Open Access Article
Open Access ArticleNon-covalent anchoring of oligonucleotides on single-walled carbon nanotubes via short bioreducible linker†
Elizaveta S. Permyakovaa,
Darya S. Novopashina ab,
Alya G. Venyaminova
ab,
Alya G. Venyaminova a and
Evgeny K. Apartsin
a and
Evgeny K. Apartsin *a
*a
aInstitute of Chemical Biology and Fundamental Medicine SB RAS, 8, Lavrentiev ave., Novosibirsk 630090, Russia. E-mail: eka@niboch.nsc.ru; Fax: +7 383 3635153; Tel: +7 383 3635129
bNovosibirsk State University, 2, Pirogov str., Novosibirsk 630090, Russia
First published on 5th June 2017
Abstract
A novel, simple and convenient approach to assemble non-covalent hybrids of carbon nanotubes with therapeutic oligonucleotides immobilized by means of biodegradable linker is described. Model RNA, DNA and 2′-O-methyl RNA oligonucleotides were anchored on the surface of single-walled carbon nanotubes using pyrene anchor groups introduced onto the 5′-termini via short linker containing bioreducible disulfide bond or via stable linker of the similar length, as a control. To optimize the conditions of non-covalent functionalization, the adsorption of oligonucleotides' pyrene conjugates on the carbon nanotubes surface has been studied. The exposure of hybrids to glutathione at physiological concentrations has been shown to cause the release of oligonucleotides anchored via bioreducible linker only. The kinetic parameters of the oligonucleotides release have been determined for the first time. The results obtained can be useful for the design of carbon nanotubes-based nanoconstructions and biomaterials.
Introduction
Nucleic acid therapy is a highly promising field of modern medicine.1 Different types of oligonucleotides, their analogues and conjugates are now under pre-clinical and clinical testing as potential drugs to treat tumors, viral infections, neurodegenerative diseases etc.; new formulations containing therapeutic nucleic acids are being elaborated.2–4 Nevertheless, some obstacles decreasing the efficiency of nucleic acid therapeutics are not yet fully overcome. One of the major challenges is the problem of a delivery of therapeutic oligonucleotides into the cytosol where they are able to cause a functional effect.5,6To date, numerous ways have been proposed to increase the efficiency of nucleic acid delivery into a cell.2 The promising way is to use nanoparticles as carriers of oligonucleotides. In particular, carbon nanotubes (CNTs) are actively used as nucleic acid carriers due to the combination of high transfection efficiency and low toxicity after modification with biomimetic organic moieties.7 CNTs and their hybrids with nucleic acids are known to penetrate across the cell membrane by a variety of mechanisms: direct spearing of cell membrane (due to the rigidity of CNT structure), endocytosis, macropinocytosis and phagocytosis.8 As a result, the effect of nucleic acid therapeutics is observed upon delivery with CNTs both in vitro and in vivo.9
Hybrids of carbon nanotubes with nucleic acids are formed by means of covalent linkage (oligonucleotide conjugation with functionalized CNTs) or non-covalent interactions (van der Waals interactions, electrostatic binding, π–π stacking, etc.).9,10 An efficient method of the latter is to use polyaromatic molecules (e.g., pyrene) interacting with CNT surface by π–π-stacking interactions as anchor groups for non-covalent immobilization of oligonucleotides on CNT surface.11–14
The use of small anchor groups, such as pyrene, for oligonucleotides immobilization on the carbon nanotube surface can simplify the construction of CNT–nucleic acids hybrid nanoparticles as well as provides a methodology to obtain multifunctional nanoparticles. Earlier, we have described the preparation of non-covalent hybrids of pyrene-modified oligonucleotides with fluorescently labeled single-walled carbon nanotubes (SWCNTs).15,16 This method is believed to be promising thanks to the relative simplicity of the synthesis of pyrene conjugates of oligonucleotides, high density of CNT functionalization and low toxicity of the hybrid constructions. However, the reported constructions lack an efficient way of release of oligonucleotides from their hybrids with CNTs. The presence of a bioreducible linkage between an anchor group and an oligonucleotide can provide the release of the oligonucleotide after the penetration of a nanoconstruction across the cell membrane.17,18
Here we report a novel, simple and convenient way to immobilize oligonucleotides on the CNT surface using pyrene anchor groups conjugated to the oligonucleotides by a short linker containing the disulfide bond. Due to the presence of the bioreducible fragment in the structure of the linker, oligonucleotide can be released from the hybrid in a controlled manner upon the interaction with cell metabolite glutathione at physiological concentrations.
Experimental section
Oxidation of carbon nanotubes
Raw single-walled carbon nanotubes (Sigma, US) were oxidized as described in ref. 19. Briefly, 500 mg raw SWCNTs were dispersed in 200 mL 70% nitric acid and heated at 115 °C for 6 h. SWCNT-COOH were recovered by filtration using PTFE filters with pore diameter of 100 nm (Millipore, US), rinsed thoroughly with deionized water and dried. The characteristics of SWCNT-COOH are given in the ESI.†Synthesis of 5′-pyrene conjugates of oligonucleotides
5′-Phosphorylated oligonucleotides bearing the 3′-“inverted” thymidine on the 3′-termini were synthesized using the solid-phase phosphoramidite method on the automatic ASM-800 RNA/DNA synthesizer (Biosset, Russia) using UltraMild β-cyanoethyl phosphoramidites and corresponding polymer supports (Glen Research, US) according to the protocols optimized for this instrument,20 deprotected using standard procedures and purified by 15% polyacrylamide gel electrophoresis (PAGE).Solutions of triphenylphosphine (6.8 mg, 25 μmol) and 2,2′-dipyridyl disulfide (5.3 mg, 25 μmol) in anhydrous DMSO (25 μL each) were added to a solution of 5′-phosphate-modified oligonucleotide (cetyltrimethylammonium salt; approx. 0.03 μmol) and 4-N,N-dimethylaminopyridine (5 mg, 41 μmol) in anhydrous DMSO (50 μL). The reaction mixture was stirred for 15 min at 37 °C. The activated oligonucleotide was precipitated with 2% NaClO4 in acetone, quickly washed with acetone, and dissolved in water (5 μL), followed by the addition of cystamine dihydrochloride (2.1 mg, 9.3 μmol) or 1,6-diaminohexane (1.1 mg, 9.4 μmol) in a DMSO (25 μL)/triethylamine (5 μL) mixture. The reaction mixture was stirred for 2 h at 37 °C. The nucleotide material was precipitated with 2% NaClO4 in acetone, washed with acetone and dried.
At the next stage, the oligonucleotide was dissolved in 0.05 M NaHCO3, pH 8.3 (5 μL)/DMF (10 μL) mixture. The solution of 1-pyrenebutyric acid N-hydroxysuccinimide ester (2.0 mg, 5.2 μmol) in DMF (20 μL) was added by 10, 5 and 5 μL portions. The reaction mixture was stirred for 30 min at 37 °C after each addition. Then nucleotide material was precipitated with 2% NaClO4 in acetone, washed with acetone and dried. The obtained conjugates were purified by 15% PAGE, and their structures were confirmed via MALDI TOF mass spectrometry and UV and fluorescence spectroscopy (for details, refer to the ESI†).
Adsorption isotherms of 5′-pyrene conjugates of oligonucleotides onto SWCNTs
To obtain solutions of the hybrids, SWCNT-COOH were added to a buffered (10 mM PBS, pH 7.4) oligonucleotide solution (1 μM) to a final concentration of 1–96 mg L−1 and sonicated for 30 min using Sonorex Super RK 31 H ultrasonic bath (Bandelin Electronic, Germany). The fluorescence spectra of samples were recorded upon excitation at 347 ± 4 nm using CLARIOstar microplate reader (BMG LABTECH, Germany). The amount of oligonucleotide adsorbed onto the SWCNTs was calculated as , where IP is the intensity of the pyrene fluorescence at 378 nm in the oligonucleotide–SWCNT hybrid solution, and I0P is the intensity of the pyrene residue fluorescence at 378 nm in the reference oligonucleotide solution at the same concentration (1 μM). A stable dispersion of carbon nanotubes was considered to be a solution for calculations.
, where IP is the intensity of the pyrene fluorescence at 378 nm in the oligonucleotide–SWCNT hybrid solution, and I0P is the intensity of the pyrene residue fluorescence at 378 nm in the reference oligonucleotide solution at the same concentration (1 μM). A stable dispersion of carbon nanotubes was considered to be a solution for calculations.
The experimental data were fitted using a model of binding with a lag:  , where nads is the amount of the adsorbed oligonucleotide [%], CSWCNT is the concentration of the SWCNT-COOH [mg L−1] in the same point, Kd is the constant of dissociation [mg L−1], nmax is a constant representing the maximum adsorption of pyrene-modified oligonucleotides on CNT surface, h is the coefficient representing the binding cooperativity. Fitting was considered satisfactory if R2 > 0.95.
, where nads is the amount of the adsorbed oligonucleotide [%], CSWCNT is the concentration of the SWCNT-COOH [mg L−1] in the same point, Kd is the constant of dissociation [mg L−1], nmax is a constant representing the maximum adsorption of pyrene-modified oligonucleotides on CNT surface, h is the coefficient representing the binding cooperativity. Fitting was considered satisfactory if R2 > 0.95.
The capacity of SWCNT-COOH for the adsorption of oligonucleotide was estimated by analogy with ref. 16 in the first point of the saturation plateau in adsorption isotherms as  , where CON is the total oligonucleotide concentration (1 μM), nads is the amount of the adsorbed oligonucleotide [%] in a given point as estimated from an isotherm, CSWCNT is the concentration of the SWCNT-COOH [mg L−1] in the same point.
, where CON is the total oligonucleotide concentration (1 μM), nads is the amount of the adsorbed oligonucleotide [%] in a given point as estimated from an isotherm, CSWCNT is the concentration of the SWCNT-COOH [mg L−1] in the same point.
Radioactive labeling of oligonucleotides
The radioactive 5′-[32P]-labeling of oligonucleotides' pyrene conjugates was performed using 4 MBq [γ-32P]-ATP (120 TBq/mol, Biosan, Russia) and T4 polynucleotide kinase (Thermo Scientific, US) by standard protocol. The resulting labeled oligonucleotides bearing [32P]-label on the 5′-terminus of the 3′-3′-“inverted” thymidine were isolated by 15% PAGE.Study of the disulfide bond cleavage kinetics
The kinetic assay of disulfide bond cleavage in hybrids of oligonucleotides dON-L1, rON-L1 and mON-L1 with SWCNT-COOH was done using [32P]-labeled oligonucleotides. Solutions of the hybrids were obtained by addition of SWCNT-COOH to a buffered (PBS: 10 mM Na2HPO4, 1.76 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl) oligonucleotide solution (1 μM) to a final concentration of 64 mg L−1 and sonicated for 30 min using Sonorex Super RK 31 H ultrasonic bath (Bandelin Electronic, Germany). Then glutathione was added to the final concentration of 1, 5 or 10 mM. Samples were incubated at room temperature for 1–1440 min, and then reaction mixtures were applied to a 15% non-denaturing polyacrylamide gel using a loading buffer (0.05% xylene cyanol FF, 0.05% bromophenol blue in 50% glycerol). Untreated free conjugates dON-L1, rON-L1 and mON-L1 were applied as controls. Additionally, oligonucleotides containing stable linker L2 were treated by 10 mM glutathione for 24 h and used as non-cleavage control. The electrophoresis was run at 50 V cm−1 voltage in a vertical electrophoresis chamber using TBE buffer (89 mM Tris-borate, pH 8.3, 10 mM Na2EDTA). All experiments were run in triplicates.The gels were radioautographed using Pharos FX (Bio-Rad Laboratories Inc., CA, US) Phosphor Imager, the images acquired were processed using Quantity One analysis software (Bio-Rad Laboratories Inc., CA, US). The percentage of disulfide bond cleavage was calculated as 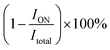 , where ION is the radioactive signal of the band of the non-cleaved oligonucleotide, and Itotal is the total radioactivity of the sample. Kinetic parameters of cleavage (kobs and S–S bond half-life) were determined from cleavage curves using the GraphPad Prism 5.0.4.533 software.
, where ION is the radioactive signal of the band of the non-cleaved oligonucleotide, and Itotal is the total radioactivity of the sample. Kinetic parameters of cleavage (kobs and S–S bond half-life) were determined from cleavage curves using the GraphPad Prism 5.0.4.533 software.
Results and discussion
The use of hybrids of carbon nanotubes with nucleic acids as therapeutic agents requires the release of oligonucleotide part from a carrier in a functional form; this requirement is especially important in the case of the delivery of siRNA or microRNA naturally existing in a duplex form. The immobilization of nucleic acids on SWCNT surface can distort their structure21,22 and consequently decrease their biological activity in a cell. That is why the development of CNT-based nucleic acid therapeutics requires the use of biodegradable linkages facilitating the release of therapeutic molecule from a hybrid upon intracellular delivery. In particular, disulfide linkage is widely used for this purpose.18,23An interesting example of CNT–oligonucleotide hybrids with biodegradable bonds was reported by Dai et al.24–26 In their work, SWCNT surface was covered by diaminopoly(ethylene glycol)–phospholipid conjugate followed by oligonucleotide conjugation via disulfide bond upon treatment with bifunctional reagent SPDP (succinimidyl 3-(2-pyridyldithio)propionate). Such a design of hybrids permits to immobilize nucleic acids on SWCNTs and to protect them from the nuclease digestion upon transporting into a cell, as well as provides their release in cytosol induced by glutathione at physiological concentrations.
In contrast, our idea is to use small anchor group and quite simple linker between this group and oligonucleotide to simplify the preparation of the nanoparticles and potentially increase the coverage of CNT surface with cargo molecules. As a platform for the formation of hybrids with oligonucleotides, carboxyl-functionalized single-walled carbon nanotubes (SWCNT-COOH) were chosen. The carboxylation step is important, since as-grown CNTs are hardly applicable for the design of the drug carriers as they may contain non-carbon impurities and large number of defects on the surface,19,27 furthermore, they generally are more toxic than chemically modified CNTs.28 To prepare SWCNT-COOH, we used the procedure that eliminates the most part of impurities, and produces 7–15 wt% carboxylic groups.19 In our hands, this procedure afforded SWCNT-COOH with good oligonucleotide binding efficiency.15,16
The approach used (Fig. 1) is based on the non-covalent functionalization of SWCNTs with pyrene conjugates of oligonucleotides that contain a bioreducible or stable linker. The use of pyrene as an anchor group provides strong immobilization of oligonucleotide on the SWCNT surface and provides the possibility to quantify in by the quenching of pyrene fluorescence. When exposed to the disulfide-reducing agents, oligonucleotides anchored via disulfide-containing linker are released from the hybrids, whereas their analogues containing stable linker remain anchored on SWCNTs.
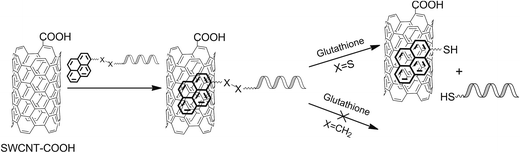 | ||
| Fig. 1 Scheme of the preparation of the SWCNT–oligonucleotides hybrids and subsequent release of oligonucleotides. | ||
As oligonucleotide components of hybrids, 18-mer model DNA, RNA and 2′-O-methyl RNA oligonucleotides of identical sequence (abbreviated here as dON, rON and mON, respectively, Fig. 2) were used. These three types of oligonucleotides were chosen to demonstrate the applicability of the proposed approach for the construction of SWCNT-based hybrids with major types of therapeutic oligonucleotides: antisense oligonucleotides (DNA oligonucleotide), small interfering RNA or microRNA (RNA oligonucleotide), and microRNA inhibitors (DNA and 2′-O-methyl RNA oligonucleotides). The oligonucleotides contained 5′-phosphate group and so-called 3′-“inverted” thymidine, the thymidine nucleotide introduced by 3′-3′-internucleotidic linkage (structures given in Fig. 2). This modification is known to increase the stability of oligonucleotides towards exonuclease digestion without decreasing the biological activity,29 however, in the present work, the 3′-“inverted” thymidine additionally provides a site for the convenient T4 polynucleotide kinase-mediated radioactive labeling that was used for the visualization of oligonucleotides by radioautography (see below).
To introduce pyrene moieties onto the 5′-termini of oligonucleotides, the 5′-phosphate group was activated by triphenylphosphine and 2,2′-dipyridyl disulfide in the presence of 4-N,N-dimethylaminopyridine as a nucleophilic catalyst30 with subsequent interaction with an aminolinker (Fig. 2). Two linkers of equal lengths were used: cystamine as bioreducible linker and 1,6-diaminohexane as stable one. 1-Pyrenebutyric acid N-hydroxysuccinimide ester was then grafted onto the 5′-amino groups to give the target conjugates (ON-L1 and ON-L2 for cystamine and 1,6-diaminohexane-containing conjugates, respectively), which were isolated by preparative gel electrophoresis. The characteristics of the 5′-pyrene conjugates of oligonucleotides are given in the ESI.†
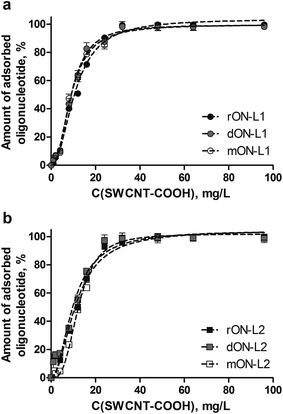
The non-covalent hybrids of the pyrene conjugates of oligonucleotides with SWCNT-COOH were formed by the adsorption of pyrene residues onto the defectless sites of the SWCNT surface. The pyrene fluorescence intensity decreases upon the increase of the SWCNT-COOH concentration due to the quenching of the fluorescence when it is adsorbed onto the SWCNTs.31 The quenching of pyrene fluorescence upon adsorption on CNTs is static,32 the same character of quenching is reported for pyrene anchor groups used for immobilization of cargo on the CNT surface.33,34 Basing on the quantitative data on the fluorescence quenching, the isotherms of adsorption of 5′-pyrene conjugates of the oligonucleotides rON, dON, mON on SWCNT-COOH were plotted (Fig. 3). The comparison of the isotherms provides the information on the effect of the oligonucleotide nature and linker structure on the oligonucleotide immobilization efficiency.
There were no considerable differences found among the isotherms of adsorption of different oligonucleotides. Neither the nature of oligonucleotide (DNA, RNA, 2′-O-methyl RNA) nor the structure of a linker (with or without disulfide bond) has the effect on the oligonucleotide adsorption efficiency. The dissociation constant Kd is approx. 10 mg L−1 in all cases. This observation is in agreement with our previous findings obtained for oligonucleotides having short stable linker between pyrene moiety and oligonucleotide.10 It should be noted that the adsorption curves have sigmoidal behavior (lag in the region 1–5 mg L−1) that suggests the partial disruption of SWCNT bundles and aggregates upon the interaction with oligonucleotides. This suggestion was confirmed by the atomic force microscopy studies (Fig. S2 in the ESI†). The capacity of the SWCNTs for oligonucleotide adsorption (i.e., the maximal amount of oligonucleotide that can be adsorbed on the CNT surface) was estimated by analogy with ref. 16 to be approx. 30 μmol of oligonucleotide per gram irrespective of the linker structure and the nature of oligonucleotide.
At the next stage, the release of oligonucleotides from the hybrids in a model system was studied. As a disulfide-reducing agent, glutathione was chosen. Glutathione is an important metabolite having multiple activities at the cellular and organismal level.35 The concentration of glutathione in a cell is quite high (generally, 0.5–10 mM in cytosol), so it is assumed to be responsible for the release of the oligonucleotide immobilized on the SWCNTs by reducing the disulfide bond in the linker L1. To confirm this assumption, SWCNT-COOH were saturated with [32P]-labeled pyrene conjugates of oligonucleotides rON-L1, dON-L1, mON-L1 (1 μM) and incubated in PBS-buffered solutions containing 1, 5, or 10 mM glutathione. The samples were analyzed by polyacrylamide gel electrophoresis assay, and the cleavage products were detected by radioautography (Fig. S5 in the ESI†). Since the concentration of oligonucleotides in the kinetic experiments is at least 1000 times lower than the concentration of glutathione, the experimental curves can be fitted using the pseudo-first order model. Kinetic parameters of the disulfide bond cleavage (apparent kinetic constants, kobs, and disulfide bond half-life time values) in pyrene conjugates of oligonucleotides dON-L1, rON-L1 and mON-L1 and their hybrids with SWCNT-COOH are given in Table 1 (for kinetic profiles, refer to the Fig. S6 in the ESI†). The immobilization of oligonucleotides on the SWCNT-COOH surface decreases the cleavage rates as compared with those of free conjugates. The cleavage rate appeared to be different for DNA, RNA and 2′-O-methyl RNA oligonucleotides: rON-L1 > dON-L1 > mON-L1. Generally, the half-life time of the disulfide bond in free conjugates ON-L1 is 1.5–9 h depending on the glutathione concentration (median approx. 3.5 h); the adsorption on SWCNTs increases the half-life time up to 2–10 h (median approx. 6 h). As expected, no release of oligonucleotides from the hybrids was observed when pyrene conjugates of oligonucleotides ON-L2 containing stable linker were subjected to the glutathione treatment (Fig. S5 in the ESI†). This finding confirms that the release of oligonucleotides from the hybrids is caused exclusively by disulfide bond reduction and not by the displacement of the pyrene moieties on the SWCNT surface by glutathione or buffer components. The typical release time values of the SWCNT-bound oligonucleotides are comparable with the typical duration of endocytosis and formation of endosomes/lysosomes8 that suggests the relevancy of described hybrids for biological experiments.
| Oligonucleotide | kobs, min−1 × 1000 | S–S bond half-life, min | ||||
|---|---|---|---|---|---|---|
| 1 mM GSH | 5 mM GSH | 10 mM GSH | 1 mM GSH | 5 mM GSH | 10 mM GSH | |
| a GSH – glutathione; +CNT – hybrid of an oligonucleotide with SWCNT-COOH; −CNT – free oligonucleotide. | ||||||
| rON-L1 (+CNT) | 1.00 ± 0.06 | 1.92 ± 0.05 | 6.6 ± 0.2 | 690 ± 29 | 361 ± 6 | 105 ± 2.4 |
| rON-L1 (−CNT) | 2.15 ± 0.05 | 3.2 ± 0.1 | 7.2 ± 0.4 | 323 ± 5 | 217 ± 4.4 | 96 ± 3.5 |
| dON-L1 (+CNT) | 1.13 ± 0.07 | 1.77 ± 0.06 | 5.1 ± 0.2 | 616 ± 26 | 392 ± 9 | 136 ± 3.8 |
| dON-L1 (−CNT) | 1.60 ± 0.07 | 3.4 ± 0.1 | 7.7 ± 0.4 | 435 ± 4 | 204 ± 4.2 | 90 ± 2.6 |
| mON-L1 (+CNT) | 1.23 ± 0.08 | 1.53 ± 0.09 | 3.6 ± 0.1 | 562 ± 25 | 455 ± 17 | 192 ± 4.4 |
| mON-L1 (−CNT) | 1.19 ± 0.05 | 2.3 ± 0.1 | 4.2 ± 0.3 | 583 ± 15 | 302 ± 8.8 | 164 ± 7.2 |
It should be noted that the rate of disulfide bond reduction in a cell can be different due to the presence of other molecules or enzymes able to cleave this bond. For instance, there is an enzyme referred to as GILT, gamma-interferon-inducible lysosomal thiol reductase. GILT is transported into endosomes and catalyzes the disulfide bond reduction both in vitro and in vivo having the highest activity at pH 4–5.36 Considering this, one can conclude that the release of oligonucleotides from hybrids can start in late endosomes/lysosomes, even before the escape of hybrid into cytosol where it can interact with glutathione.
An important consequence of the glutathione-mediated cleavage of the disulfide bond between pyrene anchor group and oligonucleotide is the fact that the residual carrier (carbon nanotube with thiol-containing pyrene molecules) is assumed to be abundantly conjugated with glutathione molecules. It is known that, being involved in many biochemical processes in a cell, glutathione takes part, in particular, in the xenobiotic excretion from a cell as an excretion marker.37,38 That is why the CNT-carrier bearing multiple glutathione fragments in principle can be eliminated from a cell after the release of oligonucleotides.
Conclusion
We have suggested a novel approach to obtain hybrids of single-walled carbon nanotubes with therapeutically relevant oligonucleotides that are released upon glutathione treatment at physiological concentrations. Thanks to being applicable for the immobilization of different types of oligonucleotides on the CNT surface (DNA, RNA, 2′-O-methyl RNA), the method can be used for the design of nanoconstructions to deliver a variety of therapeutic oligonucleotides into a cell. The formation of hybrids with oligonucleotides results in partial disaggregation of the SWCNT bundles that can be an advantage for the biomedical use, since it potentially reduces the toxicity of the hybrid material.An important advantage of the method is that the tips and defect sites on the CNT surface are not available for the oligonucleotide immobilization, so they can be functionalized orthogonally.16 The covalent functionalization of CNTs with different functional groups (reporter groups, addressing moieties etc.) followed by non-covalent anchoring of oligonucleotides by bioreducible linker can result in the formation of multifunctional CNT-oligonucleotide hybrids that are highly promising for the biomedical applications due to the possibility of the oligonucleotides release in a cell. The use of a small anchor group and short linkers for the oligonucleotide immobilization permits to avoid the steric endurance in the course of the preparation of such multifunctional hybrids.
We believe that this approach can advance the design of multifunctional constructions at nano-bio interface.
Acknowledgements
This work was supported by the RFBR grants 14-03-31691_mol_a (2014–2015) and 16-33-60152_mol_a_dk (2016–2018), as well as by the Scholarship of the President of the Russian Federation (grant 882.2016.4).References
- E. K. Apartsin and N. Y. Knauer, Genes and Cells, 2016, 11, 32–41 Search PubMed.
- V. K. Sharma and J. K. Watts, Future Med. Chem., 2015, 7, 2221–2242 CrossRef CAS PubMed.
- T. Sun, Y. S. Zhang, B. Pang, D. C. Hyun, M. Yang and Y. Xia, Angew. Chem., Int. Ed., 2014, 53, 12320–12364 CAS.
- A. Barba, G. Lamberti, C. Sardo, B. Dapas, M. Abrami, M. Grassi, R. Farra, F. Tonon, G. Forte, F. Musiani, M. Licciardi, G. Pozzato, F. Zanconati, B. Scaggiante, G. Grassi and G. Cavallaro, Curr. Drug Metab., 2015, 16, 427–452 CrossRef CAS PubMed.
- H. J. Kim, A. Kim and K. Miyata, Adv. Drug Delivery Rev., 2016, 104, 61–77 CrossRef CAS PubMed.
- W. Liao, W. Li, T. Zhang, M. Kirberger, J. Liu, P. Wang, W. Chen and Y. Wang, Biomater. Sci., 2016, 4, 1051–1061 RSC.
- R. Alshehri, A. M. Ilyas, A. Hasan, A. Arnaout, F. Ahmed and A. Memic, J. Med. Chem., 2016, 59, 8149–8167 CrossRef CAS PubMed.
- V. Raffa, G. Ciofani, O. Vittorio, C. Riggio and A. Cuschieri, Nanomedicine, 2009, 5, 89–97 CrossRef PubMed.
- E. K. Apartsin, M. Y. Buyanova, D. S. Novopashina and A. G. Venyaminova, in Nanobiophysics: Fundamentals and Applications, ed. V. Karachevtsev, Pan Stanford Publishing, Singapore, 2015, pp. 33–57 Search PubMed.
- E. K. Apartsin, M. Y. Buyanova, D. S. Novopashina, E. I. Ryabchikova and A. G. Venyaminova, in Nanomaterials Imaging Techniques, Surface Studies, and Applications, ed. O. Fesenko, L. Yatsenko and M. Brodin, Springer Science+Business Media, New York, 2013, vol. 146, pp. 291–307 Search PubMed.
- A. Di Crescenzo, V. Ettorre and A. Fontana, Beilstein J. Nanotechnol., 2014, 5, 1675–1690 CrossRef PubMed.
- B. J. Taft, A. D. Lazareck, G. D. Withey, A. Yin, J. M. Xu and S. O. Kelley, J. Am. Chem. Soc., 2004, 126, 12750–12751 CrossRef CAS PubMed.
- Y.-K. Baek, D.-H. Jung, S. M. Yoo, S. Shin, J.-H. Kim, H.-J. Jeon, Y.-K. Choi, S. Y. Lee and H.-T. Jung, J. Nanosci. Nanotechnol., 2011, 11, 4210–4216 CrossRef CAS PubMed.
- E. O. Fedorovskaya, E. K. Apartsin, D. S. Novopashina, A. G. Venyaminova, A. G. Kurenya, L. G. Bulusheva and A. V. Okotrub, Mater. Des., 2016, 100, 67–72 CrossRef CAS.
- E. K. Apartsin, D. S. Novopashina, Y. V. Nastaushev and A. G. Venyaminova, Nanotechnol. Russ., 2012, 7, 99–109 CrossRef.
- E. K. Apartsin, M. Y. Buyanova, D. S. Novopashina, E. I. Ryabchikova, A. V. Filatov, M. A. Zenkova and A. G. Venyaminova, ACS Appl. Mater. Interfaces, 2014, 6, 1454–1461 CAS.
- K. West and S. Otto, Curr. Drug Discovery Technol., 2005, 2, 123–160 CrossRef CAS.
- J. Wang, S. Li, T. Luo, C. Wang and J. Zhao, Curr. Med. Chem., 2012, 19, 2976–2983 CrossRef CAS PubMed.
- A. Gromov, S. Dittmer, J. Svensson, O. Nerushev, S. A. Perez-García, L. Licea-Jiménez, R. Rychwalski and E. E. B. Campbell, J. Mater. Chem., 2005, 15, 3334–3339 RSC.
- L. Bellon, in Current Protocols in Nucleic Acid Chemistry, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2000, pp. 3.6.1–3.6.13 Search PubMed.
- V. A. Karachevtsev, G. O. Gladchenko, M. V. Karachevtsev, V. A. Valeev, V. S. Leontiev and O. S. Lytvyn, ChemPhysChem, 2008, 9, 2010–2018 CrossRef CAS PubMed.
- M. Santosh, S. Panigrahi, D. Bhattacharyya, A. K. Sood and P. K. Maiti, J. Chem. Phys., 2012, 136, 65106–65110 CrossRef PubMed.
- C. Caoduro, E. Hervouet, C. Girard-Thernier, T. Gharbi, H. Boulahdour, R. Delage-Mourroux and M. Pudlo, Acta Biomater., 2017, 49, 36–44 CrossRef CAS PubMed.
- N. W. Kam, Z. Liu and H. Dai, J. Am. Chem. Soc., 2005, 127, 12492–12493 CrossRef CAS PubMed.
- Z. Liu, X. Sun, N. Nakayama-Ratchford and H. Dai, ACS Nano, 2007, 1, 50–56 CrossRef CAS PubMed.
- Z. Liu, S. Tabakman, K. Welsher and H. Dai, Nano Res., 2009, 2, 85–120 CrossRef CAS PubMed.
- P. V. Lakshminarayanan, H. Toghiani and C. U. Pittman Jr, Carbon, 2004, 42, 2433–2442 CrossRef CAS.
- R. Li, X. Wang, Z. Ji, B. Sun, H. Zhang, C. H. Chang, S. Lin, H. Meng, Y.-P. Liao, M. Wang, Z. Li, A. A. Hwang, T.-B. Song, R. Xu, Y. Yang, J. I. Zink, A. E. Nel and T. Xia, ACS Nano, 2013, 7, 2352–2368 CrossRef CAS PubMed.
- J. F. Ortigão, H. Rösch, H. Selter, A. Fröhlich, A. Lorenz, M. Montenarh and H. Seliger, Antisense Res. Dev., 1992, 2, 129–146 CrossRef.
- V. Zarytova, E. Ivanova and A. Venyaminova, Nucleosides Nucleotides, 1998, 17, 649–662 CAS.
- L. Wang, L. Zhang, X. Xue, G. Ge and X. Liang, Nanoscale, 2012, 4, 3983–3989 RSC.
- Y. Zhang, S. Yuan, W. Zhou, J. Xu and Y. Li, J. Nanosci. Nanotechnol., 2007, 7, 2366–2375 CrossRef CAS PubMed.
- C. Ehli, G. M. A. Rahman, N. Jux, D. Balbinot, D. M. Guldi, F. Paolucci, M. Marcaccio, D. Paolucci, M. Melle-Franco, F. Zerbetto, S. Campidelli and M. Prato, J. Am. Chem. Soc., 2006, 128, 11222–11231 CrossRef CAS PubMed.
- L. Liu, T. Wang, J. Li, Z. Guo, L. Dai, D. Zhang and D. Zhu, Chem. Phys. Lett., 2003, 367, 747–752 CrossRef CAS.
- O. W. Griffith, Free Radical Biol. Med., 1999, 27, 922–935 CrossRef CAS PubMed.
- M. P. Rausch and K. T. Hastings, Mol. Immunol., 2015, 68, 124–128 CrossRef CAS PubMed.
- S. Yadav, E. Zajac, S. S. Singhal and S. Awasthi, Cancer Metastasis Rev., 2007, 26, 59–69 CrossRef CAS PubMed.
- V. I. Lushchak, J. Amino Acids, 2012, 2012, 736837 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04933a |
| This journal is © The Royal Society of Chemistry 2017 |