Open Access Article
Open Access ArticleRecent progress in the chemistry and biology of limonoids
Yuanyuan Zhang
and
Hui Xu
 *
*
Research Institute of Pesticidal Design & Synthesis, College of Chemistry & Pharmacy, Northwest A&F University, Yangling 712100, Shaanxi Province, P. R. China. E-mail: orgxuhui@nwsuaf.edu.cn
First published on 13th July 2017
Abstract
This review covers the isolation and structure determination of limonoids reported during 2014–2016 (with 363 new compounds in 68 papers), together with the relevant biological activities and source organisms. Furthermore, the total synthesis and structural modifications of limonoids and their analogs regarding the bioactivities reported during 2011–2016 have also been summarised.
1. Introduction
Limonoids, a group of highly oxygenated triterpenoids, mainly exist in the Rutaceae and Meliaceae plant families.1 When they first attracted people's attention, limonoids were considered a major problem for the citrus juice industry due to the bitter principles through the biochemical transformation of a tasteless limonoid aglycone precursor to a bitter one.2 Tetranortriterpenoids is an alternative name for limonoids because in the process of oxidative changes of triterpenoids, the side chain is eventually oxidized to an α-substituted furyl ring by the loss of four carbon atoms.3 Basic limonoids contain the 4,4,8-trimethyl-17-furyl steroidal signature-skeleton, and all members of the family of limonoid natural products either contain this structure or are derived from such a precursor with different degrees of oxidation and skeletal rearrangement. Limonoids are classified into different subcategories such as ring-intact limonoids, ring-seco limonoids, degraded limonoids, and highly oxidatively modified limonoids.4 Limonoids exhibited a wide spectrum of biological properties including cytotoxic,5–8 antioxidant,9,10 antiinflammatory,11,12 neuroprotective,13,14 antiviral,15 antimicrobial,16,17 antiprotozoal,18 antimalarial,19–21 insect antifeedant,22–26 and insecticidal activities.27–29 The present review highlights the advances of limonoids in regard to isolation, total synthesis, and structural modifications with the relevant biological properties.2. Reviews
Some excellent reviews on various aspects of limonoids studies are listed here. Overview of the distribution and chemistry of limonoids in plants kingdom was collated in 2006.1 A comprehensive review of ‘Meliaceous limonoids: chemistry and biological activities’ has appeared in 2011.4 Topics on the chemistry and pharmacological activities of some limonoids have also been presented.30–35 In 2011, biosynthesis and total synthesis of limonoid natural products from an organic synthesis perspective were reviewed.36,37 On the other hand, it is noteworthy that some interesting limonoids with their relevant biological activities on annual reviews of ‘Marine natural products’ (covering 2011–2014)38–41 and ‘Triterpenoids’ (covering 2011–2013) have been summarised.42–443. Phytochemistry
To efficiently extract and isolate new limonoid natural products from plants, recently, several techniques have been developed. By combination of preparative high-speed countercurrent chromatography (HSCCC) and off-line LC-ESI-MS/MS analysis, Rodríguez-Rivera et al. reported a new chromatographic technique to detect very low concentrated natural products from Citrus limetta peels; moreover, four detected limonoid glucosides such as nomilinic acid glucoside, limonin glucoside, nomilin glucoside and obacunone glucoside, were easily recovered in the fast eluting.45 Haldar et al. developed the medium pressure liquid chromatography (MPLC) and LC-ESI-MS/MS-based technique to quickly isolate, identify and obtain some basic limonoids such as azadirone, epoxyazadiradione and azadiradione from neem fruits in preparative scale.46 The LC-HRMS-guided and preparative high-performance liquid chromatography (prep-HPLC)-based protocol was efficiently performed to isolate twenty-one secondary metabolites (including one limonoid, 1-O-methylclausenolide) from the leaves and stem bark extracts of Clausena anisata.47 Recently, supercritical CO2 extraction has been applied to obtain limonoid extracts from the seeds of C. aurantifolia swingle in shorter time when compared with that of the conventional methods.48 De Paula et al. reported an inexpensive and quick ultrasound-assisted extraction (UAE) and HPLC-photodiode array detector (PDA) technique to extract and determine azadirachtin from dried entire fruits of Azadirachta indica A. Juss (Meliaceae).49 More recently, Rangiah et al. have developed an ultra high performance liquid chromatography/mass spectrometry/selected reaction monitoring (UHPLC/MS/SRM) assay for quantification of five neem metabolites (e.g., azadirachtin A, nimbin, salanin, azadiradione and epoxy or hydroxy-azadiradione) from leaf extracts of Meliaceae family plants.50With the development of technology, during 2014–2016, a wide array of new limonoid natural products were isolated from different parts of plants. Recent advances on the isolation and structure determination of limonoids, together with their relevant biological activities are presented according to their source organisms such as Meliaceae, Rutaceae, Euphorbiaceae and Simaroubaceae families.
3.1. Meliaceae
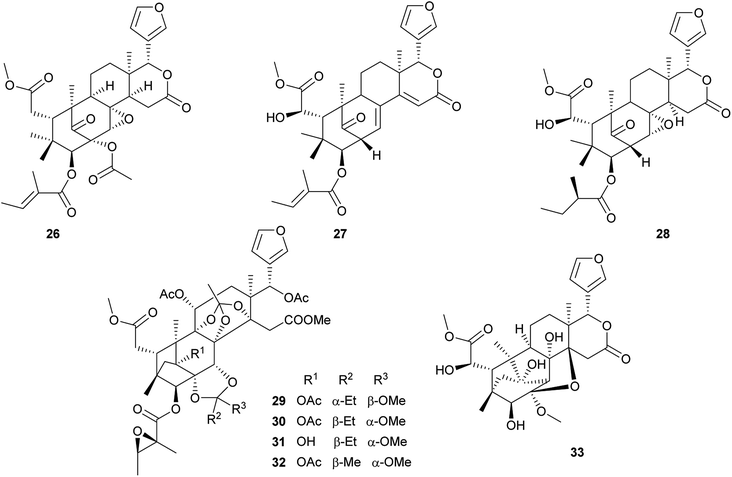
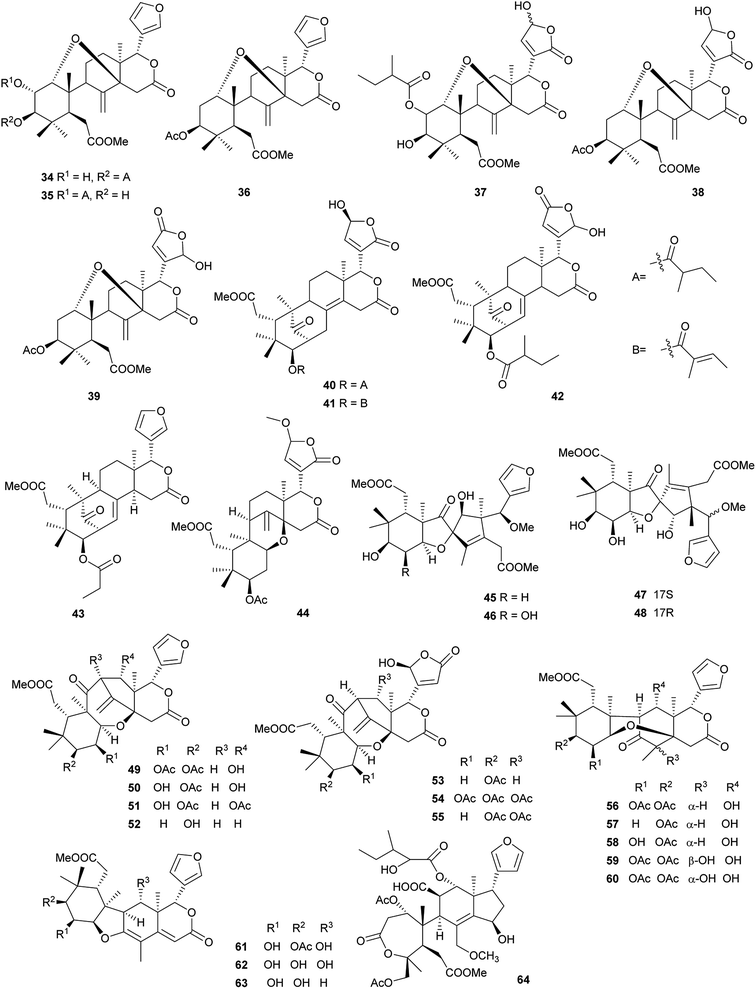
Compound 37 displayed potent cytotoxic activity against B–16 with an IC50 value of 8.51 μg mL−1.62 Compound 44 showed the potent antifeedant activity against fruit fly (Drosophila melanogaster; antifeedant index (AI) at 1 mM: 32.8%).64 Compound 45 (IC50: 16.7 μM) displayed moderate inhibition activity against protein tyrosine phosphatase 1B (PTP1B).65 Compounds 50 (EC50: 5.5 μM) and 64 (EC50: 6.1 μM) showed significant anti-HIV activities.66
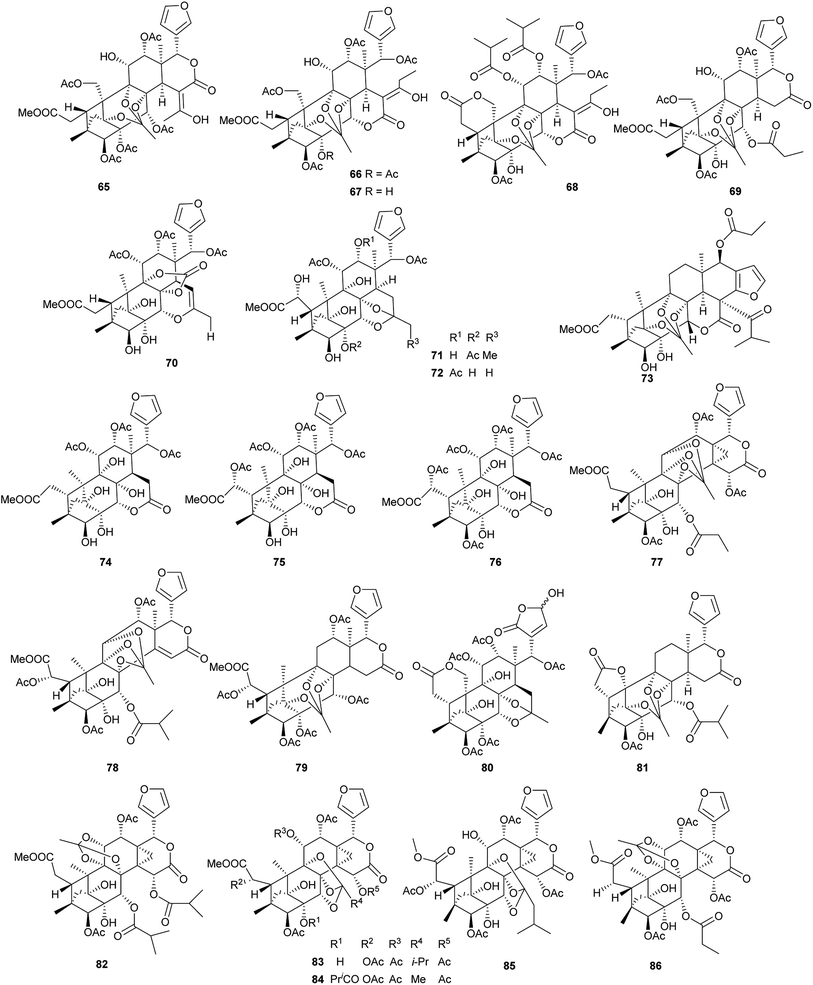
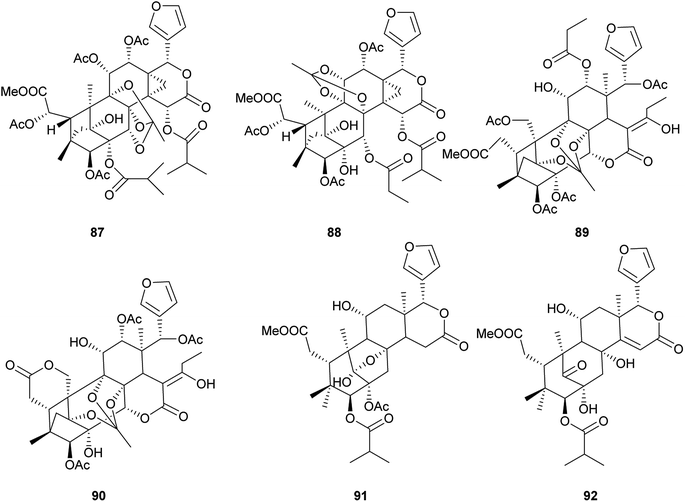
Compound 65 exhibited significant inhibition activity against LPS-induced NF-kB production. It suggested that the ortho ester group and/or the 2,7-dioxabicyclo[2.2.1]heptane moiety in these phragmalin limonoids were crucial for the activities.67 Compounds 81–84, 86, and 88 exhibited significant inhibitory activities against α-glucosidase in vitro with IC50 values of 0.06, 0.04, 0.52, 1.09, 0.15, and 0.96 mM, respectively.71–73
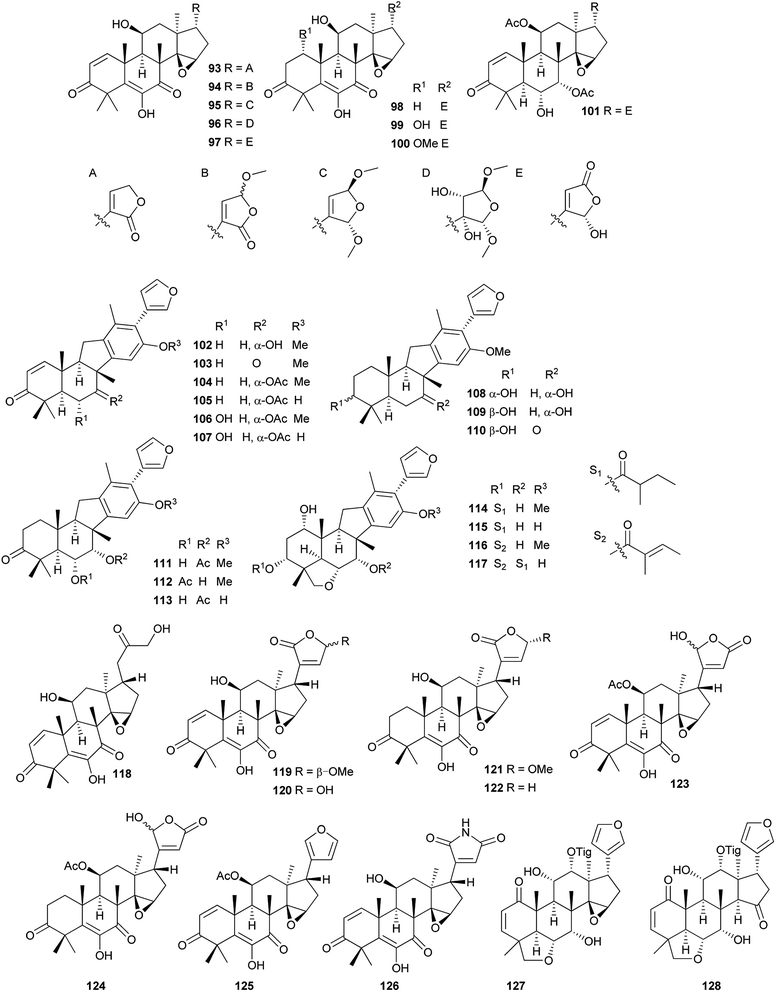
Compounds 95, 97, 127 and 128 exhibited potent cytotoxic activities against five human tumor cell lines (e.g., HL-60, SMMC-7721, A-549, MCF-7, and SW480) with IC50 values in the range of 2.2–4.5 μM.75,78 Compounds 103 and 104 exhibited mild inhibitory activities against mouse and human 11β-HSD1 with IC50 values of 13.4 and 8.25 μM, respectively.76
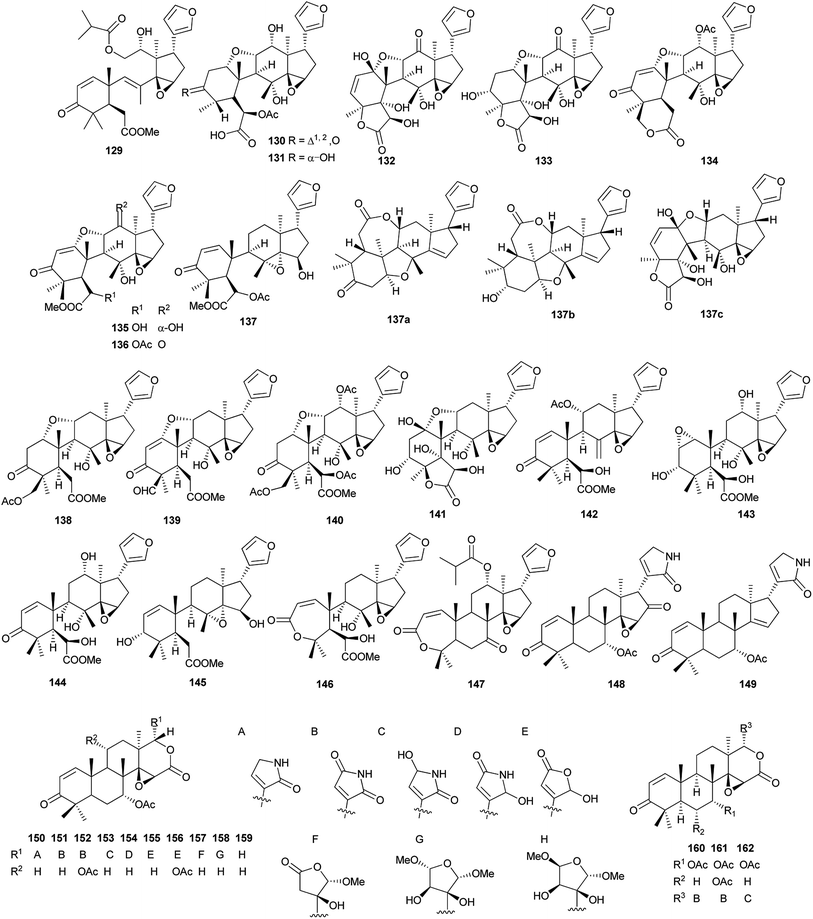
Compound 132 exhibited modest cytotoxicity against HL-60 (IC50: 5.38 μM) and HepG2 cells (IC50: 5.22 μM).80 Compounds 141, 142 and 144–147 showed potent radical scavenging activities (DPPH IC50: 51.3–104.0 μM; ABTS+ IC50: 52.2–167.3 μM); compounds 138–141 exhibited significant anti-inflammatory (selective inhibition of Cox-1 and Cox-2 at 100 μM: >88%), and cytotoxic activities against seven human tumor cell lines (IC50: 2.1–14.7 μM).81 Compounds 148 (IC50: 10.21 μM), 149 (IC50: 20.05 μM), 153 (IC50: 12.56 μM), 155 (IC50: 12.56 μM) and 156 (IC50: 20.68 μM) exhibited marked inhibitory effects on NO production in LPS-activated RAW 264.7 macrophages at nontoxic concentration.82
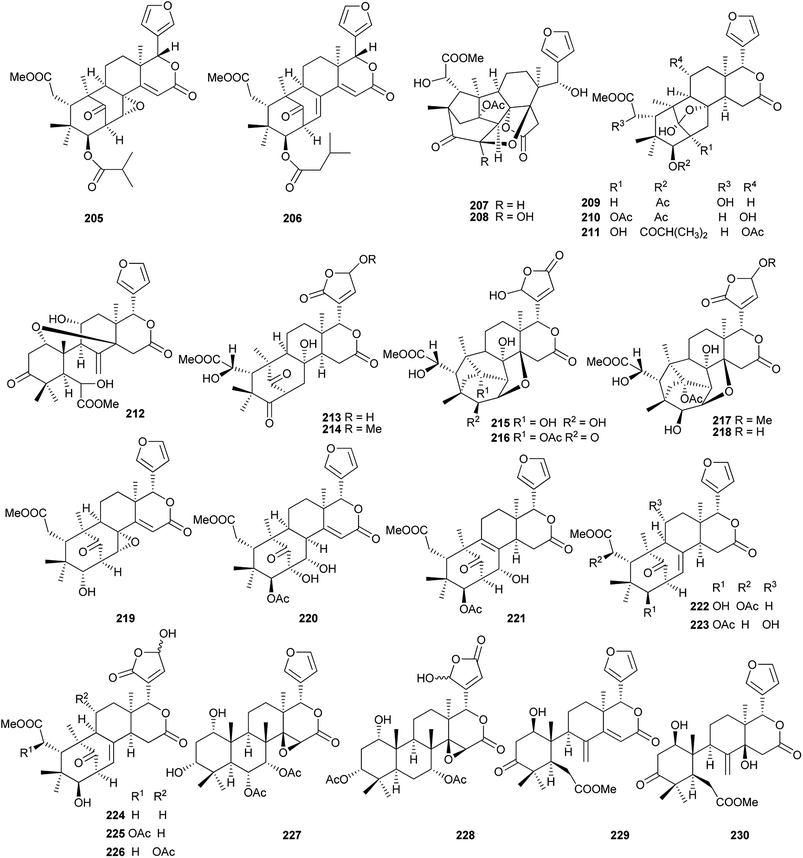
Compounds 209 (IC50: 15.3 μM) and 212 (IC50: 17.5 μM) exhibited moderate cytotoxic activity against HL-60 cell line.92 Compound 230 displayed the significant neuroprotective activity against glutamate-induced injury in primary rat cerebellar granule neuronal cells with increased viability of 83.3% at 10 μM and 80.3% at 1 μM.94
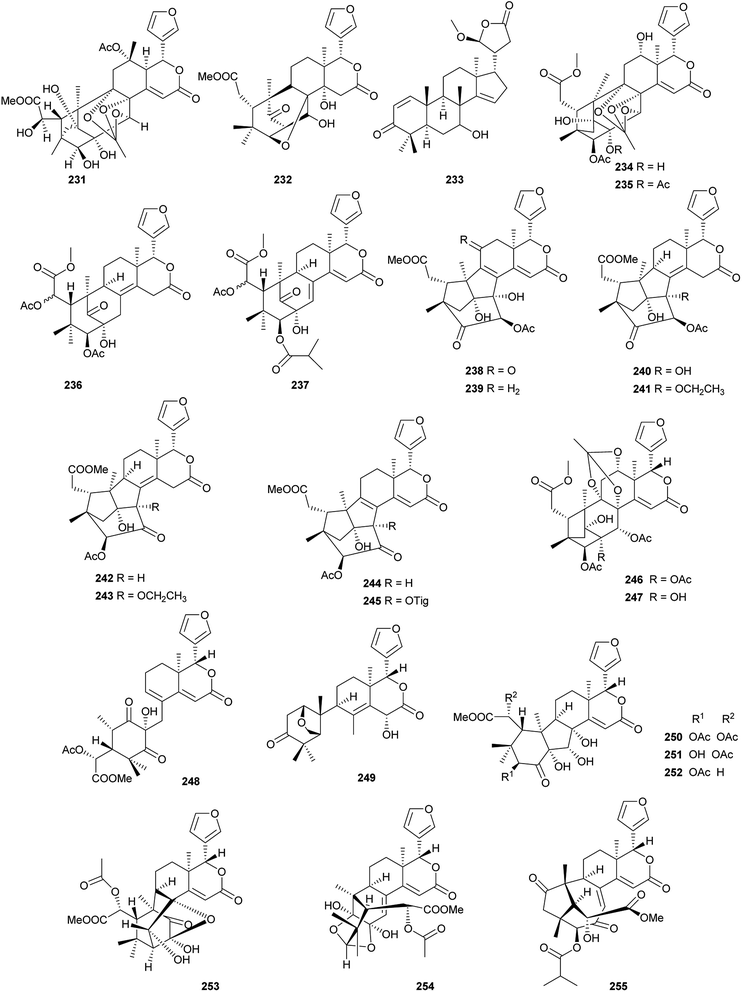
Among them, compound 240 (IC50: 77.1 μM) exhibited moderate anti-H1N1 activity;96 compound 247 showed moderate cytotoxicities against ovarian A2780 and A2780/T cells with equal IC50 values of 37.5 μM for each.97
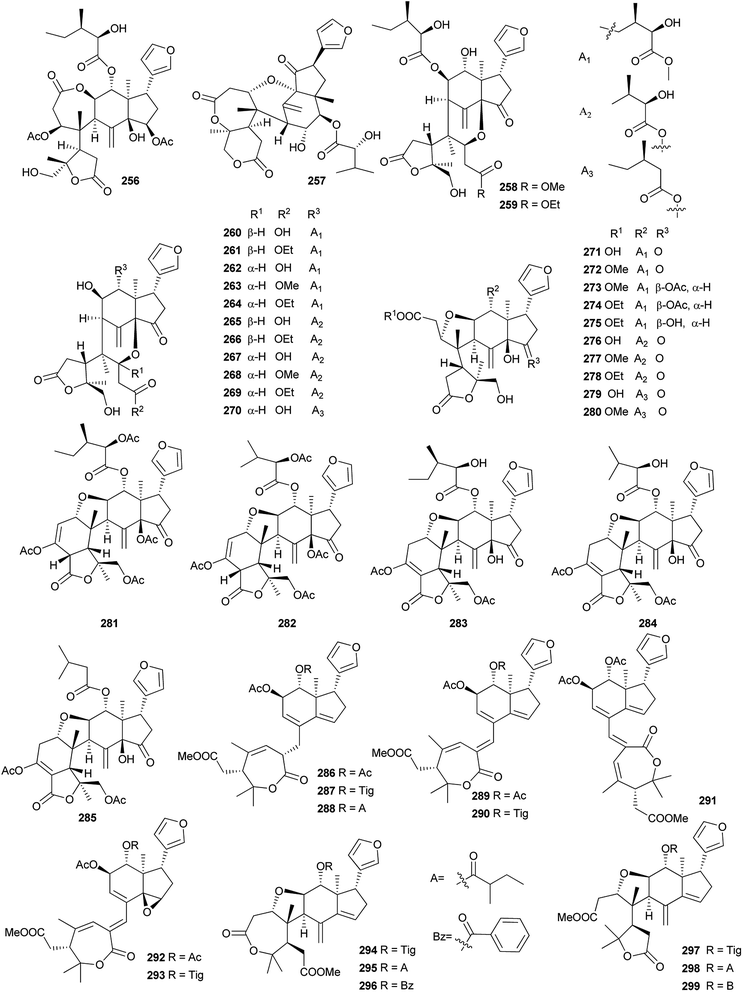
Among them, compounds 264 and 274 showed strong insecticidal activities against Plutella xylostella.99 Compounds 286, 289 and 290 exhibited potent antifeedant activities against the generalist Helicoverpa armigera with EC50 values of 0.017, 0.008, and 0.012 μmol cm−2, respectively. Preliminary structure–activity relationship indicated that Δ (ref. 2 and 30) configuration and the acetoxy substituent at C-12 position were vital to the antifeedant activity.101
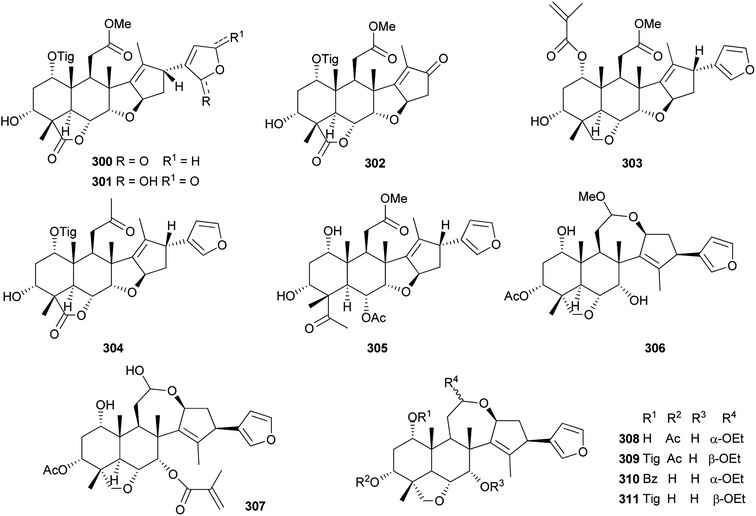
Among them, compound 300 (IC50: 86.0 μM) showed inhibitory effects against LPS-induced NO production in RAW 264.7 cell line; the IC50 values of compounds 301 and 302 against the Epstein-Barr virus early antigen (EBV-EA) were 299 and 318 molar ratio/32 pmol TPA, respectively.102 Compound 309 (MIC: 31.25 μg mL−1) exhibited the potent antibiotic activity against Porphyromonas gingivalis ATCC 33277.105
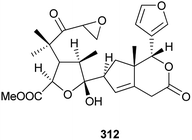
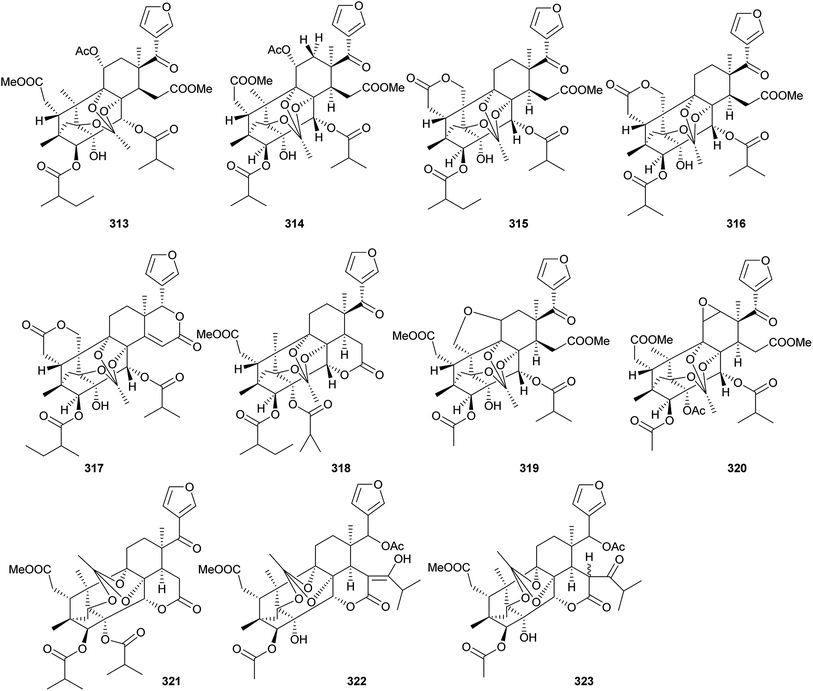
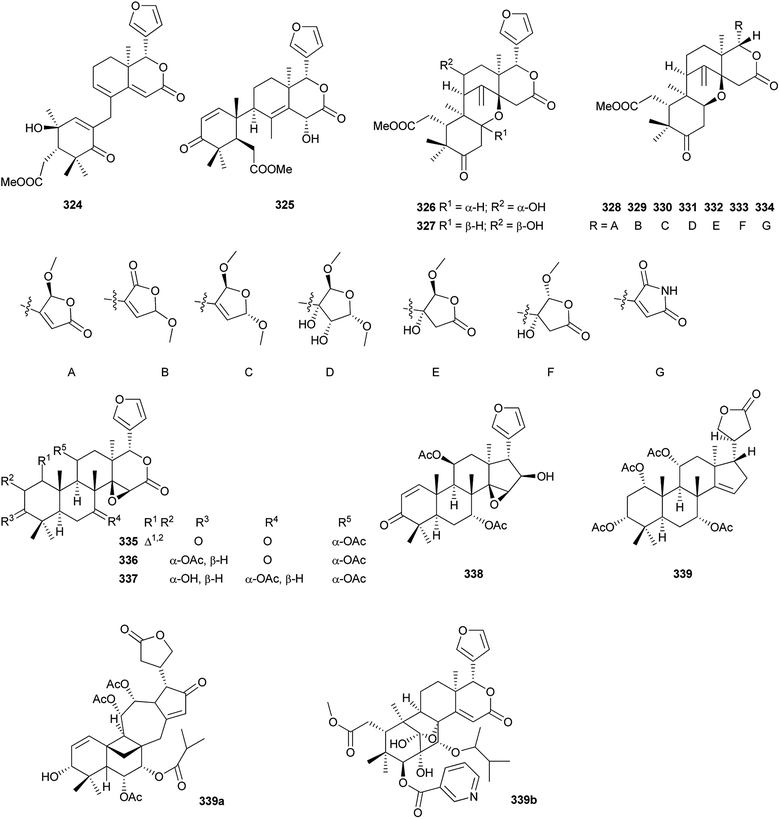
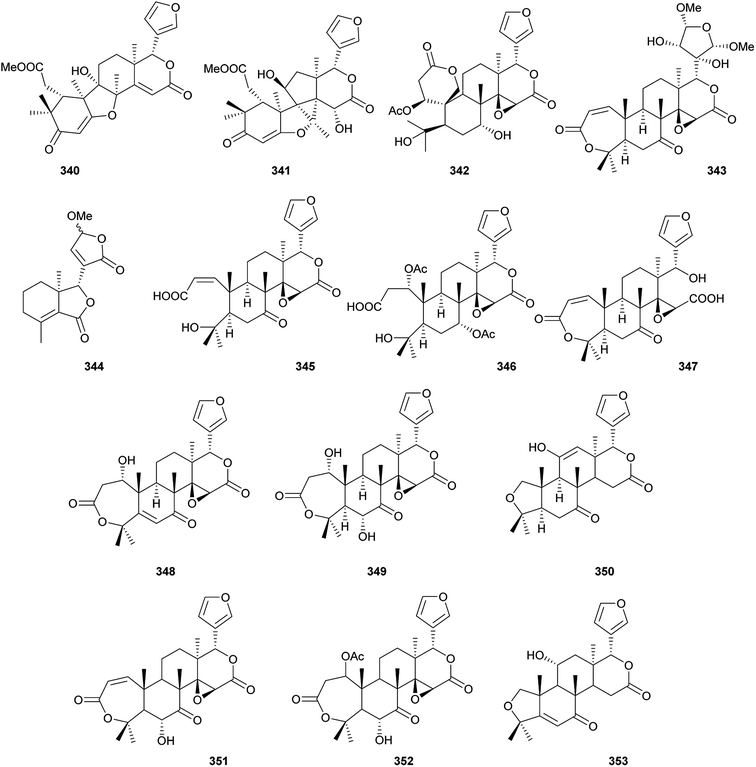
Additionally, as shown in Fig. 16, entanutilin A (339a) and B (339b) were isolated from the stem barks of Entandrophragma utile.110b Their absolute configurations were confirmed by CD exciton chirality method.
3.2. Rutaceae
3.3. Euphorbiaceae
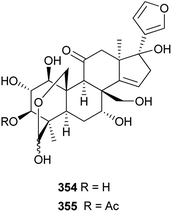
Two highly oxygenated limonoids, such as flexuosoids A 354 and B 355 (Fig. 18), with a C-19/29 lactol bridge and heptaoxygenated substituents at C-1, C-2, C-3, C-7, C-11, C-17, and C-30 positions, were isolated from the roots of Phyllanthus flexuosus.115 Compounds 354 and 355 showed antifeedant activities against the beet army worm (Spodoptera exigua) with EC50 values of 25.1 and 17.3 μg cm−2, respectively. In addition, compounds 354 (IC50: 11.5 μM) and 355 (IC50: 8.5 μM) displayed moderate cytotoxic activities against the ECA109 human esophagus cancer cell line.3.4. Simaroubaceae
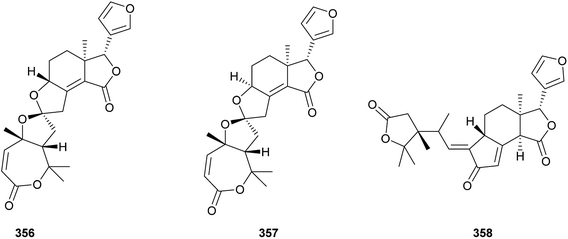
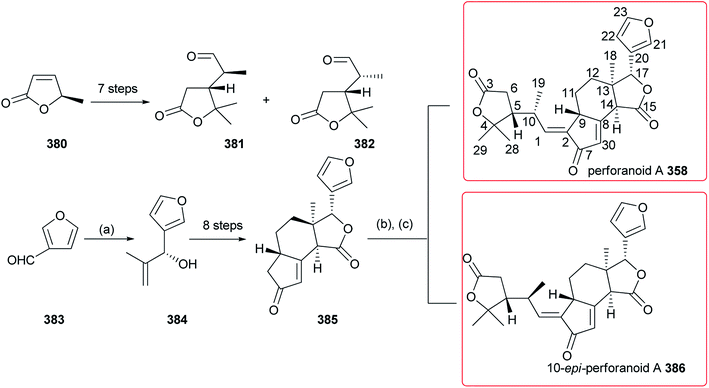
As shown in Fig. 19, two new 16-nor limonoids, harperspinoids A 356 and B 357, with a unique 7/5/5/6/5 ring system, were obtained from the leaves and branches of Harrisonia perforata.116 Especially the absolute structure of 356 was further confirmed by X-ray crystallographic analysis. Moreover, compound 356 exhibited the notable inhibitory activity against the 11β-HSD1 enzyme with an IC50 value of 0.60 μM. The biogenetic pathway of these two compounds was also proposed. Perforanoid A 358 (Fig. 19), isolated from the leaves of H. perforata, showed cytotoxic activities against HEL, K562, CB3, DP17, and WM9 tumor cell lines (IC50: 4.24–25.96 μM).1174. Total synthesis
4.1. Cipadonoid B
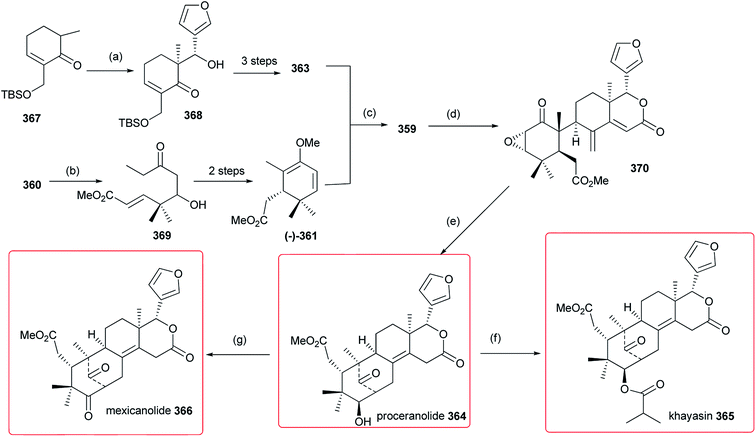
In 2011, an efficient strategy for the total synthesis of cipadonoid B 359 was reported (Scheme 1). First, compound 361 was prepared by reaction of silyl enol ether with 360. Then, azedaralide 363 reacted with 361 via the intermediate 362, to give 359 (20% yield) by a ketal-Claisen rearrangement.118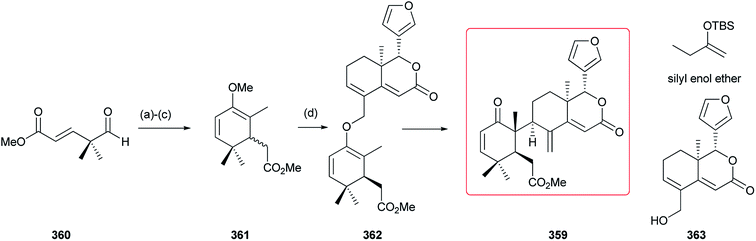 | ||
| Scheme 1 Reagents and conditions: (a) silyl enol ether, TiCl4, CH2Cl2, −78 °C; (b) KH, PhH; (c) MeOTf, CH2Cl2; (d) azedaralide 363, PTSA, xylenes, 180 °C, 4 h. | ||
4.2. Khayasin, proceranolide, and mexicanolide
Based upon cipadonoid B 359,118 Faber et al. further reported a concise and enantioselective total synthesis of proceranolide 364, khayasin 365 and mexicanolide 366 (Scheme 2). First, compound 364 was obtained from 370 by epoxidation and cyclization; then, compound 365 was afforded by acylation of 364; finally, conversion of 364 to 366 in the presence of Jones reagent was achieved.1194.3. Limonin
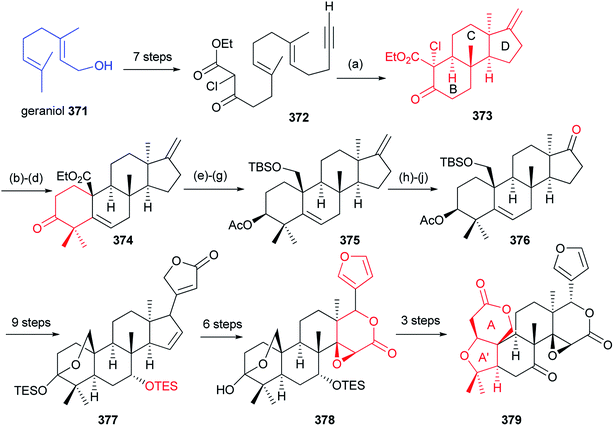
As depicted in Scheme 3, Yamashita et al. described the total synthesis of (±)-limonin 379 in 35 steps. Via an intermediate 372, a tandem radical cyclization of geraniol 371 gave 373 containing a BCD ring system with the C-13α configuration. Then, the limonoid androstane framework 374 was constructed by a Robinson annulation of 373. Subsequently, compound 376 was produced by epoxidation and nitrile addition, followed by acetonitrile elimination. Through a singlet-oxygen cycloaddition and a Baeyer–Villiger oxidation, compound 378 was obtained via the intermediate 377. Finally, construction of target compound 379 was achieved by a Suarez reaction.120 This study will lay the foundation for future synthesis of diverse limonoid skeletons.4.4. Perforanoid A and 10-epi-perforanoid A
More recently, as shown in Scheme 4, Hao et al. developed an efficient way to total synthesis of perforanoid A 358 and 10-epi-perforanoid A 386. The key steps were as follows: allylic alcohol 384 was enantioselectively obtained by alkenylation of 3-formylfuran 383 with 2-methylpropenal; then, Pd-catalyzed coupling of 384 with a vinyl ether gave the γ-lactone ring, with stereoselective construction of the C13 all carbon quaternary center, followed by formation of the cyclopentenone ring by a Rh-catalyzed Pauson-Khand reaction to give 385. Finally, reaction of 385 with 381 or 382 produced 358 (33% yield) and 386 (36% yield), respectively.117 Compound 358 showed potent cytotoxic activities against HEL, K562, and CB3 tumor cell lines with IC50 values of 6.17, 4.24, and 3.91 μM, respectively; in contrast, compound 386 did not display any cytotoxic activity.5. Structural modifications
5.1. Biocatalytic modifications
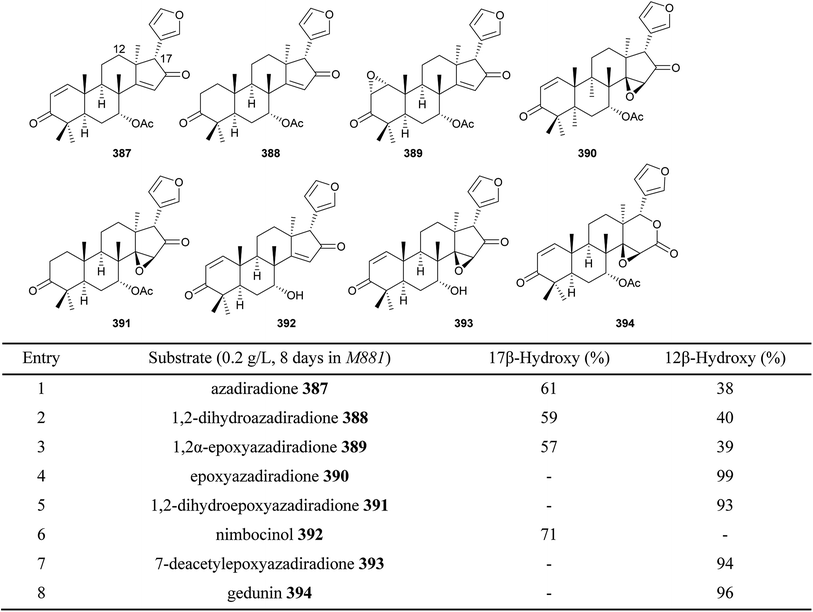
Biotransformation is a good choice for the production of sufficient amounts of scientifically and commercially valuable compounds with the advantages of strict stereo- and region-selectivity, mild reaction conditions and simple operation procedure. As shown in Fig. 20, 8 limonoids including azadiradione 387, 1,2-dihydroazadiradione 388, 1,2α-epoxyazadiradione 389, epoxyazadiradione 390, 1,2-dihydroepoxyazadiradione 391, nimbocinol 392, 7-deacetylepoxyazadiradione 393 and gedunin 394, were converted into their corresponding 12β- and/or 17β-hydroxy derivatives via fungi M881-mediated biocatalysis. Interestingly, when 14β,15β-epoxidation was on the basic limonoid skeleton (e.g., 390, 391, 393, and 394), only 12β-hydroxy derivative was produced as the single metabolite.1215.2. Chemical modifications
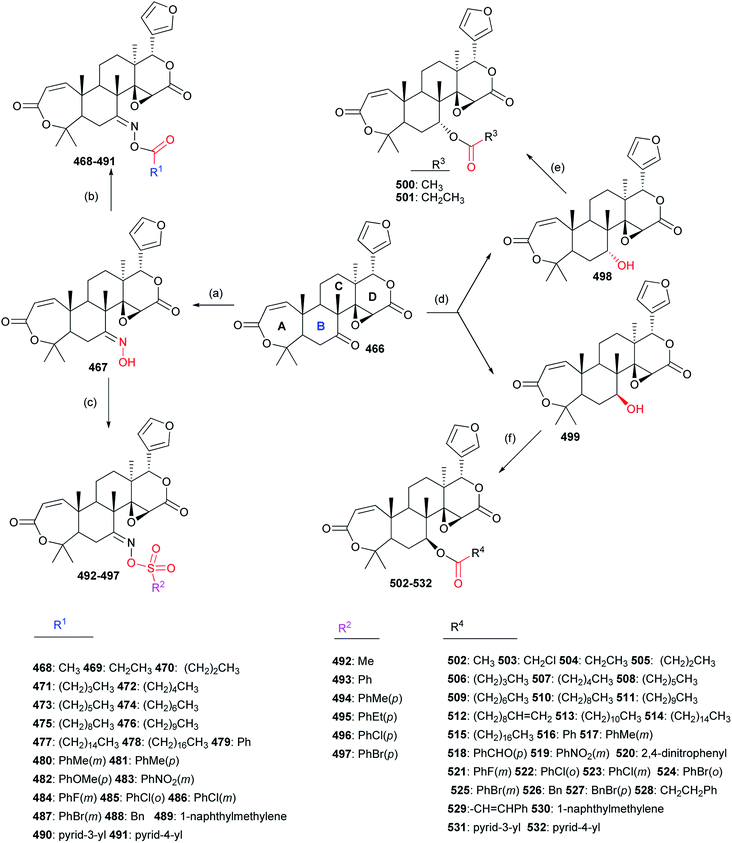
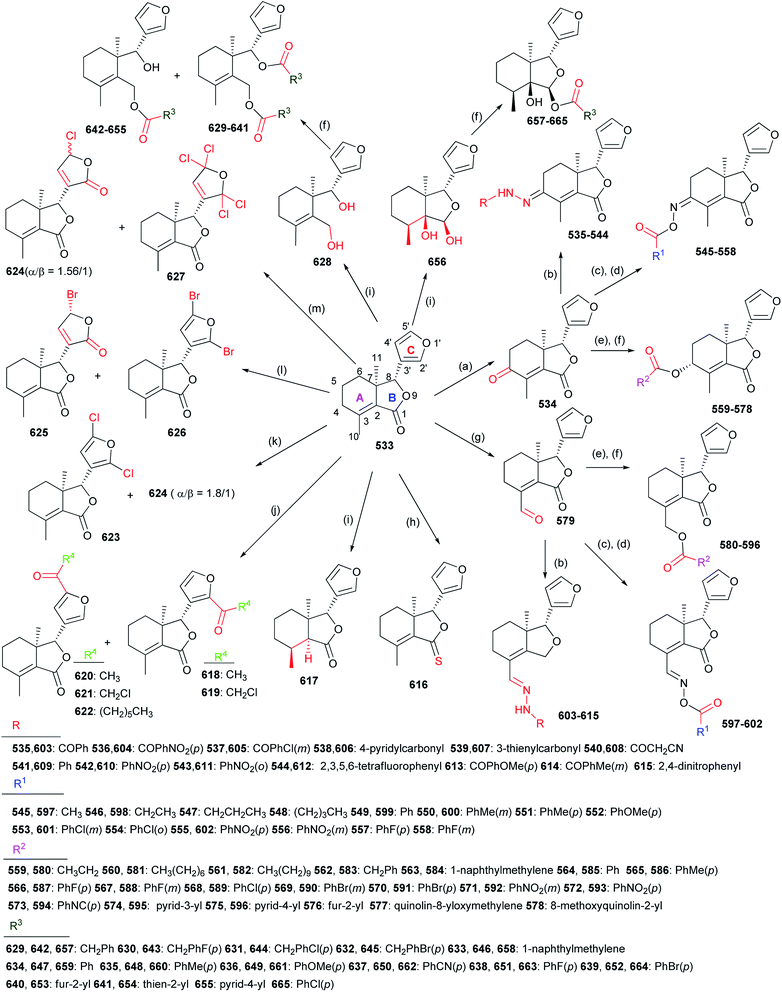
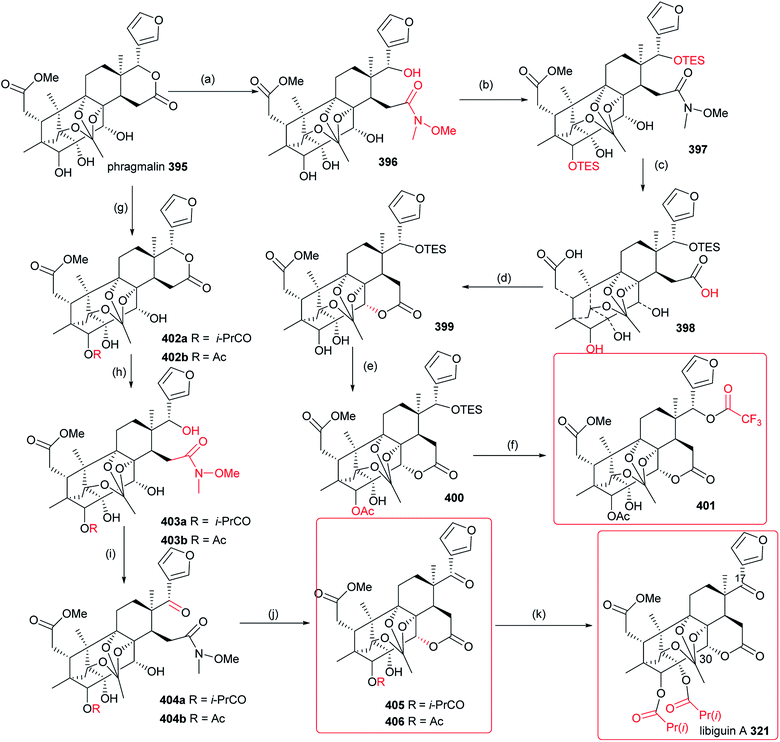
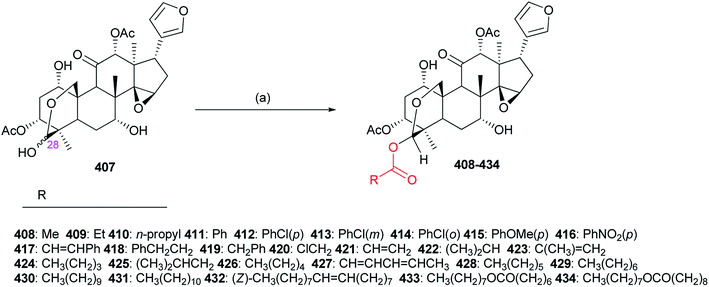
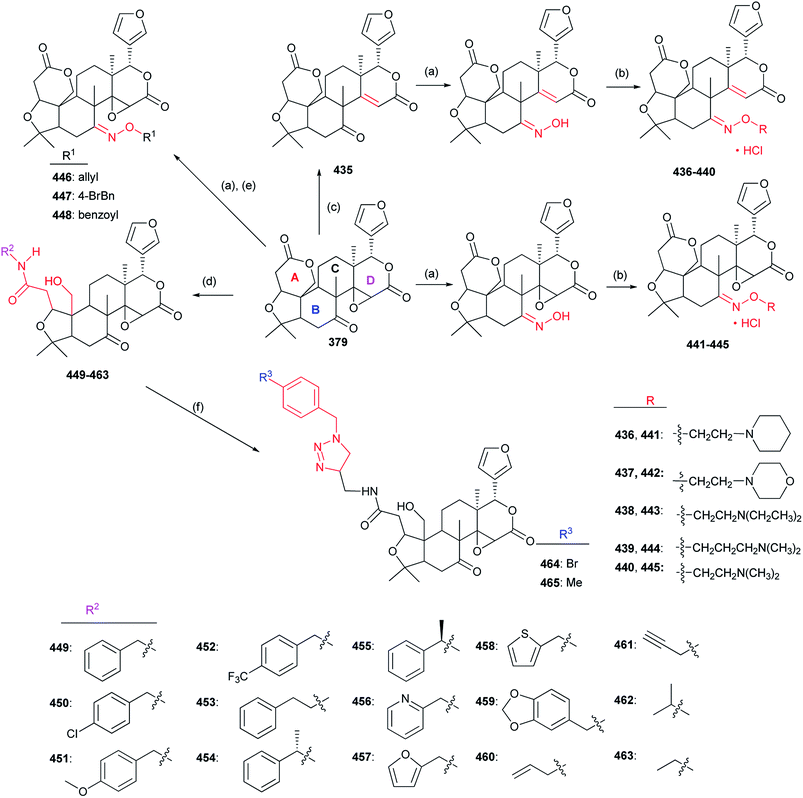
The structure–activity relationships demonstrated introduction of the carbonyl or oxime group on the C-4 position of 533 generally resulted in more promising derivatives than those containing a carbonyl or oxime one at the C-10 position; introduction of the heterocyclic fragments at C-4 or C-10 position of 533 was necessary for the insecticidal activity; the lactone (B ring) of 533 was important for the insecticidal activity; the double bond at the C-2 position of 533 was not necessary for the insecticidal activity; substitution of the oxygen atom on the carbonyl group of 533 by the sulfur one did not improve the insecticidal activity; introduction of the acyl group on the C ring of 533 could lead to more potent compounds than those containing the halogen atom at the same position.130–134
6. Biological activities of the most active limonoids
Due to exhibiting a large number of biological properties, currently, limonoids and their analogs have received much research attention in the medicinal and agricultural fields. Additionally, the most active limonoids and their analogs in each series against human cancer cell lines and insect pests were summarized in Tables 1 and 2, respectively.| Compounds | Cells | IC50 (μM) |
|---|---|---|
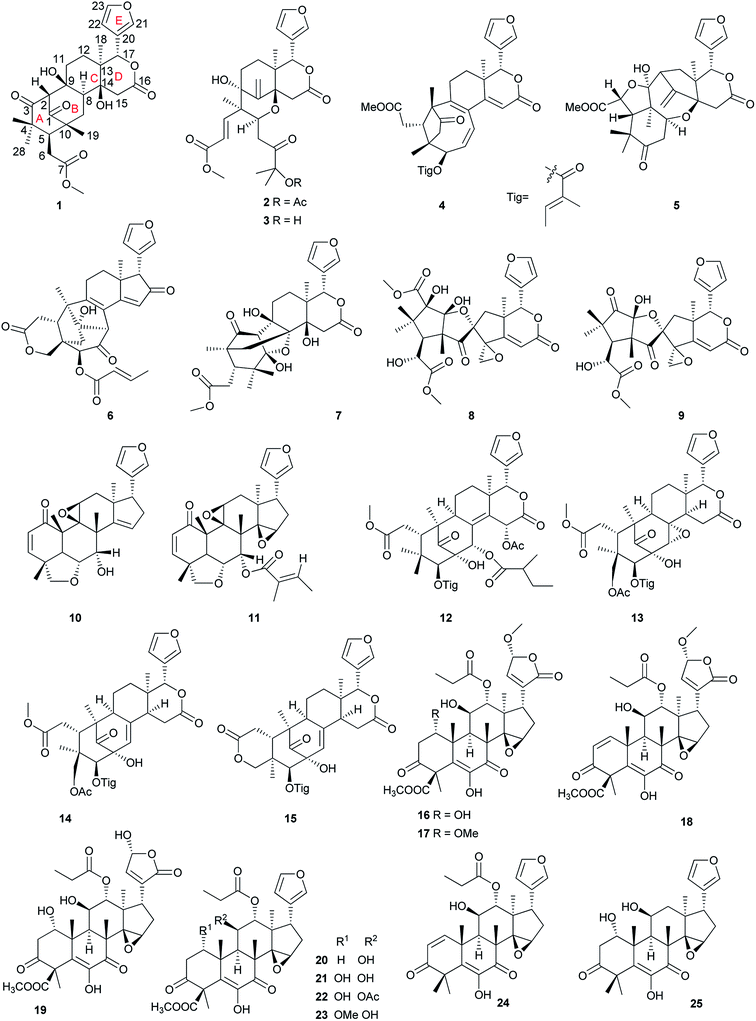
| 1,2-Dihydrodeacetylhirtin 20 (ref. 58) | HL-60 | 4.9 |
| SMMC-7721 | 3.1 | |
| A-549 | 2.9 | |
| MCF-7 | 9.8 | |
| SW480 | 9.0 | |
| 1α-Hydroxy-1,2-dihydrodeacetylhirtin 21 (ref. 58) | HL-60 | 3.1 |
| SMMC-7721 | 1.0 | |
| A-549 | 1.1 | |
| MCF-7 | 1.0 | |
| SW480 | 1.6 | |
| 1α-Methoxy-1,2-dihydrodeacetylhirtin 23 (ref. 58) | HL-60 | 5.3 |
| SMMC-7721 | 3.7 | |
| A-549 | 5.2 | |
| SMMC-7721 | 5.3 | |
| A-549 | 6.4 | |
| Cipaferen E 37 (ref. 62) | B-16 | 8.51 |
| Yunnanolide A 95 (ref. 75) | HL-60 | 3.6 |
| SMMC-7721 | 2.4 | |
| A-549 | 3.7 | |
| MCF-7 | 4.2 | |
| SW480 | 3.5 | |
| BEAS-2B | 5.0 | |
| 11β-Hydroxyisowalsuranolide 97 (ref. 75) | HL-60 | 3.1 |
| SMMC-7721 | 2.2 | |
| A-549 | 2.6 | |
| MCF-7 | 3.9 | |
| SW480 | 2.4 | |
| BEAS-2B | 9.4 | |
| Walsuronoids D 127 (ref. 78) | HL-60 | 2.7 |
| SMMC-7721 | 3.1 | |
| A-549 | 4.1 | |
| MCF-7 | 3.1 | |
| SW480 | 2.8 | |
| Walsuronoids E 128 (ref. 78) | HL-60 | 3.3 |
| SMMC-7721 | 4.1 | |
| A-549 | 4.4 | |
| MCF-7 | 4.4 | |
| SW480 | 4.5 | |
| Toonaciliatones C 132 (ref. 80) | HL-60 | 5.38 |
| HepG2 | 5.22 | |
| Toonasinenines B 139 (ref. 81) | A-549 | 5.7 |
| CHG-5 | 5.0 | |
| HCT15 | 5.7 | |
| HeLa | 6.2 | |
| HepG2 | 5.5 | |
| MDA-MB-231 | 6.0 | |
| SGC-7901 | 6.0 | |
| Toonasinenines C 140 (ref. 81) | A-549 | 9.7 |
| CHG-5 | 8.3 | |
| HepG2 | 9.1 | |
| MDA-MB-231 | 9.4 | |
| SGC-7901 | 9.4 | |
| Toonasinenines D 141 (ref. 81) | A-549 | 2.3 |
| CHG-5 | 2.8 | |
| HCT15 | 2.6 | |
| HeLa | 2.9 | |
| HepG2 | 3.0 | |
| MDA-MB-231 | 2.7 | |
| SGC-7901 | 2.1 | |
| Munronins A 188 (ref. 89) | HL-60 | 0.44 |
| SMMC-7721 | 2.3 | |
| A-549 | 1.6 | |
| MCF-7 | 1.5 | |
| SW480 | 0.86 | |
| Perforanoid A 358 (ref. 117) | HEL | 6.17 |
| K562 | 4.24 | |
| CB3 | 3.91 |
| Compounds | Insect pests | FMRs (at 1 mg mL−1) |
|---|---|---|
| a Antifeedant index: AI.b EC50 value: the effective concentration for 50% feeding reduction.c ED50 value: the effective dosage for 50% feeding reduction. | ||
| Cineracipadesin G 44 (ref. 64) | Drosophila melanogaster | AIa = 32.8% at 1 mM (nicotine: AI = 28.5% at 1 mM) |
| Aphanamixoids C 286 (ref. 101) | Helicoverpa armigera | EC50b = 9.27 μg cm−2 |
| Aphanamixoids F 289 (ref. 101) | EC50 = 4.28 μg cm−2 | |
| Aphanamixoids G 290 (ref. 101) | EC50 = 6.82 μg cm−2 (neem oil: EC50 = 2.62 μg cm−2) | |
| Flexuosoids A 354 (ref. 115) | Spodoptera exigua | ED50c = 25.1 μg cm−2 |
| Flexuosoids B 355 (ref. 115) | ED50 = 17.3 μg cm−2 | |
| 410 (ref. 123) | Pre-third-instar larvae of Mythimna separata | 73.1% toosendanin: 50.0% |
| 417 (ref. 123) | 61.5% | |
| 431 (ref. 124) | Pre-third-instar larvae of M. separata | 63.0% |
| 472 (ref. 127) | Pre-third-instar larvae of M. separata | 62.1% |
| 485 (ref. 127) | 72.4% | |
| 486 (ref. 127) | 65.5% | |
| 490 (ref. 127) | 62.1% | |
| 566 (ref. 131) | Pre-third-instar larvae of M. separata | 63.0% |
| 575 (ref. 131) | 66.7% | |
| 578 (ref. 131) | 63.0% | |
| 534 (ref. 132) | Pre-third-instar larvae of M. separata | 73.3% |
| 552 (ref. 132) | 70.0% | |
| 558 (ref. 132) | 73.3% | |
| 597 (ref. 132) | 66.7% | |
| 598 (ref. 132) | 70.0% | |
| 600 (ref. 132) | 66.7% | |
| 602 (ref. 132) | 70.0% | |
| 535 (ref. 130) | Pre-third-instar larvae of M. separata | 76.9% |
| 536 (ref. 130) | 65.4% | |
| 537 (ref. 130) | 69.2% | |
| 540 (ref. 130) | 61.5% | |
| 603 (ref. 130) | 65.4% | |
| 604 (ref. 130) | 61.5% | |
| 606 (ref. 130) | 73.1% | |
| 608 (ref. 130) | 69.2% | |
| 612 (ref. 130) | 61.5% | |
| 662 (ref. 133) | Pre-third-instar larvae of M. separata | 65.5% |
7. Conclusions
However, the availability of limonoids is very limited from natural resources because of intensive collection of plants from the wild and long plant growth period. Although total chemical synthesis of limonoids remains a challenge of significant novelty and interest, it is not a practical option from a commercial point of view. Consequently, sustainable biotechnology and tissue culture techniques may be extensively exploited to enhance production of limonoids to meet the increasing demands. Additionally, to improve the water solubility and bioactivities, structural modifications of limonoids should be further strengthened.In the present review, we summarised 363 new limonoid natural products isolated from plants during 2014–2016, together with their relevant biological activities and source organisms. Moreover, we highlighted recent developments in the total synthesis, and structural modifications of limonoids and their analogs regarding their bioactivities during 2011–2016. We hope that this review can provide necessary information for synthetic, medicinal and pesticidal chemistry, and phytochemistry researchers who are interested in the chemistry and biology of limonoids.
Acknowledgements
We thank the National Natural Science Foundation of China (31071737, 31171896, 31672071), and Special Funds of Central Colleges Basic Scientific Research Operating Expenses, Northwest A&F University (2452015096, YQ2013008) for generous financial support.References
- A. Roy and S. Saraf, Biol. Pharm. Bull., 2006, 29, 191–201 CAS.
- V. P. Maier and G. D. Beverly, J. Food Sci., 1968, 33, 488–492 CrossRef CAS.
- N. T. Kipassa, T. Iwagawa, H. Okamura, M. Doe, Y. Morimoto and M. Nakatani, Phytochemistry, 2008, 69, 1782–1787 CrossRef CAS PubMed.
- Q. G. Tan and X. D. Luo, Chem. Rev., 2011, 111, 7437–7522 CrossRef CAS PubMed.
- S. M. Poulose, E. D. Harris and B. S. Patil, J. Nutr., 2005, 135, 870–877 CAS.
- S. Ejaz, A. Ejaz, K. Matsuda and C. W. Lim, J. Sci. Food Agric., 2006, 86, 339–345 CrossRef CAS.
- B. T. Murphy, P. Brodie, C. Slebodnick, J. S. Miller, C. Birkinshaw, L. M. Randrianjanaka, R. Andriantsiferana, V. E. Rasamison, K. TenDyke, E. M. Suh and D. G. I. Kingston, J. Nat. Prod., 2008, 71, 325–329 CrossRef CAS PubMed.
- K. Awang, C. S. Lim, K. Mohamad, H. Morita, Y. Hirasawa, K. Takeya, O. Thoisone and A. H. A. Hadi, Bioorg. Med. Chem., 2007, 15, 5997–6002 CrossRef CAS PubMed.
- J. Yu, L. M. Wang, R. L. Walzem, E. G. Miller, L. M. Pike and B. S. Patil, J. Agric. Food Chem., 2005, 53, 2009–2014 CrossRef CAS PubMed.
- A. P. Breksa and G. D. Manners, J. Agric. Food Chem., 2006, 54, 3827–3831 CrossRef CAS PubMed.
- F. Xie, M. Zhang, C. F. Zhang, Z. T. Wang, B. Y. Yu and J. P. Kou, J. Ethnopharmacol., 2008, 117, 463–466 CrossRef CAS PubMed.
- J. J. Chen, S. S. Huang, C. H. Liao, D. C. Wei, P. J. Sung, T. C. Wang and M. J. Cheng, Food Chem., 2010, 120, 379–384 CrossRef CAS.
- J. S. Yoon, S. H. Sung and Y. C. Kim, J. Nat. Prod., 2008, 71, 208–211 CrossRef CAS PubMed.
- G. S. Jeong, E. Byun, B. Li, D. S. Lee, Y. C. Kim and R. B. An, Arch. Pharmacal Res., 2010, 33, 1269–1275 CrossRef CAS PubMed.
- E. Balestrieri, F. Pizzimenti, A. Ferlazzo, S. V. Giofre, D. Iannazzo, A. Piperno, R. Romeo, M. A. Chiacchio, A. Mastino and B. Macchi, Bioorg. Med. Chem., 2011, 19, 2084–2089 CrossRef CAS PubMed.
- S. A. Abdelgaleil, T. Iwagawa, M. Doe and M. Nakatani, Fitoterapia, 2004, 75, 566–572 CrossRef CAS PubMed.
- Y. Nakai, S. Pellett, W. H. Tepp, E. A. Johnson and K. D. Janda, Bioorg. Med. Chem., 2010, 18, 1280–1287 CrossRef CAS PubMed.
- A. E. Hay, J. R. Ioset, K. M. Ahua, D. Diallo, R. Brun and K. Hostettmann, J. Nat. Prod., 2007, 70, 9–13 CrossRef CAS PubMed.
- M. F. Dolabela, S. G. Oliveira, J. M. Nascimento, J. M. Peres, H. Wagner, M. M. Povoa and A. B. de Oliveira, Phytomedicine, 2008, 15, 367–372 CrossRef CAS PubMed.
- J. Bickii, N. Njifutie, J. A. Foyere, L. K. Basco and P. Ringwald, J. Ethnopharmacol., 2000, 69, 27–33 CrossRef CAS PubMed.
- K. Kaur, M. Jain, T. Kaur and R. Jain, Bioorg. Med. Chem., 2009, 17, 3229–3256 CrossRef CAS PubMed.
- M. Nakatani, S. A. M. Abdelgaleil, J. Kurawaki, H. Okamura, T. Iwagawa and M. Doe, J. Nat. Prod., 2001, 64, 1261–1265 CrossRef CAS.
- X. D. Luo, S. H. Wu, D. G. Wu, Y. B. Ma and S. H. Qi, Tetrahedron, 2002, 58, 7797–7804 CrossRef CAS.
- G. Ruberto, A. Renda, C. Tringali, E. M. Napoli and M. S. J. Simmonds, J. Agric. Food Chem., 2002, 50, 6766–6774 CrossRef CAS PubMed.
- K. Nihei, Y. Asaka, Y. Mine, Y. Yamada, M. Iigo, T. Yanagisawa and I. Kubo, J. Nat. Prod., 2006, 69, 975–977 CrossRef CAS PubMed.
- J. Wu, S. Zhang, T. Bruhn, Q. Xiao, H. X. Ding and G. Bringmann, Chem.–Eur. J., 2008, 14, 1129–1144 CrossRef CAS PubMed.
- M. Lü, W. J. Wu and H. X. Liu, Pestic. Biochem. Physiol., 2010, 98, 263–268 CrossRef.
- G. Singh, P. J. Rup and O. Koul, Bull. Entomol. Res., 2007, 97, 351–357 CrossRef CAS PubMed.
- R. Fowles, B. Mootoo, R. Ramsewak, A. Khan, A. Ramsubhag, W. Reynolds and M. Nair, Pest Manage. Sci., 2010, 66, 1298–1303 CrossRef CAS PubMed.
- R. Tundis, M. R. Loizzo and F. Menichini, Crit. Rev. Food Sci. Nutr., 2014, 54, 225–250 CrossRef CAS PubMed.
- V. Paritala, K. K. Chiruvella, C. Thammineni, R. G. Ghanta and A. Mohammed, Rev. Bras. Farmacogn., 2015, 25, 61–83 CrossRef CAS.
- R. Gualdani, M. M. Cavalluzzi, G. Lentini and S. Habtemariam, Molecules, 2016, 21, 1530 CrossRef PubMed.
- S. Z. Moghadamtousi, B. H. Goh, C. K. Chan, T. Shabab and H. A. Kadir, Molecules, 2013, 18, 10465–10483 CrossRef CAS PubMed.
- G. W. Wang, H. Z. Jin and W. D. Zhang, Phytochem. Rev., 2013, 12, 915–942 CrossRef CAS.
- J. A. Shilpi, S. Saha, S. L. Chong, L. Nahar, S. D. Sarker and K. Awang, Chem. Biodiversity, 2016, 13, 483–503 CAS.
- X. Fang, Y. T. Di and X. J. Hao, Curr. Org. Chem., 2011, 15, 1363–1391 CrossRef CAS.
- B. Heasley, Eur. J. Org. Chem., 2011, 19–46 CrossRef CAS.
- J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. Munro and M. R. Prinsep, Nat. Prod. Rep., 2013, 30, 237–323 RSC.
- J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. Munro and M. R. Prinsep, Nat. Prod. Rep., 2014, 31, 160–258 RSC.
- J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. Munro and M. R. Prinsep, Nat. Prod. Rep., 2015, 32, 116–211 RSC.
- J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. Munro and M. R. Prinsep, Nat. Prod. Rep., 2016, 33, 382–431 RSC.
- R. A. Hill and J. D. Connolly, Nat. Prod. Rep., 2013, 30, 1028–1065 RSC.
- R. A. Hill and J. D. Connolly, Nat. Prod. Rep., 2015, 32, 273–327 RSC.
- R. A. Hill and J. D. Connolly, Nat. Prod. Rep., 2017, 34, 90–122 RSC.
- M. P. Rodríguez-Rivera, E. Lugo-Cervantes, P. Winterhalter and G. Jerz, Food Chem., 2014, 158, 139–152 CrossRef PubMed.
- S. Haldar, P. B. Phapale, S. P. Kolet and H. V. Thulasiram, Anal. Methods, 2013, 5, 5386–5391 RSC.
- S. J. N. Tatsimo, J. D. D. Tamokou, M. Lamshoft, F. Mouafo, A. Lannang, P. Sarkar, P. Bag and M. Spiteller, Med. Chem. Res., 2015, 24, 1468–1479 CrossRef CAS.
- G. A. Castillo-Herrera, L. J. Farías-Álvarez, J. A. García-Fajardo, J. I. Delgado-Saucedo, A. M. Puebla-Pérez and E. Lugo-Cervantes, J. Supercrit. Fluids, 2015, 101, 81–86 CrossRef CAS.
- J. A. M. de Paula, L. F. Brito, K. L. F. N. Caetano, M. C. de Morais Rodrigues, L. L. Borges and E. C. da Conceicao, Talanta, 2016, 149, 77–84 CrossRef CAS PubMed.
- K. Rangiah, B. A. Varalaxmi and M. Gowda, Anal. Methods, 2016, 8, 2020–2031 RSC.
- C. P. Liu, J. B. Xu, Y. S. Han, M. A. Wainberg and J. M. Yue, Org. Lett., 2014, 16, 5478–5481 CrossRef CAS PubMed.
- K. L. Ji, D. H. Cao, X. F. Li, J. Guo, P. Zhang and Y. K. Xu, Phytochem. Lett., 2015, 14, 234–238 CrossRef CAS.
- F. L. An, J. Luo, X. B. Wang, M. H. Yang and L. Y. Kong, Org. Biomol. Chem., 2016, 14, 1231–1235 CAS.
- F. L. An, J. Luo, R. J. Li, J. G. Luo, X. B. Wang, M. H. Yang, L. Yang, H. Q. Yao, H. B. Sun, Y. J. Chen and L. Y. Kong, Org. Lett., 2016, 18, 1924–1927 CrossRef CAS PubMed.
- T. T. Armelle, N. K. Pamela, M. Pierre, I. B. Muller, K. Marat, G. Sass and N. A. Ephrem, Med. Chem., 2016, 12, 1–7 CrossRef.
- N. Lange, A. T. Tontsa, C. Wegscheid, P. Mkounga, A. E. Nkengfack, C. Loscher, G. Sass and G. Tiegs, PLoS One, 2016, 11, e0160843 Search PubMed.
- S. B. Liu, W. L. Mei, H. Q. Chen, Z. K. Guo, H. F. Dai and Z. N. Wang, Molecules, 2016, 21, 1152 CrossRef PubMed.
- K. L. Ji, P. Zhang, X. N. Li, J. Guo, H. B. Hu, C. F. Xiao, X. Q. Xie and Y. K. Xu, Phytochemistry, 2015, 118, 61–67 CrossRef CAS PubMed.
- L. C. Chen, H. R. Liao, P. Y. Chen, W. L. Kuo, T. H. Chang, P. J. Sung, Z. H. Wen and J. J. Chen, Molecules, 2015, 20, 18551–18564 CrossRef CAS PubMed.
- Y. B. Cheng, Y. T. Chien, J. C. Lee, C. K. Tseng, H. C. Wang, I. W. Lo, Y. H. Wu, S. Y. Wang, Y. C. Wu and F. R. Chang, J. Nat. Prod., 2014, 77, 2367–2374 CrossRef CAS PubMed.
- W. M. Zhang, J. Q. Liu, Y. Y. Deng, J. J. Xia, Z. R. Zhang, Z. R. Li and M. H. Qiu, Nat. Prod. Bioprospect., 2014, 4, 53–57 CrossRef CAS PubMed.
- B. Siva, B. Poornima, A. Venkanna, K. R. Prasad, B. Sridhar, V. L. Nayak, S. Ramakrishna and K. S. Babu, Phytochemistry, 2014, 98, 174–182 CrossRef CAS PubMed.
- X. Y. Wang, C. M. Yuan, G. H. Tang, T. Zou, F. Guo, J. H. Liao, H. Y. Zhang, G. Y. Zuo, G. X. Rao, Q. Zhao, X. J. Hao and H. P. He, J. Asian Nat. Prod. Res., 2014, 16, 795–799 CrossRef CAS PubMed.
- L. R. Fu, Q. Y. Ma, S. Z. Huang, H. F. Dai, Z. K. Guo, Z. F. Yu and Y. X. Zhao, J. Asian Nat. Prod. Res., 2014, 16, 1054–1059 CrossRef CAS PubMed.
- J. H. Yu, Q. F. Liu, L. Sheng, G. C. Wang, J. Li and J. M. Yue, Org. Lett., 2016, 18, 444–447 CrossRef CAS PubMed.
- J. H. Yu, G. C. Wang, Y. S. Han, Y. Wu, M. A. Wainberg and J. M. Yue, J. Nat. Prod., 2015, 78, 1243–1252 CrossRef CAS PubMed.
- F. Zhang, C. R. Zhang, X. Tao, J. Wang, W. S. Chen and J. M. Yue, Bioorg. Med. Chem. Lett., 2014, 24, 3791–3796 CrossRef CAS PubMed.
- H. L. Liu, X. L. Chen, W. Xiao and Y. W. Guo, Helv. Chim. Acta, 2014, 97, 1445–1451 CrossRef CAS.
- H. Li, J. Luo and L. Y. Kong, Rec. Nat. Prod., 2015, 9, 190–195 Search PubMed.
- W. X. Liu, D. Z. Chen, J. Y. Ding, X. J. Hao and S. L. Li, Helv. Chim. Acta, 2015, 98, 1403–1410 CrossRef CAS.
- J. L. Peng, J. Wang, F. D. Kong, Z. Q. Liu, P. Wang, C. J. Gai, B. Jiang, W. L. Mei and H. F. Dai, Molecules, 2016, 21, 58 CrossRef PubMed.
- J. L. Peng, J. Wang, W. L. Mei, F. D. Kong, Z. Q. Liu, P. Wang, C. J. Gai, B. Jiang and H. F. Dai, J. Asian Nat. Prod. Res., 2016, 18, 629–636 CrossRef CAS PubMed.
- J. L. Peng, J. Wang, F. D. Kong, Z. Q. Liu, P. Wang, C. J. Gai, B. Jiang, W. L. Mei and H. F. Dai, Phytochem. Lett., 2016, 15, 230–233 CrossRef CAS.
- L. Yi, H. Zhang, X. Tian, J. Luo, J. Luo and L. Kong, Phytochem. Lett., 2017, 19, 12–17 CrossRef CAS.
- K. L. Ji, P. Zhang, H. B. Hu, S. Hua, S. G. Liao and Y. K. Xu, J. Nat. Prod., 2014, 77, 1764–1769 CrossRef CAS PubMed.
- M. L. Han, Y. Shen, Y. Leng, H. Zhang and J. M. Yue, RSC Adv., 2014, 4, 19150–19158 RSC.
- G. C. Wang, J. H. Yu, Y. Shen, Y. Leng, H. Zhang and J. M. Yue, J. Nat. Prod., 2016, 79, 899–906 CrossRef CAS PubMed.
- K. L. Ji, X. N. Li, S. G. Liao, H. B. Hu, R. Li and Y. K. Xu, Phytochem. Lett., 2016, 15, 53–56 CrossRef CAS.
- J. J. Xia, X. Y. Li, S. Z. Zhang, J. Q. Liu, W. M. Zhang, Y. X. Yan, Z. T. Ding and M. H. Qiu, Tetrahedron Lett., 2014, 55, 2104–2106 CrossRef CAS.
- (a) M. S. Yang, S. M. Hu, L. Y. Kong and J. Luo, Tetrahedron, 2015, 71, 8472–8477 CrossRef CAS; (b) C. P. Liu, G. C. Wang, L. S. Gan, C. H. Xu, Q. F. Liu, J. Ding and J. M. Yue, Org. Lett., 2016, 18, 2894–2897 CrossRef CAS PubMed.
- J. Hu, Y. Song, X. Mao, Z. J. Wang and Q. J. Zhao, J. Funct. Foods, 2016, 20, 1–9 CrossRef CAS.
- J. H. Li, Y. Li, F. L. An, M. M. Zhou, J. Luo, K. L. Jian, J. Luo and L. Y. Kong, Tetrahedron, 2016, 72, 7481–7487 CrossRef CAS.
- Q. Q. Meng, X. R. Peng, S. Y. Lu, L. S. Wan, X. Wang, J. R. Dong, R. Chu, L. Zhou, X. N. Li and M. H. Qiu, Nat. Prod. Bioprospect., 2016, 6, 239–245 CrossRef CAS PubMed.
- T. Inoue, Y. Matsui, T. Kikuchi, Y. In, O. Muraoka, T. Yamada and R. Tanaka, Fitoterapia, 2014, 96, 56–64 CrossRef CAS PubMed.
- Y. Matsui, T. Kikuchi, T. Inoue, O. Muraoka, T. Yamada and R. Tanaka, Molecules, 2014, 19, 17130–17140 CrossRef PubMed.
- T. Miyake, S. Ishimoto, N. Ishimatsu, K. Higuchi, K. Minoura, T. Kikuchi, T. Yamada, O. Muraoka and R. Tanaka, Molecules, 2015, 20, 20955–20966 CrossRef CAS PubMed.
- T. Inoue, Y. Matsui, T. Kikuchi, T. Yamada, Y. In, O. Muraoka, C. Sakai, K. Ninomiya, T. Morikawa and R. Tanaka, Tetrahedron, 2015, 71, 2753–2760 CrossRef CAS.
- A. Sakamoto, Y. Tanaka, T. Yamada, T. Kikuchi, O. Muraoka, K. Ninomiya, T. Morikawa and R. Tanaka, Fitoterapia, 2015, 100, 81–87 CrossRef CAS PubMed.
- Y. Yan, J. X. Zhang, T. Huang, X. Y. Mao, W. Gu, H. P. He, Y. T. Di, S. L. Li, D. Z. Chen and Y. Zhang, J. Nat. Prod., 2015, 78, 811–821 CrossRef CAS PubMed.
- Y. Yan, C. M. Yuan, Y. T. Di, T. Huang, Y. M. Fan, Y. Ma, J. X. Zhang and X. J. Hao, Fitoterapia, 2015, 107, 29–35 CrossRef CAS PubMed.
- K. L. Ji, S. G. Liao, X. L. Zheng, Z. Na, H. B. Hu, P. Zhang and Y. K. Xu, Molecules, 2014, 19, 3004–3011 CrossRef PubMed.
- W. B. Wu, H. Zhang, H. C. Liu, S. H. Dong, Y. Wu, J. Ding and J. M. Yue, Tetrahedron, 2014, 70, 3570–3575 CrossRef CAS.
- Y. Li, Q. P. Lu, J. Luo, J. S. Wang, X. B. Wang, M. D. Zhu and L. Y. Kong, Chem. Pharm. Bull., 2015, 63, 305–310 CrossRef PubMed.
- X. Tian, H. Li, F. An, R. Li, M. Zhou, M. Yang, L. Kong and J. Luo, Planta Med., 2017, 83, 341–350 CAS.
- Y. B. Wu, D. Liu, P. Y. Liu, X. M. Yang, M. Liao, N. N. Lu, F. Sauriol, Y. C. Gu, Q. W. Shi, H. Kiyota and M. Dong, Helv. Chim. Acta, 2015, 98, 691–698 CrossRef CAS.
- W. Li, Z. Jiang, L. Shen, P. Pedpradab, T. Bruhn, J. Wu and G. Bringmann, J. Nat. Prod., 2015, 78, 1570–1578 CrossRef CAS PubMed.
- Y. G. Dai, W. S. Li, P. Pedpradab, J. Liu, J. Wu and L. Shen, RSC Adv., 2016, 6, 85978–85984 RSC.
- W. S. Li, L. Shen, T. Bruhn, P. Pedpradab, J. Wu and G. Bringmann, Chem.–Eur. J., 2016, 22, 11719–11727 CrossRef CAS PubMed.
- Y. Zhang, J. S. Wang, Y. C. Gu, X. B. Wang and L. Y. Kong, Tetrahedron, 2014, 70, 6594–6606 CrossRef CAS.
- Y. Zhang, J. S. Wang, Y. C. Gu and L. Y. Kong, Helv. Chim. Acta, 2014, 97, 1354–1364 CrossRef CAS.
- J. Y. Cai, D. Z. Chen, S. H. Luo, N. C. Kong, Y. Zhang, Y. T. Di, Q. Zhang, J. Hua, S. X. Jing, S. L. Li, S. H. Li, X. J. Hao and H. P. He, J. Nat. Prod., 2014, 77, 472–482 CrossRef CAS PubMed.
- X. Pan, M. Matsumoto, Y. Nishimoto, E. Ogihara, J. Zhang, M. Ukiya, H. Tokuda, K. Koike, M. Akihisa and T. Akihisa, Chem. Biodiversity, 2014, 11, 1121–1139 CAS.
- X. Pan, M. Matsumoto, Y. Nakamura, T. Kikuchi, J. Zhang, M. Ukiya, T. Suzuki, K. Koike, R. Akihisa and T. Akihisa, Chem. Biodiversity, 2014, 11, 987–1000 CAS.
- Q. Jin, C. Lee, J. W. Lee, J. Y. Choi, J. T. Hong, Y. S. Kim, M. Lee and B. Y. Hwang, Helv. Chim. Acta, 2014, 97, 1152–1157 CrossRef CAS.
- Q. Zhang, Q. H. Zheng, J. Y. Liang, Q. S. Li and Z. D. Min, Chin. J. Nat. Med., 2016, 14, 692–696 Search PubMed.
- Q. Zhang, Y. G. Zhang, Q. S. Li and Z. D. Min, Helv. Chim. Acta, 2016, 99, 462–465 CrossRef CAS.
- D. G. Katja, K. Farabi, V. A. Nuraini, N. Nurlelasari, A. T. Hidayat, T. Mayanti, D. Harneti and U. Supratman, Int. J. Chem., 2016, 8, 30–34 CrossRef CAS.
- T. Fossen, A. Yahorau, S. Yahorava, F. Raharinjato, S. Razafimahefa, P. Rasoanaivo and J. E. S. Wikberg, Planta Med., 2016, 82, 1087–1095 CrossRef CAS PubMed.
- S. Razafimahefa, F. Mutulis, I. Mutule, E. Liepinsh, M. Dambrova, H. Cirule, B. Svalbe, S. Yahorava, A. Yahorau, B. Rasolondratovo, P. Rasoanaivo and J. E. Wikberg, Planta Med., 2014, 80, 306–314 CrossRef CAS PubMed.
- (a) W. Y. Zhang, F. L. An, M. M. Zhou, M. H. Chen, K. L. Jian, O. Quasie, M. H. Yang, J. Luo and L. Y. Kong, RSC Adv., 2016, 6, 97160–97171 RSC; (b) J. Luo, X. Tian, H. Zhang, M. Zhou, J. Li and L. Kong, Tetrahedron Lett., 2016, 57, 5334–5337 CrossRef CAS.
- V. G. P. Severino, S. D. L. de Freitas, P. A. C. Braga, M. R. Forim, M. F. G. F. da Silva, J. B. Fernandes, P. C. Vieira and T. Venancio, Molecules, 2014, 19, 12031–12047 CrossRef PubMed.
- L. L. Wang, C. S. Jiang, Y. Fu, F. F. Chen, L. F. Lan, H. Y. Zhang and Y. W. Guo, Helv. Chim. Acta, 2014, 97, 1301–1306 CrossRef CAS.
- J. B. Sun, N. Jiang, M. Y. Lv, P. Wang, F. G. Xu, J. Y. Liang and W. Qu, RSC Adv., 2015, 5, 24750–24757 RSC.
- J. B. Sun, B. Q. Tang, Q. Li, B. Wang, J. Y. Liang and L. Chen, Fitoterapia, 2016, 115, 92–95 CrossRef CAS PubMed.
- J. Q. Zhao, Y. M. Wang, H. T. Zhu, D. Wang, S. H. Li, R. R. Cheng, C. R. Yang, Y. F. Wang, M. Xu and Y. J. Zhang, Nat. Prod. Bioprospect., 2014, 4, 233–242 CrossRef CAS PubMed.
- X. H. Yan, P. Yi, P. Cao, S. Y. Yang, X. Fang, Y. Zhang, W. Bin, Y. Leng, Y. T. Di, Y. Lv and X. J. Hao, Sci. Rep., 2016, 6, 36927 CrossRef CAS PubMed.
- C. Lv, X. Yan, Q. Tu, Y. Di, C. Yuan, X. Fang, Y. Ben-David, L. Xia, J. Gong, Y. Shen, Z. Yang and X. Hao, Angew. Chem., Int. Ed., 2016, 55, 7539–7543 CrossRef CAS PubMed.
- J. M. Faber and C. M. Williams, Chem. Commun., 2011, 47, 2258–2260 RSC.
- J. M. Faber, W. A. Eger and C. M. Williams, J. Org. Chem., 2012, 77, 8913–8921 CrossRef CAS PubMed.
- S. Yamashita, A. Naruko, Y. Nakazawa, L. Zhao, Y. Hayashi and M. Hirama, Angew. Chem., Int. Ed., 2015, 54, 8538–8541 CrossRef CAS PubMed.
- S. Haldar, S. P. Kolet and H. V. Thulasiram, Green Chem., 2013, 15, 1311–1317 RSC.
- L. Grigorjeva, E. Liepinsh, S. Razafimahefa, A. Yahorau, S. Yahorava, P. Rasoanaivo, A. Jirgensons and J. E. S. Wikberg, J. Org. Chem., 2014, 79, 4148–4153 CrossRef CAS PubMed.
- H. Xu and J. L. Zhang, Bioorg. Med. Chem. Lett., 2011, 21, 1974–1977 CrossRef CAS PubMed.
- J. L. Zhang, H. Qu, X. Yu, X. Y. Zhi, H. Chen and H. Xu, Comb. Chem. High Throughput Screening, 2013, 16, 394–399 CrossRef CAS PubMed.
- Y. Yang, X. Wang, Q. Zhu, G. Gong, D. Luo, A. Jiang, L. Yang and Y. Xu, Bioorg. Med. Chem. Lett., 2014, 24, 1851–1855 CrossRef CAS PubMed.
- L. C. Tavares, T. S. Fernandes, V. Ilha, A. T. Neto, E. W. dos Santos, R. A. Burrow, F. A. Duarte, E. M. M. Flores, U. F. Silva, M. A. Mostardeiro and A. F. Morel, J. Braz. Chem. Soc., 2016, 27, 161–178 CAS.
- X. Yu, D. F. Shi, X. Y. Zhi, Q. Li, X. J. Yao and H. Xu, RSC Adv., 2015, 5, 31700–31707 RSC.
- X. Yu, G. D. Ding, Z. N. Gao, J. Zha and H. Xu, Heterocycles, 2015, 90, 1367–1374 CrossRef CAS.
- X. Yu, G. D. Ding, X. Y. Zhi and H. Xu, Bioorg. Med. Chem. Lett., 2015, 25, 25–29 CrossRef CAS PubMed.
- Y. Guo, Y. Y. Yan, C. Yang, X. Yu, X. Y. Zhi and H. Xu, Bioorg. Med. Chem. Lett., 2012, 22, 5384–5387 CrossRef CAS PubMed.
- Y. Guo, Y. Y. Yan, X. Yu, Y. Wang, X. Y. Zhi, Y. Hu and H. Xu, J. Agric. Food Chem., 2012, 60, 7016–7021 CrossRef CAS PubMed.
- Q. Li, X. B. Huang, S. C. Li, J. C. Ma, M. Lv and H. Xu, J. Agric. Food Chem., 2016, 64, 5472–5478 CrossRef CAS PubMed.
- Y. Guo, H. Qu, X. Zhi, X. Yu, C. Yang and H. Xu, J. Agric. Food Chem., 2013, 61, 11937–11944 CrossRef CAS PubMed.
- Y. Guo, R. G. Yang and H. Xu, Sci. Rep., 2016, 6, 35321 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2017 |



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: