Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Authentication of shrimp muscle in complex foodstuff by in-solution digestion and high-resolution mass spectrometry
Qing Chen,
Xiao-Dong Pan * and
Bai-Fen Huang
* and
Bai-Fen Huang
Zhejiang Provincial Center for Disease Control and Prevention, Physical-chemistry Room No. 401, Bin-Sheng Road No. 3399, Binjiang District, Hangzhou, 310051, China. E-mail: zjupanxiaodong@hotmail.com; Fax: +86 571 87115261; Tel: +86 571 87115274
First published on 28th June 2017
Abstract
A method for shrimp muscle identification in complex foods is required to safeguard the shrimp-allergic population. This study described a method for authentication of shrimp in complex foodstuffs (fish balls) by liquid chromatography tandem QTOF mass spectrometry (UPLC-QTOF-MS). The proteins in shrimp muscle were extracted using a Tris–HCl solution and then digested using tryptic protease. The main allergen proteins, tropomyosin (TM) and arginine kinase (AK), were characterized using the ‘bottom up’ MS approach. After analysis of their peptide mass fingerprinting based on the UniProt database, two specific heat-stable peptides, ALSNAEGEVAALNR for TM and VSSTLSSLEGELK for AK, were screened as surrogate (signature) peptides. The detection limit, expressed as shrimp meat per kilogram of food, was 8 g kg−1 (usage of TM) or 5 g kg−1 (usage of AK). The developed method is suitable to screen potential addition of shrimp meat in foodstuffs by detection of allergen proteins.
Introduction
The consumption of shrimp continuously increases worldwide due to its well-established health benefits. For the majority of world's population, the growing interest can be considered a nutritional advantage. However, for a small but rather significant group of food-allergic individuals, the consumption can pose a severe health problem.1 Many cases of shellfish allergies have been frequently reported, being currently viewed as an emergent issue of public health.2,3In the European Union (EU), food labeling regulations have been revised and labeling of several allergenic ingredients is now mandatory (Directives 2003/89/EC and 2007/68/EC).4 But different countries mandate a different selection of allergens for food labeling. Despite this regulation, total avoidance might be difficult for the allergic consumer. Furthermore, little is known on threshold doses, i.e. the minimum amount of an allergenic food which is able to cause an allergic reaction. Hence, the sensitive and selective analytical methods for allergens are required to protect certain consumers.
The most frequently used analytical methods for allergen detection are either immunological based on antibodies or polymerase chain reactions (PCR).2 Many disadvantages of current established methods for allergen analyses are discussed, such as cross-reactivity and potential false-negative results.5–8 Recently, LC/MS technique using protease digestion was developed to characterize and identify proteins.9–12 Different mass analyzer including triple quadrupole (QQQ), time-of-flight (TOF), orbitrap and ion trap (IT) are applied for the allergen test.13,14 Because MS identification is a direct type of detection using different principles, it is expected to be an effective method for allergen confirmation. Noticeably, identifying and detecting allergens using MS systems, sample preparation is a critical step. Allergen proteins are usually extracted from food matrice and digested with enzymes generating peptides15
In this study, we aimed to analysis shrimp allergen protein in complex foodstuffs (fish balls) by liquid chromatography tandem QToF mass spectrometry (UPLC-QToF-MS). The allergen protein was prepared by in-solution digestion and (solid phase extraction) SPE clean-up. The main allergen protein in shrimp, tropomyosin (TM) or arginine kinase (AK) was tested by their optimized signature peptides.
Experimental
Chemicals and materials
Ammonium bicarbonate (NH4HCO3), dithiotheritol (DTT), iodoacetamide (IAA) and hydrochloric acid (HCl, 37%) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile (ACN) and formic acid (FA) were purchased from Merck (Darmstadt, Germany). All the reagents used were analytical or HPLC grade. Sequencing grade modified trypsin was from Shanghai Yaxin Biotechnology Co., Ltd (Shanghai, China). All chemical agents were prepared using ultrapure water and without further purification. Ultrapure water was obtained by a Milli-Q Gradient A10 water purification system (Millipore, Bedford, MA, USA) during all the experiments.Samples
The samples (fish balls) were purchased from local supermarket (TESCO, Hangzhou, China). Fish balls are mainly made from fish meat, starch, pork meat or shrimp meat. For validation of developed method, the home-made fish balls containing 10, 50, 100, 150, 200 g kg−1 shrimp meat were prepared by mixture of fish meat (Anguilla japonica), starch (Dioscorea esculenta Burkill), pork meat (Yorkshire), shrimp meat (Penaeus vannamei) and water. The home-made fish balls are cooked in boiled water for 30 min. The samples were stored at −20 °C before experimental analysis.Sample extraction and digestion
Allergen proteins were extracted from 5 g ground matrix in 15 mL Tris–HCl (200 mM, pH 9.2) with 2 M urea by shaking for 30 min. Then, the mixture was sonicated for 15 min at 4 °C to avoid carbamidomethylation with urea. The samples were centrifuged at 5000 g for 15 min at 10 °C. 100 μL supernatant, 100 μL 500 mM NH4HCO3 and 665 μL deionized water were mixed in an Eppendorf tube. 10 μL 50 mM DTT solution were added to the mixtures and reduced in 40 °C water bath for 30 min at this stage. In the next step an alkylation was performed by adding 10 μL of 150 mM IAA in the dark for 30 min at room temperature. Immediately prior to the incubation, 100 μL of 500 mM NH4HCO3 and 10 μL of 400 μg m L−1 trypsin (freshly prepared) were added and incubated 6 h at 37 °C. The reaction was terminated by addition of 5 μL formic acid. The insoluble substances in tryptic hydrolysates were removed by centrifuging at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. Before analysis of Q-TOF, the supernatant was further cleaned up by SPE.
000g for 10 min. Before analysis of Q-TOF, the supernatant was further cleaned up by SPE.
For screening of the surrogated peptides for the allergens, 0.5 g shrimp muscle sample was directly selected, and prepared by the above method.
Peptides clean-up
SPE purification of digested proteins was performed on Oasis® HLB SPE columns (3 cm3/60 mg, 30 μm). Cartridge was pre-conditioned with 1 mL MeOH followed by equilibration with 1 mL water of 0.1% formic acid. The digested samples were loaded on the column. The samples were washed with 1 mL of 20% MeOH in water. The analytes were eluted with 1 mL MeOH and then dried down under nitrogen. The dried samples were reconstituted into 1 mL of 2% acetonitrile in water with 0.1% formic acid. The mixture was analyzed by UPLC-QToF-MS after passing through a 0.22 μm nylon filter (Agela Technologies).Instrument conditions
Tryptic hydrolysates were separated using an ACQUITY UPLC System equipped with ACQUITY UPLC binary solvent manager, sample manager, and column manager (Waters, Milford, MA, USA). Chromatographic separation was carried out on a narrow-bore Acquity UPLC BEH 300C18 column (1.7 μm, 2.1 mm × 100 mm) maintained at 35 °C, equipped with a guard column of the same material (Waters, Milford, MA, USA). The 0.1% FA aqueous solution (solvent A) and 0.1% FA ACN solution (solvent B) were used for the mobile phases. Gradient elution was: 2% B to 40% B for 20 min; 100% B for 1 min; re-equilibration at the initial conditions for 2 min. The flow rate for separations was maintained at 0.3 mL min−1 and a 10.0 μL injection volume was used for all standards and samples.Quadruple time-of-flight tandem mass spectrometry (QToF-MS) detection was performed on a Synapt G2 HDMS equipped with an electrospray ion (ESI) source (Waters). All data were acquired in the electrospray positive ion (ESI+) mode with MSE mode. Details of TOF conditions were as follows: capillary voltage, 3 kV; sampling cone voltage, 25 V; extraction cone voltage, 4 V; source temperature, 100 °C; desolvation temperature, 400 °C; cone gas flow, 30 L h−1; desolvation gas flow, 800 L h−1; ramp trap collision energy, 15–35 V; and lockspray reference compound, leucine-enkephalin (m/z 556.2771 Da).
Allergen database and data processing
The MSE data were searched against the shrimp allergen database containing TM and AK of Litopenaeus vannamei (whiteleg shrimp) (downloaded from http://www.uniprot.org/ on Jan. 10, 2015) using the IDENTITYE search algorithm within the ProteinLynx Global Server v. 2.5 (PLGS 2.5; Waters, U.K.). Search parameters included the “automatic” setting for mass accuracy (50 ppm for precursor ions and 0.1 Da for product ions), a minimum of one peptide match per protein, a minimum of three consecutive product ion matches per peptide, and a minimum of seven total product ion matches per protein. The maximum false positive rate (FPR) against the randomized forward database was set to 4%. Only one missed tryptic cleavage site was allowed during the search. Modifications included the following: fixed, carbamidomethylation of Cys; variable, deamidation of Asn and Gln, oxidation of Met, and dehydration of Ser and Thr.Method validation
Method validation was performed based on the international conference on harmonization (ICH) guidelines16 for validation of bio-analytical procedures. For linearity measurements, fish balls containing shrimp meat (Penaeus vannamei) with six different concentrations (10, 50, 100, 150, 200 g kg−1) along with blank samples were performed for calibration curves over 3 days.Method acceptance criteria states that the precision of the calibration curve and QC samples (containing 1.5% shrimp meat) are considered to be acceptable if RSD ≤ 15% for intra and inter day precision. Furthermore, the accuracy compared with the nominal value needs to be no more than 15%. Finally, the calibration curves must meet the above criteria and have a correlation coefficient r of at least 0.99.
Results and discussion
Analysis of tryptic peptides
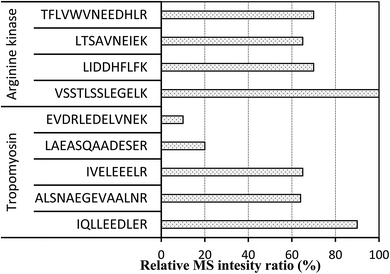
Selection of suitable signature peptides for accurate quantitation of targeted protein is a crucial challenge for developing LC-MS/MS approach. Bioinformatics tool is usually adopted to assist the computational prediction of tryptic products. The theoretical tryptic cleavage peptides of allergen proteins were obtained by computational prediction by Waters Biolynx softwares and online PeptideMass tools provided by UniProt (http://web.expasy.org/peptide_mass).For further confirmation of tryptic peptides, UPLC-QToF-MS was applied for comparing the endogenous and theoretical peptides from tryptic proteins. These peptides were identified detected in tryptic shrimp muscles after comparing the acquired data and sequence database search (Table 1). The coverage ratios of the searched peptides to targeted protein were all more than 50%. Theoretically, tryptic peptides numbers of TM and AK from Litopenaeus vannamei (whiteleg shrimp) were 32 and 29. But, actual detected peptides in MS showed in Table 1 were less than those. Not all the obtained peptides can be used as the surrogate (signature) peptides. The candidate peptides were selected based on several critical factors such as specificity of amino acid sequences, reproducibility in sample preparation, intensity of their MS signal.17
| Protein | Peptides (*recommended signature peptides) | Matched products string | Precursor intensity | Precursor (m/z) | Mass error (ppm) |
|---|---|---|---|---|---|
| B4YAH6 LITVA Lit v 1 tropomyosin | IQLLEEDLER | b2b2b3y1y3y5y6y7y8y9y10 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 938 938 |
629.3392 | 3.5958 |
| *ALSNAEGEVAALNR | b9b14y5y6y7y8y9y10y14 | 8017 | 707.8684 | 2.0720 | |
| IVELEEELR | y2y4y6y7y7y8y9 | 6999 | 565.3094 | 1.8585 | |
| LAEASQAADESER | y4y5y7y9y10y13 | 6492 | 688.8181 | 2.8633 | |
| SITDELDQTFSELSGY | b10b13b16y10y11y16 | 5227 | 902.9098 | 3.1328 | |
| LAMVEADLER | y5y6y7y8 | 1250 | 573.7949 | 5.1705 | |
| EVDRLEDELVNEK | b7b8y4y5y7y11y12 | 1040 | 794.3971 | 5.6077 | |
| Q004B5 LITVA arginine kinase | LGFLTFCPTNLGTTVR | b16y5y7y9y10y11y12y16 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790 790 |
898.9724 | 3.3503 |
| *VSSTLSSLEGELK | b5b6b8b13b13y4y6y8y9y10y11y12y13 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 996 996 |
675.3613 | 5.8746 | |
| DFGDVNSFVNVDPEGK | b11y4y5y7y10y16 | 9333 | 869.8987 | 3.5018 | |
| *LIDDHFLFK | b6y5y6y7y9 | 3888 | 574.3119 | 4.2465 | |
| LTSAVNEIEK | y5y6y7y9y10 | 3506 | 552.3025 | 4.9357 | |
| EMQDGILELIK | y5y7y8y9 | 2755 | 644.8461 | 5.3712 | |
| TFLVWVNEEDHLR | y3y7y8y9y10y11y13 | 2641 | 829.4190 | 3.0708 | |
| FLQAANACR | b5y1y5y6y7 | 2093 | 1050.5166 | 1.3153 | |
| GEHTEAEGGIYDISNK | b8b9b10y4y5y9 | 1279 | 860.3935 | 3.2045 |
Selection of signature peptides
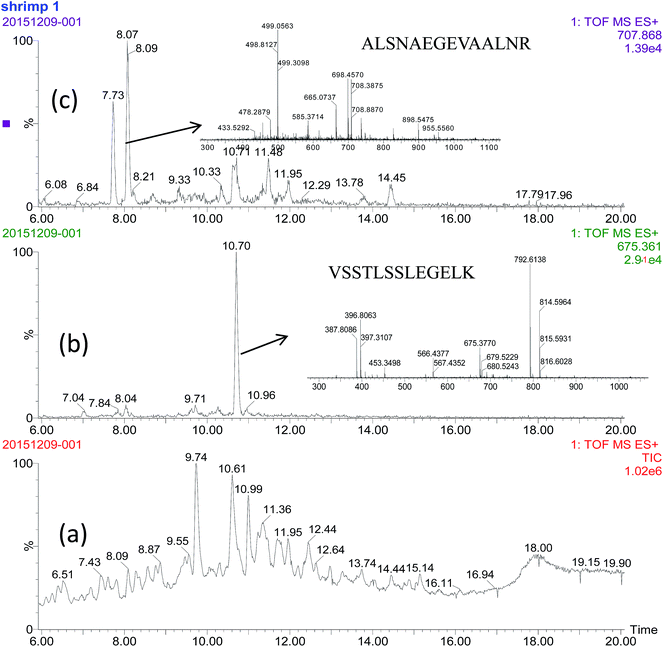
Applied criteria for signature peptides are the absence of cysteine and methionine, peptide size between seven and 20 amino acids, and no modification of amino acids. Furthermore, the peptides with more amino acids (>14) may be not used due to their expensive synthesize and unfavorable LC properties.17,18 Therefore, we selected the peptide VSSTLSSLEGELK, LIDDHFLFK, LTSAVNEIEK and TFLVWVNEEDHLR for AK, and IQLLEEDLER, ALSNAEGEVAALNR, IVELEEELR, LAEASQAADESER and EVDRLEDELVNEK for TM.With the aim to screening these peptides with the property of easy tryptic digestion and high MS intensity, we reduced the digestion time to 2 h and investigated the MS intensity of these peptides. As showed in Fig. 1, different intensities of peptides were obtained. The signature peptides, IQLLEEDLER, IVELEEELR and ALSNAEGEVAALNR for TM and VSSTLSSLEGELK and LIDDHFLFK for AK were selected. The difference of tryptic hydrolysis degree within peptides is hard to explain. Although disulfide bonds of proteins were sheared by DTT and IAA in the pretreatment process, the secondary structure main composed of hydrogen bond and van der Waals forces still existed.
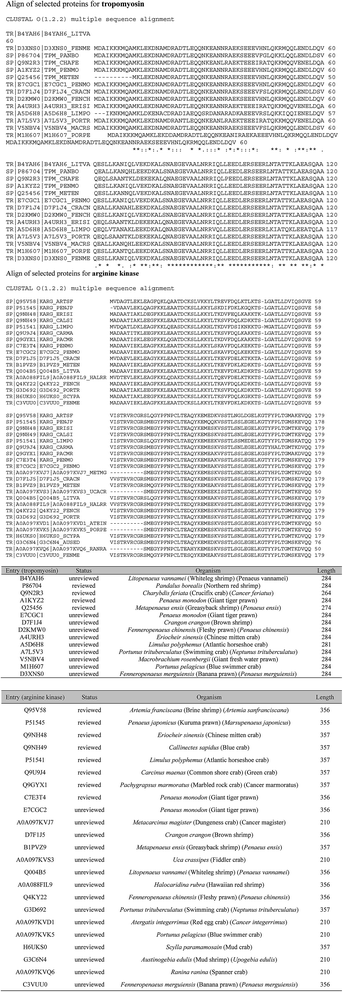
For further optimization of signature peptides, the sequences of TM and AK in some crabs and shrimps were aligned (Fig. 2). We selected the peptides, ALSNAEGEVAALNR for TM and VSSTLSSLEGELK for AK, which are not presented in most crabs. In addition, the intensities of the two peptides were not significantly changed after treatment with boiled water for 30 min (n = 8, P < 0.05).
The specificity of targeted signature peptides was confirmed by the Basic Local Alignment Search Tool (BLAST) of Uniprot (http://www.uniprot.org/blast). It is critical that signature peptides are unique to the target protein and detectable by the MS systems of choice.10 The more information is available, the higher the certainty that the peptide represents the allergen and species of interest. Unfortunately, because of the sequence homology of allergens in different species, it is not always possible that signature peptides are species specific. The results of cross-reaction with TM and AK from other organisms by BLAST were listed in Table 2.
| Protein | Surrogate peptide | Cross-reaction from other organism by BLAST |
|---|---|---|
| Tropomyosin | ALSNAEGEVAALNR | Sinonovacula constricta, Tyrophagus putrescentiae, Portunus pelagicus, Paralithodes camtschaticus, et al. |
| Arginine kinase | VSSTLSSLEGELK | Drosophila mojavensis, Portunus trituberculatus, Macrophthalmus japonicas, Portunus trituberculatus, Larinus sp. BHJ-2011, Calcinus laevimanus, Aegla neuquensis, Gomeza bicornis, Dotilla myctiroides, Eplumula phalangium, Hylaeus elegans, Orithyia sinica, et al. |
The selected signature peptides do not need to be the most intense signals found in MS spectra, but they do need to be sufficiently intense to allow clear separation from other peptides or MS background (Fig. 3). The heat stability of the selected signature peptides were tested at 100 °C for 0.5 h. There is no significantly change (P < 0.05, data not shown here) to the intensity of each peptide.
Digestion by trypsin
When identifying and detecting allergen proteins using MS systems, sample preparation is a critical step. Allergen proteins are usually required to be extracted from food matrice and digested with enzymes generating peptides. For comparison, molecular cut-off filter was used after the protein extraction with Tris–HCl solution as described by previous reports.19–21 However, there is no obvious difference in the numbers of obtained peptides. For clean-up of obtained peptides, SPE was adopted. A systematic investigation of orthogonal SPE clean-up of digested samples was performed by Yuan et al.22 In this study, Oasis® HLB SPE column was used for purification.Various enzymes are available with specific cleavage sites. Trypsin was adopted in our experiment. It is the most commonly used enzyme due to the well-known cleavage sites between the amino acid arginine (R) and lysine (K). Carrera et al.19 has approved that more peptides could be identified when fish species were digested with trypsin compared with Glu-C.
Method validation
For validation of the detection ability, the home-made fish balls containing 1.5% shrimp meat were selected as the QC sample. As shown in Table 3, the RSDs of inter- and intra-day were all less than 15%, and the recoveries were more than 80% (Table 3). Usually, the level owning mass response of a signal-to-noise ratio (S/N) of 3 is considered as limit of detection. In this study, the detection limit, expressed of shrimp meat per kilogram of food, was 8 g kg−1 (usage of TM) or 5 g kg−1 (AK).| Protein | Surrogate peptide | Precursor (2+)/product ion (1+) | Correlation coefficient (r) | Intraday (n = 3) | Interday (n = 3) | ||
|---|---|---|---|---|---|---|---|
| R | RSD | R | RSD | ||||
| Tropomyosin | ALSNAEGEVAALNR | 707.868/829.452 | 0.9912 | 86 | 9.5 | 90 | 7.5 |
| Arginine kinase | VSSTLSSLEGELK | 675.361/862.451 | 0.9909 | 88 | 11.3 | 92 | 8.1 |
Conclusion
The shrimp allergen proteins (tropomyosin and arginine kinase) in complex foodstuffs can be fast detected by in-solution digestion and UPLC-QTQF-MS. The surrogate peptides, ALSNAEGEVAALNR for TM and VSSTLSSLEGELK for AK were screened. The developed method is suitable to screen potential addition of shrimp muscle in foodstuffs by detection of allergen proteins.Furthermore, it should be noted that the present method was only used for screening shrimp muscle via allergenic proteins in complex foodstuffs. Although different crustacean have species-specific peptides for TM, AK or other proteins,23 selected surrogate peptides of ALSNAEGEVAALNR and VSSTLSSLEGELK have cross-reaction with other organisms, especially for some crabs (for example Portunus trituberculatus and Calcinus laevimanus) (Table 2). Our further work will focus on the screening of a generic surrogate peptide in TM or AK for quantifying most of crustacean foods.
Author contributions statement
X.-D. P, Q. C and B.-F. H conceived the experiment(s), X.-D. P, conducted the experiment(s), X.-D. P and B.-F. H analyzed the results. All authors reviewed the manuscript.Conflicts of interest
The authors declare no competing financial interests.References
- R. N. Bauer, M. Manohar, A. M. Singh, D. C. Jay and K. C. Nadeau, J. Allergy Clin. Immunol., 2015, 135, 312–323 CrossRef PubMed.
- T. J. Fernandes, J. Costa, M. B. P. Oliveira and I. Mafra, Food Agric. Immunol., 2015, 26, 848–869 CrossRef CAS.
- M. Pedrosa, T. Boyano-Martínez, C. García-Ara and S. Quirce, Clin. Rev. Allergy Immunol., 2015, 49, 203–216 CrossRef CAS PubMed.
- L. Monaci, R. Pilolli, E. De Angelis, M. Godula and A. Visconti, J. Chromatogr. A, 2014, 1358, 136–144 CrossRef CAS PubMed.
- P. E. Johnson, N. M. Rigby, J. R. Dainty, A. R. Mackie, U. U. Immer, A. Rogers, P. Titchener, M. Shoji, A. Ryan, L. Mata, H. Brown, T. Holzhauser, V. Dumont, J. A. Wykes, M. Walker, J. Griffin, J. White, G. Taylor, B. Popping, R. Crevel, S. Miguel, P. Lutter, F. Gaskin, T. B. Koerner, D. Clarke, R. Sherlock, A. Flanagan, C. H. Chan and E. N. Mills, Food Chem., 2014, 148, 30–36 CrossRef CAS PubMed.
- S. D. Kamath, A. M. Abdel Rahman, T. Komoda and A. L. Lopata, Food Chem., 2013, 141, 4031–4039 CrossRef CAS PubMed.
- S. D. Kamath, A. M. Rahman, A. Voskamp, T. Komoda, J. M. Rolland, R. E. O'Hehir and A. L. Lopata, Mol. Nutr. Food Res., 2014, 58, 1144–1155 CAS.
- S. Sakai, R. Matsuda, R. Adachi, H. Akiyama, T. Maitani, Y. Ohno, M. Oka, A. Abe, K. Seiki, H. Oda, K. Shiomi and A. Urisu, J. AOAC Int., 2008, 91, 123–129 CAS.
- J. Heick, M. Fischer and B. Popping, J. Chromatogr. A, 2011, 1218, 938–943 CrossRef CAS PubMed.
- G. Picariello, G. Mamone, F. Addeo and P. Ferranti, J. Chromatogr. A, 2011, 1218, 7386–7398 CrossRef CAS PubMed.
- J. Sealey-Voyksner, J. Zweigenbaum and R. Voyksner, Food Chem., 2016, 194, 201–211 CrossRef CAS PubMed.
- H. Nagai, T. Minatani and K. Goto, J. AOAC Int., 2015, 98, 1355–1365 CrossRef CAS PubMed.
- M. Koeberl, D. Clarke and A. L. Lopata, J. Proteome Res., 2014, 13, 3499–3509 CrossRef CAS PubMed.
- Z. Huang and X.-D. Pan, RSC Adv., 2017, 7, 20212–20218 RSC.
- A. I. Sancho and E. N. Mills, Regul. Toxicol. Pharmacol., 2010, 58, S42–S46 CrossRef CAS PubMed.
- Department of Health Human Services Food and Drug Administration, Methodology ICH, 1996.
- M. Rauh, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2012, 883–884, 59–67 CrossRef CAS PubMed.
- L. V. DeSouza and K. W. Siu, Clin. Biochem., 2013, 46, 421–431 CrossRef CAS PubMed.
- M. Carrera, B. Canas, J. Vázquez and J. M. Gallardo, J. Proteome Res., 2010, 9, 4393–4406 CrossRef CAS PubMed.
- J. Heick, M. Fischer, S. Kerbach, U. Tamm and B. Popping, J. AOAC Int., 2011, 94, 1060–1068 CAS.
- D. Weber, P. Raymond, S. Ben-Rejeb and B. Lau, J. Agric. Food Chem., 2006, 54, 1604–1610 CrossRef CAS PubMed.
- L. Yuan, A. F. Aubry, M. E. Arnold and Q. C. Ji, Bioanalysis, 2013, 5, 2379–2391 CrossRef CAS PubMed.
- I. Ortea, B. Canas and J. M. Gallardo, J. Proteome Res., 2009, 8, 5356–5362 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |