaw and A. M. A. El-Rasheed, Measurement, 2008, 41, 412–423 CrossRef.
- M. L. Martínez, M. C. Penci, V. Ixtaina, P. D. Ribotta and D. Maestri, LWT--Food Sci. Technol., 2013, 51, 44–50 CrossRef.
- E. Bakkalbasi, O. M. Yilmaz, I. Javidipour and N. Artik, LWT--Food Sci. Technol., 2012, 46, 203–209 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2017 |