Open Access Article
Open Access ArticleA nucleic acid logic gate system that distinguishes different sets of inputs from one miRNA collection with shared members†
You Zhanga,
Xiangjiang Zhengb,
Lian Xia a,
Jinmao You*a and
Rui Ren
a,
Jinmao You*a and
Rui Ren *ab
*ab
aKey Laboratory of Life-Organic Analysis of Shandong Province, Qufu Normal University, Qufu 273165, P. R. China. E-mail: greeneese@163.com
bShandong Province Key Laboratory of Detection Technology for Tumor Makers, School of Chemistry and Chemical Engineering, Linyi University, Linyi 276000, P. R. China
First published on 31st August 2017
Abstract
A proof-of-principle logic system was established to process multi-input and multi-output logic relationships with the aim of identifying the tissue origins of cancer in light of their established relationships with miRNA distributions. The system detected two characteristic miRNA sets from a collection with two gates, each identifying a set of three input miRNAs, with miRNAs members shared among the two sets. The design of the logic system featured three stages: the initialization stage, the logic stage, and the signal stage. The initialization stage transduced each inputted miRNA into a messenger DNA in multiplied quantities. In the logic stage, the two logic gates were implemented; each picked up one set of inputted miRNA and produced the according RCA product. In the signal stage, the RCA products from the two gates were labelled with respective Raman tags, and the according SERS signal for each gate was produced. The logic system provided amplification for the miRNA inputs and thus was capable of processing miRNA with low abundance. The designed logic behavior was also verified to ensure that each gate only reacted to its specific input set, and the shared miRNA inputs could be processed by both gates. It was also verified that mismatched miRNA inputs did not interfere with the designed logic behaviors.
Introduction
Identifying the tissue origins of cancer is of great importance in the diagnosis and treatment of cancer, especially for cancers of unknown primary origin (CUP). The tissue origins of CUPs are difficult to identify because of metastasis, which resulted in cancer tumors occurring in places other than where they originated.1 Recently, relationships were established between miRNA distributions and the tissue origins of cancer.2,3 The expression distributions of miRNAs depend on the specific organism in which they occur. The miRNAs in cancer cells maintain the distribution patterns of the tissues or organisms where the cancer originated, regardless of metastasis. New approaches are thus expected for identifying the origins of cancer based on this relationship; this expectation has led to the following considerations and challenges.In the established relationship, one cancer origin usually corresponds to more than one miRNA, and one miRNA is often related to more than one cancer origin.3 Thus, multi-input, multi-output systems are required. Such systems have been reported using techniques involving DNA logic gates and linear threshold gates; in these systems, a single gate usually accepts more than one input and accordingly produces one on/off output.4,5 Combination of multiple gates addressed the above requirement. For example, Qian et al.6,7 developed linear threshold complexes with the aim of providing small-scale artificial molecular models for the neural network algorithm. While this work was a prominent achievement at the fundamental level, it lacked efficiency and feasibility from the practical point of view. Notably, it required prolonged reaction time and high-concentration input. A system with practical improvements is thus needed.
The involvement of miRNA raises additional concerns.8 The abundance of miRNA in cells is usually very low, and miRNA is prone to degradation. Therefore, sensitive and degradation-resistant techniques are required. Such techniques are not often encountered in previous studies on logic gates, such as Qian's report.6 In addition, miRNAs often occur in organisms in families with similar lengths and sequences (typically the let7 family) but with different functions. Thus, it is of vital importance for any miRNA assay to distinguish among such miRNAs. Various approaches have been developed to address these challenges, including transduction and/or amplification based on nucleic acid molecular machines9 and signal enhancement based on surface-enhanced Raman scattering (SERS).10
Principle and design
The main purpose of the current study is to establish a proof-of-principle logic system that is capable of processing the aforementioned multi-input/multi-output logic relationship in the hope that it could be used in the detection of the characteristic miRNA sets from a collection. The overall aim is to identify the tissue origins of cancer in light of their established relationships with miRNA distributions.The principle of this study featured a logic gate system that processed a collection of miRNA inputs. The collection contained four input miRNAs. These inputs fell into two sets, with each set corresponding to one origin according to the established relationship. Each set consisted of three members, and two of the miRNA inputs were shared among the two sets. The logic gate system consisted of two gates. Based on its own logic, each gate picked one characteristic set of miRNAs from the collection as its specific input and produced the corresponding on–off output, which served as the indicator of the corresponding cancer origin.
The shared miRNA members acted as the inputs of the two gates simultaneously. Considering this and the low abundances of miRNAs in organisms, at least these two members must be recycled or subjected to signal amplification. As miRNAs are prone to degradation, it is necessary to transduce the target into more stable forms, such as messenger DNA. Finally, signal production is also necessary to make the result of the logic gates and the amplifications readable.
The main purpose and principle of this study are outlined in Scheme 1. The scheme also shows the relationship between miRNA distribution and cancer origin. The design consisted of three stages: the initialization stage, gate stage, and signal stage. For the convenience of description, each stage includes “nodes” and “wires.” Each node consisted of all the materials needed for the stage. The wires, which usually linked nodes, consisted of the designed motions (e.g., reactions) needed for the stage to be carried out (Scheme 2).
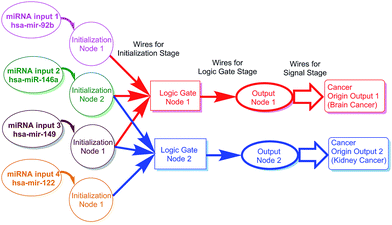 | ||
| Scheme 1 Outline of the logic gate system in the study. The scheme also illustrates the relationship between miRNA distribution and cancer origin. | ||
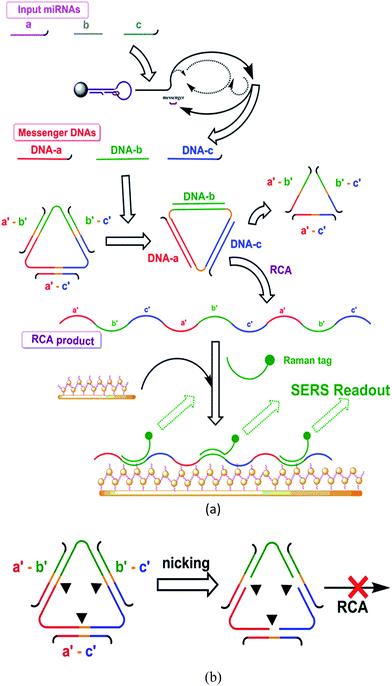 | ||
| Scheme 2 Implementation of the logic system. The two most typical reaction paths are illustrated. (a) “On” path producing “on” output and (b) “off” path producing “off” output. | ||
Implementation
The implementation of the current study was shown in Scheme 2.Initialization stage
The initialization stage was designed to recognize the input miRNAs and transduce them into messenger DNAs with multiplied quantities. The messenger DNAs, as the representatives for the input miRNAs, then served as the inputs for the following logic stage.This stage was implemented with a protocol based on one of our previous reports.11 For the node, one probe and one primer were prepared for each input miRNA. Each probe consisted of a hairpin and a messenger DNA bound to the hairpin. For the wire, this stage started with the recognition of the target miRNA by the hairpin and resulted in the messenger DNA strand being released in multiplied quantities. The details of this stage are given in the ESI.†
Each node centered on a single-stranded circular DNA template, and three “blocking” strands were also included. The structures of the probes were designed to support the operation of the wire. This stage was also assisted by nicking endonuclease, DNA polymerase, and other needed materials.
Because of the nature of the logic operations, each wire had at least two paths depending on the inputted messenger DNAs and the binding statuses, “off” and “on”, for producing the on/off outputs. Each path involved three steps: variation of the binding statuses, on/off nicking and on/off RCA. And each based on certain designs in the node.
Each path started from different binding statuses of the circular template with the messenger DNAs or with the binding strands. Various binding status might occur according to the input. The different binding statuses led to different behaviors regarding the nicking and RCA, thereby resulting in different outputs:
“Off” path: the template bound with the three “blocking strands” (as in the initial status). This typically occurred in the case that no messenger DNA (no input miRNA) was present. The binding of any blocking strand(s) on the circular template formed a functional nicking site, thus leading to the breaking of the template. Once the template was nicked at any point, it became unavailable for the RCA. Nicking and damaging of the template in massive quantities were expected to prominently lower the reaction level of RCA. In summary, from “off” binding status (where the circular template bound with three “blocking” strands), the three nicking sites were functional, caused “off” nicking, and led to the following “off” RCA (low content RCA product or none).
“On” path: typically, the template bound with the three messenger DNAs. This occurred when the entire set of characteristic miRNA inputs were present. Only the binding of the entire set of characteristic miRNA inputs (three) displaces and thus removed all the previously bound blocking strands, disabling the nicking sites, and thus the circular template would remain intact. The circular template remains intact, and RCA could carry on. In summary, from “on” binding status (i.e. when the whole set of messenger DNAs, as the substitutes of the pre-designed set of input miRNAs, were present and bound on the circular template), the blocking strands would be removed through hybridization displacements, causing “on” nicking, and leading to the following “on” RCA (producing RCA products).
The absence of one member in the set resulted in the inefficient removal of at least one blocking strand, leaving at least one nicking site functional. This resulted in “off” nicking and “off” RCA.
The nodes of this stage included the SERS matrix for the enhancement of the Raman signal and Raman tags for the production of the signal. The matrix was an assembly of the gold thin film and gold nanoparticles and was able to bind the RCA products through “capturing strand.” Two Raman tags were also prepared, each for the signal production of one gate. Each tag was a DNA segment that could bind the corresponding RCA product through hybridization. Each tag was labelled with different Raman functional groups corresponding to different cancer origins.
For the wire: the RCA product was labelled with Raman tags, then captured on the enhancing matrix; and the entire complex was then quantified through SERS measurements. The Raman scattering signals were recorded on a Renishaw inVia Raman microscope.
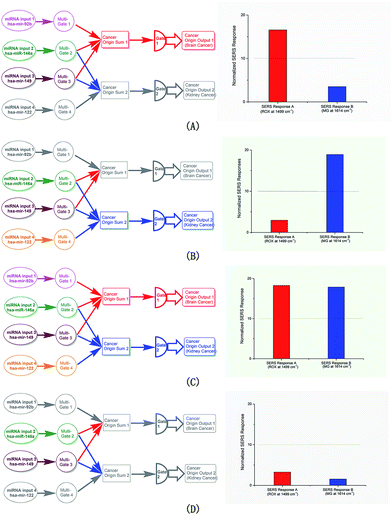
The proof-of-principle miRNA collection contained hsa-mir-92b (a), hsa-miR-146a (b), hsa-mir-149 (c) and hsa-mir-122a (d). According to the molecular biology report, they are related to two cancer origins in sets: the set of (a), (b) and (c) for brain cancer and the set of (b), (c) and (d) for kidney cancer. The collection of the four miRNAs was thus used as the input in this proof-of-principle study.
The outputs of the two gates associated in principle with the two cancers were labelled with two Raman tags, ROX (for brain cancer, characteristic Raman peak at 1499 cm−1) and malachite green (for kidney cancer, characteristic Raman peak at 1614 cm−1).
Results and discussion
Amplification performance
The amplification capacity (i.e., the ability of the system to sense miRNA inputs with low abundance) was first tested. The reaction system contained two major amplification mechanisms, exponential amplification in the initialization stage and RCA in the logic stage. These amplifications are both based on well-developed DNA molecular machine techniques.The following investigations of amplification were all exemplified by one of the two gates, the one that produced readouts through ROX tag (since the performances of the two gates were assumed to be equal), with its input miRNA set served as target as a whole. The three miRNA members in the input set were always input with the same concentration, which was recorded as the target concentration for the following calibration curve and other quantitative experiments.
The optimal reaction conditions for amplification were determined to be a reaction time of 2 h, a Phi29 DNA polymerase concentration of 0.60 IU μL−1, and a Nb.BbvCI (nicking endonuclease) concentration of 0.40 IU μL−1 (see Section S2.1 “Optimization of reaction conditions” and Fig. S4 in the ESI†). Under the optimal conditions, the calibration curve was measured by plotting the normalized ΔI, the SERS response at 1499 cm−1 (after subtracting the blank response) against the concentration of the target miRNA (cTarget). A linear relationship was observed between the SERS response and the logarithm of target miRNA concentration across a concentration range of 10−16 to 10−14 M. In this range, the regression equation was ΔI = 4.78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg(cTarget) + 76.86, and the limit of the detection (LOD) was determined to be 7.6 × 10−17 M according to the 3σ rule.
lg(cTarget) + 76.86, and the limit of the detection (LOD) was determined to be 7.6 × 10−17 M according to the 3σ rule.
To explore the contributions of the two mechanisms of amplification, comparative amplification experiments were carried out. The SERS responses of the following modes were investigated across the respective concentration gradients:
(a) The complete reaction system was carried out; (b) “RCA only” mode: The logic stage and signal stage were carried out using prepared messenger strands of known concentration as inputs instead of those produced in the initialization stage; (c) “No amplification” mode: as a basepoint for the comparison of amplification, the SERS responses of ROX-tagged strands of known concentration were measured (i.e., in the case where no amplification was applied).
The results of the comparison experiments are shown in Fig. 1, while the SERS responses can be found in Fig. S6 and S7 in the ESI.† The comparison of mode (b) and mode (c) indicated that RCA in the logic stage provided an amplification of about 103 times. The comparison of mode (a) and mode (b) indicated that extra amplification was provided by the initialization stage: to reach the same SERS response, the target miRNA concentration at 10−14 M grade was required in mode (a), while 10−11 M was required in mode (b), thus at this response level, the initialization stage superposed the amplification of about 103 times. The comparison of mode (a) and mode (c) showed that the total reaction system provided an amplification of about 106 times. Thus, the system was verified to be capable of sensing miRNA inputs with low abundance.
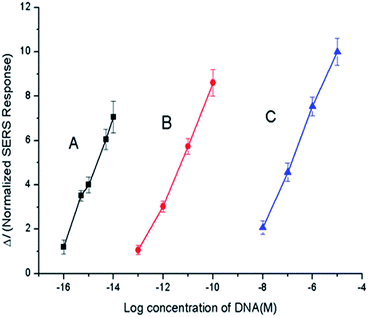 | ||
| Fig. 1 Calibration curves of the amplification performances under different modes: (A) entire reaction system; (B) “RCA only” mode; and (C) “no amplification” mode. | ||
The calibration curve for the “non-amplification” case (Fig. 1, curve C, details shown in the ESI†) was defined by I = 4.56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg(c) + 47.01 in the concentration range of 10−16 to 10−14 M. In contrast, the “RCA only” mode produced a calibration curve of I = 4.24
lg(c) + 47.01 in the concentration range of 10−16 to 10−14 M. In contrast, the “RCA only” mode produced a calibration curve of I = 4.24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg(c) + 56.46.
lg(c) + 56.46.
Logic behaviors
The responses of the linear gates under different input configurations are the key proof for the system. Various cases were tested. The input sets of the two gates were tested (Fig. 2A and B, respectively). Only the gate that received its corresponding input set produced the positive SERS readout, while the other only showed the negative readout. The input of the entire collection containing all four miRNA members activated both gates, producing two positive readouts (Fig. 2C). Finally, as a control, the two shared miRNAs were input, and no positive readout was observed in either gate (Fig. 2D); although the two members can react in both gates, no input set was complete in this case.Interference of the logic behaviors
One key requirement for the logic gate system is that a false-positive readout should not be brought about by incorrect input miRNAs, especially in the case that miRNAs with similar sequences are present, which might occur in real organism due to the nature of the miRNAs. This was examined with modified selectivity experiments: one of the three miRNA members in an input set was replaced with mismatched miRNA, and the output of the logic system was examined.The results (Fig. S12–S14†) confirmed the design: only the correct set of input miRNA produced positive output, while the mismatch of one or more members resulted in negative output. Thus, the specificity of the approach was verified, and the specificity would not be affected under high amplification. The details of the experiments are shown in the ESI.†
Conclusions
In summary, in this study, a proof-of-principle logic system was established to deal with the multi-input, multi-output logic relationship with the aim of identifying the tissue origins of cancer in light of their established relationships with miRNA distributions. The system did this by detecting two characteristic miRNA sets from a collection with two gates, each identifying a set of three input miRNAs with miRNA members shared among the two sets.To fulfil its task, the system centered on logic judgement, which was implemented with a multi-input and multi-output logic gate complex. Signal amplification was also incorporated, providing the system with the practical capacity and efficiency to deal with miRNA collection. This system successfully addressed the multiple-input and multiple-output logic relationship between miRNA distribution and cancer origin. At the same time, other concerns related to the nature of miRNA (e.g., low concentration and instability) were also addressed.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21205066).Notes and references
- (a) H. Blaszyk, A. Hartmann and J. Bjornsson, APMIS, 2003, 111, 1089–1094 CrossRef PubMed; (b) J. D. Hainsworth and F. A. Greco, N. Engl. J. Med., 1993, 329, 257–263 CrossRef CAS PubMed; (c) J. M. Pimiento, D. Teso, A. Malkan, S. J. Dudrick and J. A. Palesty, Am. J. Surg., 2007, 194, 833–838 CrossRef PubMed; (d) P. H. S. Shaw, R. Adams, C. Jordan and T. D. L. Crosby, J. Clin. Oncol., 2007, 19, 87–95 CrossRef CAS; (e) G. R. Varadhachary, J. L. Abbruzzese and R. Lenzi, Cancer, 2004, 100, 1776–1785 CrossRef CAS PubMed.
- (a) S. Baskerville and D. P. Bartel, RNA, 2005, 11, 241–247 CrossRef CAS PubMed; (b) J. Lu, G. Getz, E. A. Miska, E. A. Saavedra, J. Lamb, D. Peck, A. S. Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–838 CrossRef CAS PubMed; (c) S. Volinia, G. A. Calin and C. G. Liu, et al., Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 2257–2261 CrossRef CAS PubMed.
- N. Rosenfeld, R. Aharonov and E. Meiri, et al., Nat. Biotechnol., 2008, 26, 462–469 CrossRef CAS PubMed.
- S. Bi, Y. Yan, S. Hao and S. S. Zhang, Angew. Chem., Int. Ed., 2010, 49, 4438–4442 CrossRef CAS PubMed.
- R. Pei, E. Matamoros, M. Liu, D. Stefanovic and M. N. Stojanovic, Nat. Nanotechnol., 2010, 5, 773–777 CrossRef CAS PubMed.
- (a) L. Qian, E. Winfree and J. Bruck, Nature, 2011, 475, 368–372 CrossRef CAS PubMed; (b) L. Qian and E. Winfree, Science, 2011, 332, 1196–1201 CrossRef CAS PubMed.
- W. Li, Y. Yang, H. Yan and Y. Liu, Nano Lett., 2013, 13, 2980–2988 CrossRef CAS PubMed.
- (a) I. Lee, S. S. Ajay, H. Chen, A. Maruyama, N. Wang, M. G. McInnis and B. D. Athey, Nucleic Acids Res., 2008, 36, e27 CrossRef PubMed; (b) A. Valoczi, C. Hornyik, N. Varga, J. Burgyan, S. Kauppinen and Z. Havelda, Nucleic Acids Res., 2004, 32, e175 CrossRef PubMed.
- (a) H. M. Chan, L. S. Chan, N. S. Wong and H. W. Li, Anal. Chem., 2010, 82, 6911–6918 CrossRef CAS PubMed; (b) S. C. Chapin and P. S. Doyle, Anal. Chem., 2011, 83, 7179–7185 CrossRef CAS PubMed; (c) Y. Cheng, X. Zhang, Z. Li, X. Jiao, Y. Wang and Y. Zhang, Angew. Chem., Int. Ed., 2009, 48, 3268–3272 CrossRef CAS PubMed; (d) F. Degliangeli, P. Kshirsagar and V. J. Brunetti, J. Am. Chem. Soc., 2014, 136, 2264–2267 CrossRef CAS PubMed; (e) R. J. Deng, L. H. Tang, Q. Q. Tian, Y. Wang, L. Lin and J. H. Li, Angew. Chem., Int. Ed., 2014, 53, 2389–2393 CrossRef CAS PubMed; (f) H. X. Jia, Z. P. Li, C. H. Liu and Y. Q. Cheng, Angew. Chem., Int. Ed., 2010, 49, 5498–5501 CrossRef CAS PubMed; (g) S. Neubacher and C. Arenz, ChemBioChem, 2009, 10, 1289–1291 CrossRef CAS PubMed; (h) L. Yang, C. Liu, W. Ren and Z. Li, ACS Appl. Mater. Interfaces, 2012, 4, 6450–6453 CrossRef CAS PubMed.
- (a) X. Kang, S. J. Ye, X. Y. Li, Y. Ma, C. Y. Zhang and B. Tang, Chem. Commun., 2016, 52, 5432–5435 RSC; (b) S. Lai, S. Wang, J. Luo, L. J. Lee, S. T. Yang and M. J. Madou, Anal. Chem., 2004, 76, 1832–1837 CrossRef CAS PubMed; (c) M. Li, J. Zhang, S. Suri, L. J. Sooter, D. Ma and N. Wu, Anal. Chem., 2012, 84, 2837–2842 CrossRef CAS PubMed; (d) X. Li, S. Ye and X. Luo, Chem. Commun., 2016, 52, 10269–10272 RSC; (e) Y. Li, C. Lei, Y. Zeng, X. Ji and S. Zhang, Chem. Commun., 2012, 48, 10892–10894 RSC; (f) Y. Li, X. Qi, C. Lei, Q. Yue and S. Zhang, Chem. Commun., 2014, 50, 9907–9909 RSC; (g) G. Wang and R. J. Lipert, Anal. Chem., 2011, 83, 2554–2561 CrossRef CAS PubMed; (h) S. J. Ye, Y. X. Yang, J. Xiao and S. S. Zhang, Chem. Commun., 2012, 48, 8535–8537 RSC; (i) S. J. Ye, Y. Y. Guo, J. Xiao and S. S. Zhang, Chem. Commun., 2013, 49, 3643–3645 RSC; (j) S. J. Ye, Y. Y. Wu, W. Zhang, N. Lia and B. Tang, Chem. Commun., 2014, 50, 9409–9412 RSC; (k) Z. Zhang, Y. Y. Wang, F. W. Zheng, R. Ren and S. S. Zhang, Chem. Commun., 2014, 51, 907–910 RSC.
- R. Ren, Z. T. Yu, Y. M. Zou and S. S. Zhang, Chem.–Eur. J., 2012, 18, 14201–14209 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra05807a |
| This journal is © The Royal Society of Chemistry 2017 |