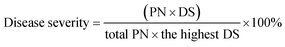Open Access Article
Open Access ArticleGraphene oxide loaded with copper oxide nanoparticles as an antibacterial agent against Pseudomonas syringae pv. tomato
Yadong Li a,
Desong Yang*a and
Jianghu Cui
a,
Desong Yang*a and
Jianghu Cui *b
*b
aCollege of Agriculture, Shihezi University, Shihezi 832000, Xinjiang, P. R. China. E-mail: yds_agr@shzu.edu.cn
bGuangdong Key Laboratory of Integrated Agro-Environmental Pollution Control and Management, Guangdong Institute of Eco-Environmental Science & Technology, Guangzhou 510650, China. E-mail: lantian9426@163.com
First published on 8th August 2017
Abstract
Bacterial speck caused by Pseudomonas syringae pv. tomato (Pst) is a major disease of tomatoes. Infection by Pst in the seedling stage could cause 75% of total yield loss and quality losses in tomatoes. Applying massive chemical bactericides to control this disease has caused undesirable pathogen resistance, environmental pollution, and threats to humans. Here, we developed a candidate antibacterial agent made from copper oxide nanoparticles loaded onto the surfaces of graphene oxide sheets (GO–Cu NPs). A series of characterization measurements showed that the nanocomposite had been successfully prepared. Antibacterial activity results indicated that GO–Cu NPs had a 16-times higher antibacterial activity than Kocide 3000. From further investigation, the GO–Cu NPs composite could lead to damaging the cell structure, increasing the level of reactive oxygen species, and decreasing the content of DNA in Pst cells. From an in vivo test, GO–Cu NPs at 4 and 8 μg mL−1 significantly reduced the severity of bacterial speck below 25% and the tomatoes had no phytotoxicity. Comparatively, 125 and 250 μg mL−1 were required for Kocide 3000 to achieve similar effects. Therefore, GO–Cu NPs composites as a high-efficiency biocide have great potential for managing crop diseases.
1. Introduction
Tomato (Lycopersicon esculentum) is the second most important vegetable crop. Food and Agriculture Organization (FAO) statistics show the current world production of tomatoes was about 170 million tons in 2014. Bacterial speck is a serious bacterial disease in the tomato industry caused by Pseudomonas syringae pv. tomato (Pst). Pst can result in small, sunken, and black lesions on leaves, stems, and fruits.1 According to previous literature, infection of Pst in the seedling stage could cause 75% total yield loss and significant reduction of tomato quality.2 Due to this significant harmfulness, bacterial speck has been the target of numerous studies on control measures for protecting tomato, including biological control and chemical control. It has been reported that some bacteria, such as Bacillus pumilus and Bacillus amyloliquefacies, could efficiently control tomato bacterial speck.3 However, environmental conditions are always adverse for a biocontrol organism to exert antibacterial activity. On the other hand, implementation of biocontrol requires more labour and economic costs. Therefore, it is difficult for biocontrol to be widely applied in the agricultural industry. Chemical agents provide convenient and quick methods to control bacterial speck in tomatoes. Both streptomycin and copper biocide are regarded as the most effective and commonly used agents for control of bacterial speck with tomatoes.4,5 However, long term use of these biocides has induced undesirable pathogen resistance.6,7 Besides, residues of these agents in soil and food are harmful to the environment and human health.8 Therefore, it is urgent to develop alternative bactericides to protect tomatoes from Pst infection.Recently, inorganic nano-biocides, such as silver,9,10 titanium dioxide,11 and zinc oxide,12 have been of great interest in applications for plant disease management. Especially silver-based antibacterial agents have been widely studied due to their good antibacterial activity.13,14 In our previous studies, we prepared some silver-based composites with graphene oxide (GO),15 silicon oxide nanospheres,16 and porous carbon.17 These nanocomposites showed excellent antibacterial activity against Escherichia coli and Xanthomonas oryzae pv. oryzae. However, the practical application of silver composites has been limited due to their high cost and phytotoxicity. Thus, many researchers are searching for an inorganic alternative with low cost and toxicity. Copper biocides also have been used widely in crop disease control as a result of their wide antimicrobial spectrum and low cost. For example, Young et al. and Bogdanović et al. found that Cu nanoparticles (Cu NPs) had greatly enhanced antibacterial activity against bacteria.18,19 In addition, it was reported that Cu NPs were beneficial to growth and nutriment accumulation of Vigna radiate and maize.20,21 However, pure Cu NPs are easy to agglomerate, leading to decrease of antibacterial activity.22 In order to address this problem, we are searching for a suitably-based material to reduce the agglomeration of Cu NPs.
With excellent physical and chemical properties, GO has been widely used in biomedicine, the environment, and agriculture.23–26 As a result of hydrophilic reactive oxygen functional groups on its surface, GO tends to readily disperse in water.27 Chen et al. reported that GO had a broad-spectrum bactericidal efficacy toward plant pathogenic bacteria and fungi.23 And Ocsoy et al. prepared DNA-directed Ag NPs grown on GO which efficiently could control bacterial spots with tomatoes.13 Our previous study showed also that GO and silver nanowire composites could have improved antibacterial activity with better cell compatibility.15 In addition, Zhang et al. reported that graphene had a positive impact on promoting seed germination and seedling growth of tomatoes.28 Thus, GO was believed to be an ideal host material to load with Cu NPs. Here, we report a Cu-based nano-biocide where Cu nanoparticles (Cu NPs) were immobilized on the surfaces of GO sheets (GO–Cu NPs) with good antibacterial activity toward Pst. A series of characterization tests were conducted for particle size, morphology, element composition, and surface structure of the prepared GO–Cu NPs. We further propose that GO–Cu NPs have the ability to induce cell membrane damage, oxidative stress mediated by reactive oxygen species (ROS), and degradation of DNA. Finally, with tomato transplants, GO–Cu NPs at an extremely low concentration reduced the severity of bacterial speck without phytotoxicity, similarly with the current standard treatment.
2. Experimental section
2.1 Materials
Cupric chloride was obtained from Guangzhou Chemical Reagent Factory (Guangzhou, China). Graphite and KMnO4 were purchased from Shanghai Chemical Factory (Shanghai, China). 25% glutaraldehyde was obtained from Chengdu Kelong Chemical Factory (Chengdu, China). 2,7-Dichlorofluorescein-diacetate (DCFH-DA) was purchased from BD Biosciences (CA, USA). Kocide 3000 (46.1 wt%, cupric hydroxide) is a bactericide product of DuPont (Wilmington, DE, USA). Other chemical reagents in this study were analytical grade and used without further purification. Water was purified by using an ultrapure water system (Pine-Tree, China). Pseudomonas syringae pv. tomato (Pst) strain was obtained from an agricultural college, Shihezi University (Shihezi, China). This strain was originally derived from an infected tomato leaf with bacterial speck in an experimental field at Shihezi University.2.2 Preparation of graphene oxide sheets
GO powder was prepared through oxidation of natural flake graphite powder according to a modified Hummers' method.29 Briefly, graphite powder (1 g) was loaded into concentrated H2SO4 (98%, 23 mL) with vigorous stirring for 30 min in an ice bath. Subsequently, KMnO4 (3 g) was added gradually to the mixture with continuous stirring for 2 h below 10 °C. Then the mixture was warmed to 35 °C and stirred continuously for another 2 h, followed by slow addition of 46 mL distilled water and stirring for another 20 min. Finally, 140 mL distilled water was added to terminate the reaction. Then, 30% H2O2 was added and the colour of the mixture changed to bright yellow. The sample was washed with repeated centrifugation with an HCl solution and distilled water. The sediment was dried in vacuum and GO powder was obtained.2.3 Preparation and characterization of GO–Cu NPs composites
Cupric chloride was reduced and immobilized on the surfaces of GO sheets by ammonia water. Briefly, 40 mg GO powder was dissolved in 40 mL distilled water and flaked into a homogeneous GO suspension by sonication for 30 min. Then, 2 g cupric chloride was added into the GO suspension and kept stirring for 30 min. Then, 1 mL ammonia water was quickly added into the mixture with vigorous stirring for 1 h. After that, the obtained samples were washed three times with deionized water with centrifugation. The sample was dried in vacuum for further measurements.Morphology of the composites was investigated with a Philips TECNAI 10 transmission electron microscope (TEM). Samples for TEM tests were prepared by placing a drop of solution dispersion onto carbon-coated copper grids and drying at room temperature. Elemental composition was analyzed with a JEOL JEM-2100F high-resolution TEM (HRTEM) equipped with an Oxford INCA energy-dispersive X-ray spectroscopy (EDS) device. Microstructures of the obtained samples were recorded on a MSAL-XD2 X-ray diffractometer (XRD) employing Cu target in the 2θ range from 5° to 80° (40 kV, 30 mA, λ = 1.54051 Å). Interactions of GO with Cu NPs were carried out with a Nicolet Avatar-300 Fourier transform infrared (FTIR) spectrophotometer.
2.4 Antibacterial assays
To explore the antibacterial properties of GO–Cu NPs, Pst was introduced for measurements of agar disk diffusion and minimum inhibitory concentration (MIC) tests. GO and sterile water served as negative controls. The presence of antibacterial activity of GO–Cu NPs was affirmed through agar disk diffusion tests.30 Briefly, Pst cells were incubated in King's B broth with shaking at 25 °C and 180 rpm overnight. Then the bacteria suspension was diluted to an optical density at 600 nm (OD600) of 0.1 (∼1 × 108 cfu mL−1). A 100 μL bacterial suspension was swabbed over a King's B agar plate. Filter paper discs, beforehand immersed in sterile water, GO, GO–Cu NPs, and Kocide 3000, respectively, were placed on a plate. After incubating at 25 °C for 48 h, the size of the inhibition zone around the filter papers was measured. To further evaluate the antibacterial activity level of GO–Cu NPs, MIC was measured according to the broth dilution method.31 Kocide 3000 was tested synchronously as a reference. First, the Pst suspension was incubated with different concentrations of GO–Cu NPs or Kocide 3000 (0 to 250 μg mL−1) at 25 °C and 180 rpm for 2 h. Then, 100 μL from each treatment was evenly coated on King's B agar plates. The MIC was recorded as the lowest concentration at which no visible colonies grew after being inoculated at 25 °C for 48 h.16 All experiments were repeated three times.2.5 Cell morphology observation
To observe morphological changes of bacterial cells after treatment with the GO–Cu NPs composites, Pst cells were exposed to the GO–Cu NPs composites (32 μg mL−1). Sterile water served as a control treatment. All the samples were harvested by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min and washed three times with phosphate buffered saline. After that, the condensed bacteria were fixed with 2.5% glutaraldehyde overnight at 4 °C and post-fixed with 1% aqueous osmic acid for 2 h. Then the samples were rinsed with buffer and dehydrated in an ascending ethanol series (50%, 70%, 80%, 90%, and 100%) for 15 min, respectively. Subsequently, the samples were embedded in Epon/Araldite resin according to a standard protocol. Finally, thin sections containing the cells were stained with 4% uranyl acetate and 0.2% Reynolds lead citrate and then air-dried for TEM detection.
000 rpm for 3 min and washed three times with phosphate buffered saline. After that, the condensed bacteria were fixed with 2.5% glutaraldehyde overnight at 4 °C and post-fixed with 1% aqueous osmic acid for 2 h. Then the samples were rinsed with buffer and dehydrated in an ascending ethanol series (50%, 70%, 80%, 90%, and 100%) for 15 min, respectively. Subsequently, the samples were embedded in Epon/Araldite resin according to a standard protocol. Finally, thin sections containing the cells were stained with 4% uranyl acetate and 0.2% Reynolds lead citrate and then air-dried for TEM detection.
2.6 Measurement of intracellular reactive oxygen species (ROS)
To determine generation of ROS in Pst cells, an indicator of ROS, 2,7-dichlorofluorescein diacetate (DCFH-DA), was introduced into this experiment. DCFH-DA can easily enter a cell and be hydrolysed by cellular esterase into DCFH. Subsequently, DCFH reacts with intracellular ROS to produce highly fluorescent dichlorofluorescein (DCF). Fluorescence intensity is proportional to the amount of ROS.32 Herein, Pst suspensions were treated with GO–Cu NPs, GO, and sterile water respectively for 2 h at 25 °C and 180 rpm. Then the Pst cells were strained with DCFH-DA for 30 min and washed twice with phosphate buffered saline in darkness. Fluorescent intensity of DCF was measured by flow cytometry with an excitation wavelength of 488 nm. For each sample, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were measured for the mean fluorescent intensity to present the intracellular generation of ROS.
000 cells were measured for the mean fluorescent intensity to present the intracellular generation of ROS.
2.7 Analysis of DNA content
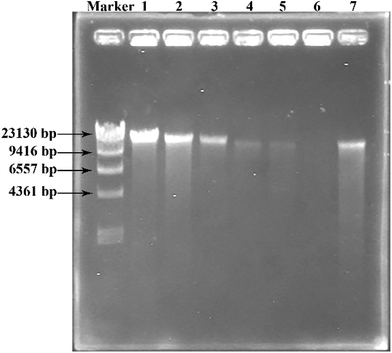
To investigate the degradation of DNA in Pst cells treated with GO–Cu NPs, DNA kits (Tiangen, Beijing, China) were employed to extract the DNA from Pst cells. Before the operation, Pst was allowed to inoculate in King's B broth and incubated under 25 °C and 180 rpm overnight. The experiments were carried out in Pst suspensions (OD600 = 0.3) containing 0, 8, 16, 32, 62.5, and 125 μg mL−1 of GO–Cu NPs. After being inoculated at 25 °C and 180 rpm for 2 h, 2 mL of cell suspension from each treatment was pelleted in a microcentrifuge. The DNA was isolated and dissolved in 50 μL buffer according to specifications of the kits. Subsequently, 10 μL of each DNA solution was mixed thoroughly with 2 μL 6× DNA loading buffer (Beijing TransGen Biotech Co., Ltd., China) and electrophoresis was conducted in 1% agarose gel using ethidium bromide to stain. DNA concentration was determined by the intensity of the DNA band observed from the gel imaging and analysis system (Bio-Rad, USA).2.8 Greenhouse experiment
To evaluate the control effect of GO–Cu NPs against the bacterial speck in tomatoes, an in vivo study was conducted on tomato transplants of the cultivar “Shi Hong 9”. Tomato plants were grown in plastic pots containing cultivated soil in a greenhouse. After growing to a five-leaf stage, GO–Cu NPs at 4 and 8 μg mL−1 were sprayed onto the tomato leaves. Plants treated with Kocide 3000 (125 and 250 μg mL−1) and sterile water were kept as controls. Each treatment was repeated five times. Two hours after treatment, Pst was inoculated on these treated plants by spraying. All the treated leaves of the tomatoes were bagged with transparent plastic bags after the infection of Pst. After 48 h, the bags were opened and all plants were placed in a greenhouse at 20–25 °C and ≥70% relative humidity. Then the percentage of bacterial speck severity was recorded at the 4th, 7th, and 10th days according to a method reported by Ocsoy.13 The disease severity was calculated according to the following formula:Note: PN represents plant number; DS represents disease scale.
2.9 Statistical analysis
One-way analysis of variance (ANOVA) and Duncan's multiple range test with a significant level of 95% (p < 0.05) were performed to confirm the statistical differences by using IBM SPSS Statistics 19.3. Results and discussion
3.1 Characterization of GO–Cu NPs
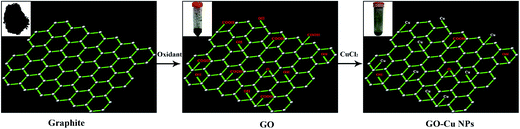
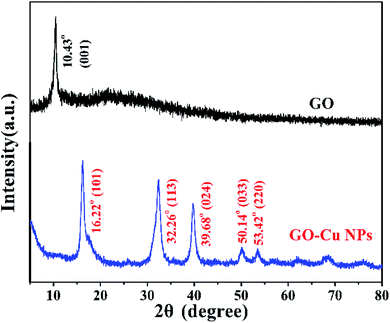
Cu NPs were immobilized on the surface of GO by ammonia reductant and stabilizer. The detailed synthesis process is illustrated in Fig. 1. TEM was employed to observe the morphologies of GO and GO–Cu NPs. GO was observed as a thin and semitransparent sheet in Fig. 2A. After the reaction, many dark spots were firmly immobilized on GO sheets (Fig. 2B and C). From the HRTEM image (Fig. 2D), the lattice fringes of these dark spots on GO were measured as 0.209 nm and assigned to the (111) lattice plane of Cu NPs.33 The size of Cu NPs, as measured through TEM, was approximately 21.28 nm. Such small particles may enhance antibacterial properties due to more interaction chances with bacteria.9,34 The elemental component of as-prepared GO–Cu NPs was determined by EDS and indicated that the GO–Cu NPs composites contained carbon, oxygen, chlorine, and copper elements (Fig. 2E). Among all the elements, copper content accounted for 57.46 wt% of the GO–Cu NPs. The microstructures of GO and GO–Cu NPs were further revealed by XRD patterns (Fig. 3). A specific XRD pattern with a characteristic peak concerting at 2θ = 10.43° was in agreement with the crystal structure of GO.35 However, this peak disappeared in the pattern of GO–Cu NPs indicating Cu NPs changed the surface structure of GO. The diffraction peaks of GO–Cu NPs at diffraction angles of 16.22°, 32.26°, 39.68°, 50.14°, and 53.42° were indexed as (101), (113), (024), (033), and (220) crystalline planes, which was similar to the paratacamite crystal (Cu2(OH)3Cl, JCPDS file no. 87-0679).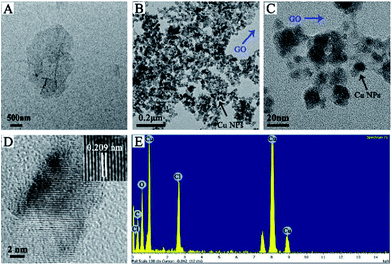 | ||
| Fig. 2 TEM images of (A) GO and (B and C) GO–Cu NPs; (D) HRTEM image of an individual Cu NPs on GO; (E) EDS pattern of GO–Cu NPs. | ||
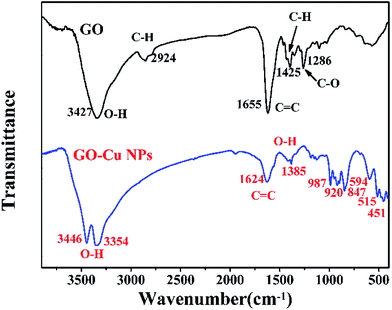
FTIR spectroscopy was conducted to detect changes in functional groups. The results of FTIR showed that the spectrum of GO displayed numerous functional groups peaks, including the stretching vibration peaks of O–H (3427 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (1655 cm−1) and C–O (1286 cm−1), the asymmetric stretching vibration of C–H bonds in –CH3 and –CH2 (2924 cm−1)36 and the in-plane bending vibration of C–H (1425 cm−1) (Fig. 4). After the reaction between GO and Cu NPs, several changes took place in the function groups. The strong peak at 3427 cm−1 was divided into two adsorptions of O–H stretching vibration at 3446 cm−1 and 3354 cm−1.37 The adsorption band of C
C (1655 cm−1) and C–O (1286 cm−1), the asymmetric stretching vibration of C–H bonds in –CH3 and –CH2 (2924 cm−1)36 and the in-plane bending vibration of C–H (1425 cm−1) (Fig. 4). After the reaction between GO and Cu NPs, several changes took place in the function groups. The strong peak at 3427 cm−1 was divided into two adsorptions of O–H stretching vibration at 3446 cm−1 and 3354 cm−1.37 The adsorption band of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C at 1655 cm−1 drifted to 1624 cm−1.38 The adsorption bonds at 2924 cm−1, 1425 cm−1 and 1286 cm−1 disappeared.
C at 1655 cm−1 drifted to 1624 cm−1.38 The adsorption bonds at 2924 cm−1, 1425 cm−1 and 1286 cm−1 disappeared.
However, some new peaks appeared, including the O–H stretching vibration at 1385 cm−1,39 the C–H out-plane bending vibrations at 987, 920, and 847 cm−1 and Cu–O stretching vibrations in monoclinic CuO below 600 cm−1.40 These changes in functional groups suggested that Cu NPs were successfully immobilized on the surface of GO.
3.2 Antibacterial assay
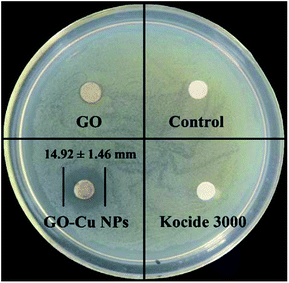
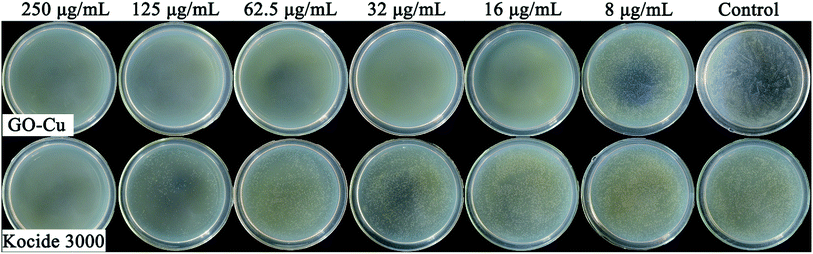
Antibacterial activity of GO–Cu NPs on Pst was qualitatively investigated by an agar disk diffusion test. As shown in Fig. 5, GO–Cu NPs were observed by a clear and significant inhibitory zone, while the other treatments had no inhibitory zone. The average diameter value of inhibition zone for GO–Cu NPs was (14.92 ± 1.46) mm. These results indicated that Kocide 3000 and GO separately have no considerable antibacterial activity at tested concentrations. However, GO–Cu NPs composites have good antibacterial property against Pst.To study the antibacterial effect of GO–Cu NPs further, the value of MIC was measured using a microdilution broth method. After 48 h incubation, GO–Cu NPs and Kocide 3000 exhibited good antibacterial activity in a dose-dependent manner (Fig. 6). No colony growth was observed with treatment of GO–Cu NPs (16 μg mL−1) and Kocide 3000 (250 μg mL−1). Therefore, the MIC value of GO–Cu NPs was almost 16 times lower than that of Kocide 3000, indicating GO–Cu NPs composites had better antibacterial property. The enhanced antibacterial activity of GO–Cu NPs may be derived from congregated Cu NPs tending to disperse in the process of application. In our study, Cu NPs were loaded onto the surface of GO, which could effectively prevent Cu NPs from aggregation.
Our result was consistent with a previously reported study.41 On the other hand, FTIR results showed GO–Cu NPs composites had some functional groups, such as C–O, which could greatly improve the dispersive capacity of GO–Cu NPs composites in solution. In addition, it has been reported that GO has strong antibacterial activity against plant pathogenic bacterium.23 In our study, GO had no significant antibacterial ability due to its low concentration. Consequently, the enhanced antibacterial effect of GO–Cu NPs might be caused by a synergistic effect between GO and Cu NPs.
3.3 Observation of the cell morphology change
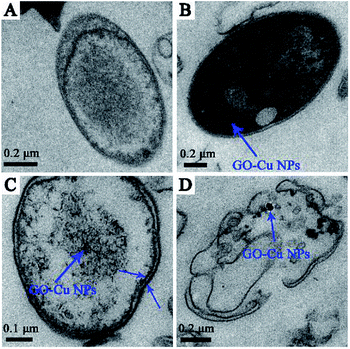
According to the previous study, GO sheets tended to non-specifically wrap the bacteria cells.42 Meanwhile, the positive charge of Cu NPs can bind to the negative charge of bacteria cell surfaces and react with phosphorus- and sulfur-containing compounds.34,43,44 Based on the previous antibacterial results, we further investigated the antibacterial behaviour of GO–Cu NPs throughout using TEM. Results showed that the untreated cells displayed intact structures without morphological changes (Fig. 7A). However, after incubation with GO–Cu NPs, the nanoparticles were adsorbed onto the cell membrane/wall (Fig. 7B) and then penetrated the cell Fig. 7C which was likely due to a change of cell permeability.45 Cu NPs inside the cells could react with the intercellular structure, and then damaged it. Consequently, disorganization of the bacterial membrane and damage of the cellular structure leads to bacteria cell death (Fig. 7D).3.4 Intracellular reactive oxygen species (ROS)
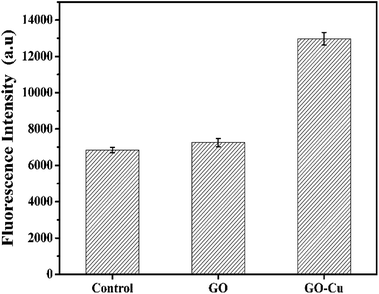
ROS generation is regarded as one of the critical antibacterial mechanisms of Cu NPs.46,47 A high level of ROS inside cells can perturb the redox potential equilibrium, produce a pro-oxidant environment, and further result in a range of adverse biological responses.48 In order to investigate the level of ROS inside the GO–Cu NPs-exposed Pst cells, DCFH-DA was introduced as a visual indicator for the oxidative status of a cell; fluorescent intensity of DCF is positively correlated with the amount of ROS. Compared with the control, GO-treated cells had no noticeable increase of ROS (Fig. 8), which was in agreement with a previous report.42 However, the level of ROS in the GO–Cu NPs treated cells was significantly increased (almost 1.9 times more than the control). Such a significant increase of intercellular ROS can lead to a strong oxidative environment inside the cells, which mediates oxidative damage to membrane lipids, proteins, DNA, and other compounds.483.5 DNA damage caused by GO–Cu NPs
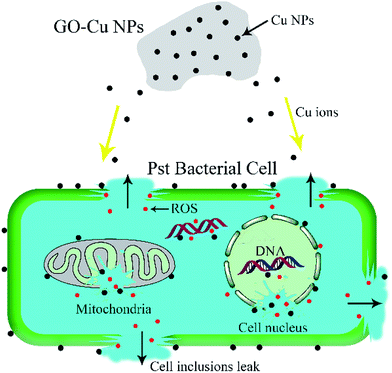
Both Cu NPs and intercellular ROS are known to target cellular DNA, leading to DNA strand breaks and blocking DNA replication.46,49 Released Cu ions from Cu NPs inside bacteria cells can bind strongly to DNA at binding sites and react with cellular ROS, producing hydroxyl radicals which immediately attacked the DNA bases.49,50 In order to investigate the fate of DNA from Pst cells with exposure to GO–Cu NPs, the DNA was extracted and analysed by electrophoresis. The intensity of a DNA band in agarose gel has a positive relation with the DNA concentration. From the result (Fig. 9), a strong DNA band was observed from the control treatment (lane 1). However, the DNA concentration decreased with an increase of GO–Cu NPs concentration (lanes 2 to 6) as compared to the control treatment (lane 1). No DNA band was observed in the samples treated with GO–Cu NPs at 32 μg mL−1 and above due to their complete inhibition to Pst cells growth (lanes 4 to 6). In the case of the cells incubated with Kocide 3000 at 250 μg mL−1 (lane 7), the DNA concentration was similar to that of GO–Cu NPs at 16 μg mL−1 (lane 3), due to their similar effects in the MIC test.According to the above results, the antibacterial mechanism of GO–Cu NPs was proposed as follows (Fig. 10): when GO–Cu NPs were exposed to Pst cells, the Cu NPs were released from GO–Cu NPs and adhered to the bacteria cell surfaces. Then the Cu NPs reacted with the membrane protein, leading to increased cell permeability which damaged the integrity of cell membrane/wall. Therefore, Cu NPs on a cell surface can penetrate inside the cells, which causes serious damage by reacting with DNA and other intracellular compounds. These reactions could produce a high level of ROS which mediated strong oxidative stress in the cell. The membrane lipids, proteins, DNA, and other compounds were subsequently degraded under the oxidative environment. Finally, bacteria cells were thoroughly killed.
3.6 Effectiveness of GO–Cu NPs against bacterial speck on tomato
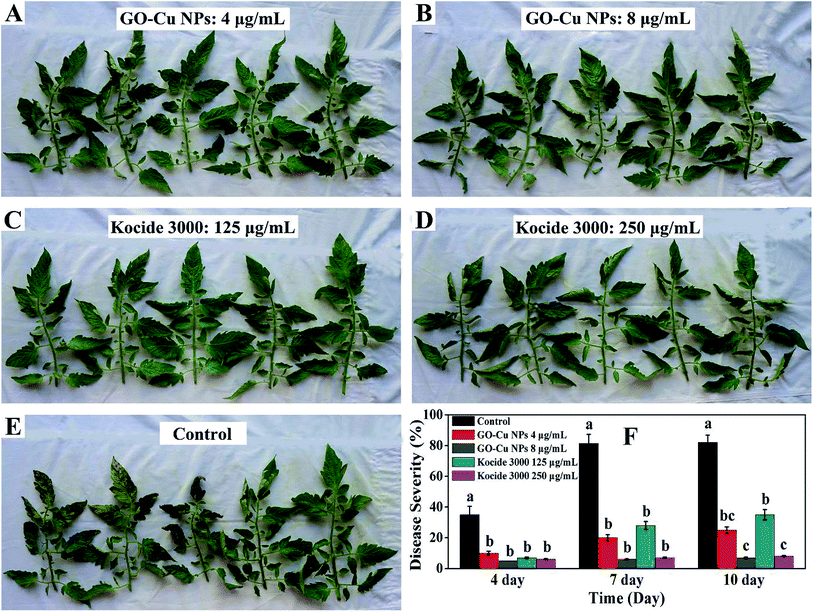
To further investigate practical efficiency, GO–Cu NPs were applied to tomato transplants to control bacterial speck. The severity of the disease was recorded after 4, 7, and 10 days (Fig. 11). Results indicated that tomatoes treated with GO–Cu NPs had no obvious bacterial speck after the fourth day (Fig. 11A and B). Even after 10 days, the disease severity on tomato leaves treated with GO–Cu NPs was still below 25% (Fig. 11F). When treated with Kocide 3000 at 125 and 250 μg mL−1, the disease severity was below 35% at the 10th day (Fig. 11C, D and F). However, with the control treatment, a large number of bacterial specks were observed (Fig. 11E). Bacterial speck severity increased from 35% at the fourth day to over 80% at the tenth day (Fig. 11F). In addition, treated tomatoes with GO–Cu NPs had no phytotoxicity. Although there was no significant difference between the control effects of the two agents, the concentration of Kocide 3000 was more than 32 times higher than GO–Cu NPs. It was concluded that the GO–Cu NPs have better antibacterial activity than Kocide 3000.4. Conclusions
Copper oxide nanoparticles were successfully loaded on the surface of GO by a facile chemical reduction method in aqueous solutions with the assistance of ammonia water. It was found that GO–Cu NPs have excellent antibacterial activity at extremely low concentrations. Investigation of the antibacterial mechanism showed that a GO–Cu NPs composite could lead to damaging the cell structure, increasing the level of reactive oxygen species, and decreasing the content of DNA in bacterial cells. The in vivo test on tomato plants indicated that GO–Cu NPs could reduce the severity of bacterial speck without phytotoxicity. Therefore, it is important and valuable to explore potential applications of this composite as a biocide for managing crop diseases.Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 41401564), Guangdong Science and Technology Plan Program (2014A030313706), Science and Technology Program of Guangzhou, China (1563000751) and Opening Fund of Key Laboratory of Integrated Pest Management on Crops in Northwestern Oasis of Ministry of Agriculture (KFJJ20170105).Notes and references
- M. K. Bryan, Phytopathology, 1933, 23, 897–904 Search PubMed
.
- H. Yunis, Y. Bashan, Y. Okon and Y. Henis, Plant Dis., 1980, 64, 937–939 CrossRef
.
- R. Lanna Filho, R. M. de Souza, A. Ferreira, M. C. Quecine, E. Alves and J. L. de Azevedo, Australas. Plant Pathol., 2013, 42, 643–651 CrossRef
.
- K. C. Conlin, Plant Dis., 1983, 67, 639–644 CrossRef CAS
.
- W. G. Bonn and S. Lesage, J. Environ. Sci. Health, Part B, 1984, 19, 29–38 CrossRef CAS PubMed
.
- C. K. Bower and M. A. Daeschel, Int. J. Food Microbiol., 1999, 50, 33–44 CrossRef CAS PubMed
.
- F. J. Louws, M. Wilson, H. L. Campbell, D. A. Cuppels, J. B. Jones, P. B. Shoemaker, F. Sahin and S. A. Miller, Plant Dis., 2001, 85, 481–488 CrossRef CAS
.
- W. Zhang, F. Jiang and J. Ou, Proc. Int. Acad. Ecol. Environ. Sci., 2011, 1, 125–144 CAS
.
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramírez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS PubMed
.
- M. A. Salam, A. Y. Obaid, R. El-Shishtawy and S. A. Mohamed, RSC Adv., 2017, 7, 16878–16884 RSC
.
- K. Sunada, Y. Kikuchi, K. Hashimoto and A. Fujishima, Environ. Sci. Technol., 1998, 32, 726–728 CrossRef CAS
.
- C. O. Dimkpa, J. E. McLean, D. W. Britt and A. J. Anderson, BioMetals, 2013, 26, 913–924 CrossRef CAS PubMed
.
- I. Ocsoy, M. L. Paret, M. A. Ocsoy, S. Kunwar, T. Chen, M. You and W. Tan, ACS Nano, 2013, 7, 8972–8980 CrossRef CAS PubMed
.
- C. Marambio-Jones and E. M. V. Hoek, J. Nanopart. Res., 2010, 12, 1531–1551 CrossRef CAS
.
- J. Cui and Y. Liu, RSC Adv., 2015, 5, 85748–85755 RSC
.
- J. Cui, Y. Liang, D. Yang and Y. Liu, Sci. Rep., 2016, 6, 21423 CrossRef CAS PubMed
.
- J. Cui, Y. Yang, Y. Hu and F. Li, J. Colloid Interface Sci., 2015, 455, 117–124 CrossRef CAS PubMed
.
- M. Young and S. Santra, J. Agric. Food Chem., 2014, 62, 6043–6052 CrossRef CAS PubMed
.
- U. Bogdanović, V. Lazić, V. Vodnik, M. Budimir, Z. Marković and S. Dimitrijević, Mater. Lett., 2014, 128, 75–78 CrossRef
.
- S. Pradhan, P. Patra, S. Mitra, K. K. Dey, S. Basu, S. Chandra, P. Palit and A. Goswami, J. Agric. Food Chem., 2015, 63, 2606–2617 CrossRef CAS PubMed
.
- V. Saharan, R. V. Kumaraswamy, R. C. Choudhary, S. Kumari, A. Pal, R. Raliya and P. Biswas, J. Agric. Food Chem., 2016, 64, 6148–6155 CrossRef CAS PubMed
.
- H. L. Karlsson, P. Cronholm, J. Gustafsson and L. Moller, Chem. Res. Toxicol., 2008, 21, 1726–1732 CrossRef CAS PubMed
.
- J. Chen, H. Peng, X. Wang, F. Shao, Z. Yuan and H. Han, Nanoscale, 2014, 6, 1879–1889 RSC
.
- V. Chabot, D. Higgins, A. Yu, X. Xiao, Z. Chen and J. Zhang, Energy Environ. Sci., 2014, 7, 1564–1596 CAS
.
- W. Liu, C. Sun, C. Liao, L. Cui, H. Li, G. Qu, W. Yu, N. Song, Y. Cui, Z. Wang, W. Xie, H. Chen and Q. Zhou, J. Agric. Food Chem., 2016, 64, 5909–5918 CrossRef CAS PubMed
.
- R. Karthik, M. Govindasamy, S.-M. Chen, T.-W. Chen, J. Vinoth kumar, A. Elangovan, V. Muthuraj and M.-C. Yu, RSC Adv., 2017, 7, 25702–25709 RSC
.
- D. R. Dreyer, S. Park, C. W. Bielawski and R. S. Ruoff, Chem. Soc. Rev., 2010, 39, 228–240 RSC
.
- M. Zhang, B. Gao, J. Chen and Y. Li, J. Nanopart. Res., 2015, 17, 1–8 CrossRef CAS
.
- W. S. Hummers and R. E. Offeman, J. Am. Chem. Soc., 1958, 80, 1339 CrossRef CAS
.
- K. Fiebelkorn, S. Crawford, M. McElmeel and J. Jorgensen, J. Clin. Microbiol., 2003, 41, 4740–4744 CrossRef CAS PubMed
.
- I. Wiegand, K. Hilpert and R. E. W. Hancock, Nat. Protoc., 2008, 3, 163–175 CrossRef CAS PubMed
.
- C. P. LeBel, H. Ischiropoulos and S. C. Bondy, Chem. Res. Toxicol., 1992, 5, 227–231 CrossRef CAS PubMed
.
- V. A. Phillips and J. A. Hugo, Micron, 1969, 1971(3), 212–223 Search PubMed
.
- M. Raffi, S. Mehrwan, T. M. Bhatti, J. I. Akhter, A. Hameed, W. Yawar and M. M. ul Hasan, Ann. Microbiol., 2010, 60, 75–80 CrossRef CAS
.
- J. Ma, J. Zhang, Z. Xiong, Y. Yong and X. Zhao, J. Mater. Chem., 2011, 21, 3350–3352 RSC
.
- J. Liu, H. Bai, Y. Wang, Z. Liu, X. Zhang and D. D. Sun, Adv. Funct. Mater., 2010, 20, 4175–4181 CrossRef CAS
.
- J. Chen, X. Wang and H. Han, J. Nanopart. Res., 2013, 15, 1658 CrossRef
.
- Q. Chen, L. Zhang and G. Chen, Anal. Chem., 2011, 84, 171–178 CrossRef PubMed
.
- Y. Huang, Y. Qin, Y. Zhou, H. Niu, Z.-Z. Yu and J.-Y. Dong, Chem. Mater., 2010, 22, 4096–4102 CrossRef CAS
.
- Y. Xu, D. Chen and X. Jiao, J. Phys. Chem. B, 2005, 109, 13561–13566 CrossRef CAS PubMed
.
- D. Zhang, X. Liu and X. Wang, J. Inorg. Biochem., 2011, 105, 1181–1186 CrossRef CAS PubMed
.
- S. Liu, T. H. Zeng, M. Hofmann, E. Burcombe, J. Wei, R. Jiang, J. Kong and Y. Chen, ACS Nano, 2011, 5, 6971–6980 CrossRef CAS PubMed
.
- P. K. Stoimenov, R. L. Klinger, G. L. Marchin and K. J. Klabunde, Langmuir, 2002, 18, 6679–6686 CrossRef CAS
.
- T. J. Beveridge and R. G. Murray, J. Bacteriol., 1980, 141, 876–887 CAS
.
- Y. Ohsumi, K. Kitamoto and Y. Anraku, J. Bacteriol., 1988, 170, 2676–2682 CrossRef CAS PubMed
.
- M. Shi, H. S. Kwon, Z. Peng, A. Elder and H. Yang, ACS Nano, 2012, 6, 2157–2164 CrossRef CAS PubMed
.
- M. Valko, H. Morris and M. T. D. Cronin, Curr. Med. Chem., 2005, 12, 1161–1208 CrossRef CAS PubMed
.
- N. Ercal, H. Gurer-Orhan and N. Aykin-Burns, Curr. Trends Med. Chem., 2001, 1, 529–539 CrossRef CAS
.
- J.-L. Sagripanti, P. L. Goering and A. Lamanna, Toxicol. Appl. Pharmacol., 1991, 110, 477–485 CrossRef CAS PubMed
.
- O. I. Aruoma, B. Halliwell, E. Gajewski and M. Dizdaroglu, Biochem. J., 1991, 273, 601–604 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2017 |