Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optimization of culturing conditions for isolated Arthrobacter sp. ZXY-2, an effective atrazine-degrading and salt-adaptive bacterium†
Xinyue Zhaoab,
Li Wanga,
Linna Duc,
Jixian Yanga,
Jing Donga and
Fang Ma *a
*a
aState Key Laboratory of Urban Water Resource and Environment, School of Municipal and Environmental Engineering, Harbin Institute of Technology, Harbin 150090, China. E-mail: mafang@hit.edu.cn; Fax: +86-045186283008; Tel: +86-045186283008
bSection of Sanitary Engineering, Department of Water Management, Delft University of Technology, Delft, 2628CN, The Netherlands. E-mail: X.Zhao-3@tudelft.nl
cDepartment of Agriculture and Biotechnology, Wenzhou Vocational College of Science and Technology, Wenzhou 325006, China
First published on 30th June 2017
Abstract
The increasing salinity in aquatic environments has had a negative impact on the biodegradation of atrazine, an extensively used herbicide which has been proven to pollute soil and water ecosystems. In the present study, a novel atrazine-degrading strain (ZXY-2) was isolated from industrial wastewater and identified as the Arthrobacter genus with the 16S rRNA gene. Results indicated that the strain showed a high salinity tolerance, and was able to tolerate NaCl concentrations up to 10% (w/w). Plackett–Burman (PB) multifactorial design and response surface methodology (RSM) were then employed to optimize the culturing conditions. Results showed that among the selected fifteen factors, six contributing factors were obtained. Subsequently, by employing the RSM to model and optimize atrazine degradation, a biodegradation efficiency of 12.73 mg L−1 h−1 was reached under optimal conditions (34.04 °C, pH 9.0, inoculum size 10% (v/v), 2.212 g L−1 of sucrose, 6 g L−1 of Na2HPO4·12H2O, and 50 mg L−1 of atrazine). In addition, a statistically quadratic polynomial mathematical model was suggested (R2 = 0.9873). In contrast to other atrazine-degrading bacteria, ZXY-2 appears to be adapted to life under high salinity conditions and sustains excellent atrazine degradation performance. Therefore it could potentially be applied in atrazine bioremediation.
1. Introduction
Atrazine, an environmentally ubiquitous organic herbicide, has been extensively used to control a variety of broadleaf and grass weeds in agriculture and forestry.1,2 Atrazine has been reported to be a possible human carcinogen and is a suspected endocrine disrupter. It is commonly detected in soils, surface water, and ground water near where it is used.3,4 Moreover, atrazine has been classified as a major toxicological contaminant, causing increasing concern regarding its presence in the environment. It is therefore critical that improved methods to remove atrazine from the environment are found.The fate of atrazine in contaminated sites largely depends on microbial metabolism.5 Bioaugmentation using atrazine degraders could be an attractive and cost-effective method. However, since ground water salinity has been increasing due to climate change in recent years, the increasing salinity has led to a low biodegradability of atrazine.6 This necessitates the isolation of atrazine-degrading microorganisms with salt adaptation. Various atrazine-degrading strains, belonging to diverse genera, have been reported to metabolize atrazine as a sole source of carbon and energy (e.g., Pseudomonas, Acinetobacter, Rhodococcus, Arthrobacter, Bacillus, and Variovorax2,4,7–10). Although these reported strains seem to show varying abilities to degrade atrazine, with the highest degradation efficiency of 19.03 mg L−1 h−1, no salt-adaptive capacity has been identified. To the best of our knowledge, only Pseudomonas sp. ADP and Ochrobactrum oryzae have been reported to be capable of degrading atrazine under different salinity conditions; they achieved limited atrazine-degrading efficiencies of 1.04 mg L−1 h−1 and 0.04 mg L−1 h−1, respectively.6,11 Therefore, it is essential to isolate an atrazine-degrading strain with dual capacities of removing atrazine effectively and tolerating salinity efficiently.
Numerous studies show that microbial growth and biodegradation efficiency are influenced by the various media-components (e.g., C sources and N sources) and growth conditions (e.g., temperature, pH, and shaking speed).4,12,13 For example, a low level of nitrate has no effect on atrazine degradation, while high levels of nitrate would inhibit the removal process;14 and sucrose promotes atrazine degradation by Arthrobacter sp. strain DAT1, while it inhibits atrazine degradation by Arthrobacter sp. strain HB-5.13,15 Various media-component factors influence the growth of atrazine-degrading strains in different ways. To better understand the relationship between a strain and its growth environment, it is necessary to identify which factors are more influential and to adopt an effective methodology to separate these critical factors from all other factors. Additionally, it is likely that the effects of these factors are interrelated rather than independent. Therefore, investigating the combined effect of these factors is significant for understanding atrazine biodegradation. Considering these factors that vary simultaneously, the statistical Plackett–Burman (PB) design and the central composite design (CCD) based on response surface methodology (RSM) are effective methods for optimization of growth conditions for the biodegradation of atrazine. The PB design provides a way to screen for operational parameters that have significant effects on atrazine degradation.16–18 RSM is the statistical technique applied to seek the optimal conditions and understand the interactive effects on the atrazine biodegradation process.19,20
In the present study, an atrazine-degrading strain with effective atrazine-degrading abilities and high salinity tolerance was newly isolated. The culturing conditions that had a significant effect on atrazine degradation were then determined using the PB design. Following, CCD was used to identify the optimal culturing conditions for achieving enhanced atrazine removal efficiency. The combined effect of different environmental factors on atrazine degradation was also investigated.
2. Experimental
2.1 Isolation and enrichment
Industrial wastewater and soil samples were collected from Jilin Pesticide Plant, China. The enrichment medium consisted of mineral salts medium (MSM), 100 mg L−1 atrazine as the sole nitrogen source, and 3 mg L−1 sucrose as the carbon source. It was autoclaved at 121 °C for 15 minutes. The MSM contained (per liter) 0.9 g of KH2PO4, 6.5 g of Na2HPO4·12H2O, 0.15 g of MgSO4·7H2O, 0.01 g of FeSO4·7H2O and 2 mL of trace element solution. The trace element solution contained (per liter) 0.1 g of CoCl2·6H2O, 0.425 g of MnCl2·4H2O, 0.05 g of ZnCl2, 0.01 g of NiCl2·6H2O, 0.015 g of CuSO4·5H2O, 0.01 g of Na2MoO4·2H2O and 0.01 g of Na2SeO4·2H2O. The pH of the medium was adjusted to 7.0 with 1 M NaOH solution. Enrichments were carried out at 30 °C and 150 rpm for seven days. The partially enriched biomass was cultivated in fresh medium for another seven days and then repeated for three times. After one month, the enrichment cultures were plated onto agar plates. Colonies forming a clear zone were identified as possible atrazine-degrading isolates. An isolate named ZXY-2 showed high atrazine biodegradation ability and was selected for further study.2.2 Cloning and sequencing of 16S rRNA sequence
Bacterial DNA was extracted from ZXY-2 using a genomic DNA extraction kit (Sangon, China) following the manufacturer's instructions. Polymerase chain reaction (PCR) was used to amplify the 16S rRNA gene from total bacterial DNA. The PCR conditions consisted of an initial denaturation at 94 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1.5 min, and a final 5 min extension at 72 °C. PCR products were sequenced by the TaKaRa Biotechnology Company (Dalian, China). The sequence was compared with those available in the NCBI database using the BLAST program.2.3 Incubation of Arthrobacter sp. ZXY-2 under different salinities
10 mL of the enrichment cultures after the log phase were inoculated into flasks with 90 mL sterilized MSM medium. Each flask was supplemented with 100 mg L−1 atrazine and 0–10% NaCl (w/w). Samples were withdrawn each hour for analysis of atrazine concentration and cell growth.2.4 Plackett–Burman design for factors screening
The PB design was introduced as the first optimization step to identify the culture factors that had significant effects on atrazine degradation by strain ZXY-2. The factors tested included temperature, pH, shaking speed, inoculum size, glucose, sucrose, sodium citrate, KNO3, NH4Cl, isopropylamine, KH2PO4, Na2HPO4·12H2O, FeSO4·7H2O, MgSO4·7H2O, and atrazine concentrations. The independent variables were converted to coded values for computational convenience: the upper limit of a factor was coded as +1, the lower limit as −1, and the center level as 0 (Table 1). Fifty-four runs were performed according to the experimental layout (Table S1†). All the runs were carried out in triplicate, and the average atrazine degradation for each run was used as the response. Atrazine degradation percentages were assessed after 8 hours of incubation. Minitab 17.1 (Minitab Inc., State College, PA, USA) was employed for statistical analysis of the PB design. Regression analysis determined the factors that had a significant (p < 0.01) effect on atrazine degradation, and these factors were evaluated in further optimization experiments.| No. | Factors | +1 level | −1 level | Center level | Effect | T-Value | p-Value |
|---|---|---|---|---|---|---|---|
| a Note: R-Sq = 96.06%; R-Sq (adj.) = 94.35%.b Significant at “p-value” less than 0.01. | |||||||
| X1 | Temperature (°C) | 40 | 20 | 30 | 13.192 | 7.85 | <0.0001b |
| X2 | pH | 9 | 5 | 7 | 12.058 | 7.18 | <0.0001b |
| X3 | Shaking speed (rpm) | 200 | 100 | 150 | −4.067 | −2.42 | 0.021 |
| X4 | Inoculum size (% (v/v)) | 10 | 1 | 5.5 | 37.008 | 22.03 | <0.0001b |
| X5 | Glucose (g L−1) | 2 | 0 | 1 | −6.567 | −3.91 | <0.0001b |
| X6 | Sucrose (g L−1) | 2 | 0 | 1 | 7.992 | 4.76 | <0.0001b |
| X7 | Sodium citrate (g L−1) | 2 | 0 | 1 | −0.350 | −0.21 | 0.836 |
| X8 | KNO3 (g L−1) | 0.5 | 0 | 0.25 | −2.158 | −1.28 | 0.207 |
| X9 | NH4Cl (g L−1) | 0.5 | 0 | 0.25 | −6.658 | −3.96 | <0.0001b |
| X10 | Isopropylamine (mL L−1) | 5 | 0 | 2.5 | −0.758 | −0.45 | 0.654 |
| X11 | KH2PO4 (g L−1) | 1.5 | 0.5 | 1 | 4.067 | 2.42 | 0.021 |
| X12 | Na2HPO4·12H2O (g L−1) | 12 | 2 | 7 | 20.500 | 12.20 | <0.0001b |
| X13 | FeSO4·7H2O (g L−1) | 0.02 | 0 | 0.01 | 2.975 | 1.77 | 0.085 |
| X14 | MgSO4·7H2O (g L−1) | 0.2 | 0 | 0.1 | −2.358 | −1.40 | 0.169 |
| X15 | Atrazine concentration (mg L−1) | 150 | 50 | 100 | −15.192 | −9.04 | <0.0001b |
2.5 Response surface method for optimizing the biodegradation ability
RSM based on CCD was used to further optimize the culturing conditions. In the present work, fifty-four experimental runs with ten replicates at the center point were designed for the selected six of the eight variables found to be significant in the PB design. The specified ranges of the variables were as follows: temperature 30–40 °C, pH 7–9, inoculum size 5–10% (v/v), sucrose 1–3 g L−1, Na2HPO4·12H2O 6–12 g L−1 and atrazine concentration 50–100 mg L−1. The analyzed response was the atrazine degradation after 3 hours of incubation. All experiments were performed in triplicate and their response and predicted values were given in Table S2.†The experimental data was further analyzed by assuming a second order polynomial with linear, quadratic and interaction effects as shown in eqn (1):
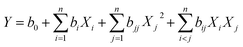 | (1) |
Minitab 17.1 (Minitab Inc., State College, PA, USA) was used to run an analysis of variance (ANOVA), as well as regression and graphical analyses.
2.6 Analytical methods
The atrazine was extracted from the growth medium samples with dichloromethane and then filtered using a sterile filter with 0.22 μm pore size (Sartorius Stedim, Germany). The analysis was performed by high-performance liquid chromatography (HPLC, Shimadzu, Japan) equipped with a C18 column (length 25 cm, internal diameter 4.6 mm; Varian) and a UV detector at 220 nm wavelength. The mobile phase consisted of acetonitrile/water (6/4, v/v) at a flow rate of 1.0 mL min−1. Samples were injected with a constant injection volume of 20 μL. Atrazine concentration was calculated by comparison to a standard curve.3. Results and discussion
3.1 Isolation and characterization of the atrazine-degrading strain ZXY-2
After four rounds of enrichment, a total of fourteen pure isolates were obtained. Among these isolates, a strain designated as ZXY-2 was selected as it completely degraded 100 mg L−1 atrazine within 14 h (7.14 mg L−1 h−1) and 50 mg L−1 atrazine within 8 h (6.25 mg L−1 h−1). The colonies of strain ZXY-2 were yellowish, wet, circular with semitransparent, and smooth surfaces on MSM agar plates. The 16S rRNA sequences exhibited a high sequence similarity with the members of the genus Arthrobacter and had maximum similarity with Arthrobacter TBD185 (99%). The sequence was deposited in GenBank under accession number KT778822 and strain ZXY-2 was deposited in China General Microbiological Culture Collection Center (CGMCC) with the deposited number CGMCC no. 10937.3.2 Degradation and growth of ZXY-2 at different salinities
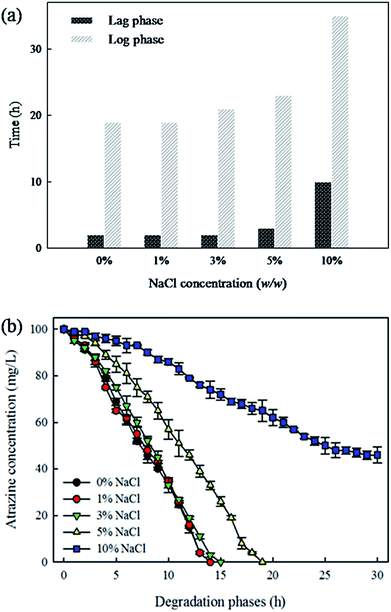
Fig. 1a shows the growth of ZXY-2 grown at a series of salinities: 0%, 1%, 3%, 5%, and 10%. The shortest lag phase (less than 1.5 h) was observed at the NaCl concentration of 0%, 1% and 3%. The lag phase of ZXY-2 was prolonged with increasing salinity. When the NaCl concentration reached 10%, the lag phase of strain ZXY-2 lasted for 10 h. As shown in Fig. 1b, more than 99% of atrazine was degraded with 20 h when the media were supplemented with 0–5% NaCl. Even though the concentration of NaCl reached 10%, strain ZXY-2 could still degrade up to 50% of atrazine after 30 h. There is limited information about salt-tolerance atrazine-degrading strains. To date, only Pseudomonas sp. ADP and Ochrobactrum oryzae have been reported to degrade atrazine under salinity conditions, and the two strains could only completely degrade atrazine at 4% and 1% salinities with efficiencies of 1.04 mg L−1 h−1 and 0.04 mg L−1 h−1, respectively.6,11 Therefore, compared with the reported isolates, strain ZXY-2 exhibited higher tolerance toward salts and better degradation efficiency.3.3 Plackett–Burman design analysis: critical factors for growth
Fifteen growth factors including temperature, pH, shaking speed, inoculum size, glucose, sucrose, sodium citrate, KNO3, NH4Cl, isopropylamine, KH2PO4, Na2HPO4·12H2O, FeSO4·7H2O, MgSO4·7H2O and atrazine concentrations were used to identify the significant effects on atrazine degradation. The experimental design and the values of atrazine degradation for the fifty-four runs are presented in Tables 1 and S1.† Degradation percentages of atrazine at different conditions ranged from 5.1% to 98.3%, indicating that the selected factors had significant effects on atrazine degradation. The regression analysis and p-values of the experimental factors are given in Table 1. The high values of R2 (0.9606) and adjusted R2 (0.9435) indicated that the observed values were in good fit with the model-predicted values. Additionally, setting a confidence level of 99% for factor screening, temperature (X1), pH (X2), inoculum size (X4), glucose (X5), sucrose (X6), NH4Cl (X9), Na2HPO4·12H2O (X12), and atrazine concentration (X15), were identified as the most influential factors on atrazine degradation.Previous studies have shown that additional nitrogen or carbon sources have different effects on atrazine degradation.12–14 Among the above significant factors, both glucose (as a carbon source) and NH4Cl (as a nitrogen source) had negative coefficients. However, sucrose was found to have a positive coefficient on atrazine degradation (Table 1), suggesting that a relatively high sucrose concentration was needed for atrazine degradation by strain ZXY-2. Sucrose supplementation might promote the growth of strain ZXY-2 and thus enhanced the atrazine degradation. This trend was consistent with previous study that: when atrazine was the sole carbon source for Arthrobacter sp. DAT1, degradation was slow whereas the addition of sucrose as supplemental carbon source resulted in higher degradation rate.13
In this study pH and temperature had significant positive effects within the experimental region with a lower pH and temperature value resulting in lower atrazine degradation. Higher ones might be beneficial for the bacterial growth, thus resulting in higher atrazine degradation. Previously, many studies had shown that the pH and temperature are the two important variables concerned and strongly influenced the degradation of toxic pollutants by microorganisms.21–23 Additionally, the relatively high pH (pH = 9) in this study might correspond to the optimal cell growth of the strain, as has been suggested for other strain in previous research.4
Among these significant factors (p < 0.01), the concentrations of glucose and NH4Cl were eliminated due to their negative effect on atrazine degradation. Hence, six factors (temperature, pH, inoculum size, sucrose, NaHPO4·12H2O and atrazine concentration) were chosen for further optimization by RSM.
3.4 Response surface methodology model analysis
The CCD design was employed for further optimization of atrazine degradation. Based on the results of the PB design, a six-factor CCD with fifty-four experimental runs was carried out (Table S2†). The observed degradation percentages varied from 21.55% to 72.25%. The analysis of variance (ANOVA) was used to test the fit of the model (Table 2). The ANOVA of the quadratic regression model demonstrated that the model was highly significant, as was evident from the F-test (F-value = 69.58) with a very low probability value (p < 0.0001). The adequacy of the model was indicated by the determination coefficient (R2 = 0.9873), which suggested that 98.73% of the variability in the response could be attributed to the independent variables. The value of the adjusted determination coefficient (adj. R2 = 0.9731) was also high, again supporting the significance of the model. The lack of fit test (p > 0.05) revealed that the quadratic model was statistically significant and could be used for further analysis.| Source | Coefficient | DF | SS | MS | F-Value | p-Value |
|---|---|---|---|---|---|---|
| a Note: R2 = 0.9873; coefficient of variation (CV) = 1.97; adj. R2 = 0.9731. DF = degrees of freedom; SS = sum of squares; MS = mean square. X1 = temperature; X2 = pH; X3 = inoculum size; X4 = sucrose; X5 = Na2HPO4·12H2O; X6 = atrazine concentration.b Significant at “p-value” less than 0.05.c Insignificant at “p-value” more than 0.05. | ||||||
| Model | 28 | 7560.13 | 270.00 | 69.58 | <0.0001b | |
| X1 | 0.479 | 1 | 7.80 | 7.80 | 2.01 | 0.169c |
| X2 | 2.811 | 1 | 268.64 | 268.64 | 69.23 | <0.0001b |
| X3 | 8.255 | 1 | 2316.77 | 2316.77 | 597.07 | <0.0001b |
| X4 | 2.235 | 1 | 169.88 | 169.88 | 43.78 | <0.0001b |
| X5 | 0.584 | 1 | 11.58 | 11.58 | 2.98 | 0.096c |
| X6 | −9.964 | 1 | 3375.25 | 3375.25 | 869.85 | <0.0001b |
| X12 | −3.21 | 1 | 31.16 | 31.16 | 6.30 | 0.019b |
| X22 | 1.58 | 1 | 51.88 | 51.88 | 1.52 | 0.229c |
| X32 | 2.53 | 1 | 32.25 | 32.25 | 3.92 | 0.059c |
| X42 | −2.53 | 1 | 4.45 | 4.45 | 3.92 | 0.059c |
| X52 | 1.34 | 1 | 10.66 | 10.66 | 1.10 | 0.304c |
| X62 | 3.94 | 1 | 36.89 | 36.89 | 9.51 | 0.005b |
| X1 × X2 | 0.320 | 1 | 3.28 | 3.28 | 0.84 | 0.367c |
| X1 × X3 | −1.020 | 1 | 33.29 | 33.29 | 8.58 | 0.007b |
| X1 × X4 | −1.427 | 1 | 65.15 | 65.15 | 16.79 | <0.0001b |
| X1 × X5 | −0.384 | 1 | 4.73 | 4.73 | 1.22 | 0.280c |
| X1 × X6 | 1.137 | 1 | 41.36 | 41.36 | 10.66 | 0.003b |
| X2 × X3 | 0.114 | 1 | 0.42 | 0.42 | 0.11 | 0.745c |
| X2 × X4 | 0.502 | 1 | 8.08 | 8.08 | 2.08 | 0.161c |
| X2 × X5 | −1.307 | 1 | 54.71 | 54.71 | 14.10 | 0.001b |
| X2 × X6 | 0.425 | 1 | 5.78 | 5.78 | 1.49 | 0.234c |
| X3 × X4 | −0.781 | 1 | 19.53 | 19.53 | 5.03 | 0.034b |
| X3 × X5 | −0.829 | 1 | 21.98 | 21.98 | 5.66 | 0.025b |
| X3 × X6 | 4.923 | 1 | 775.39 | 775.39 | 199.83 | <0.0001b |
| X4 × X5 | 0.248 | 1 | 1.97 | 1.97 | 0.51 | 0.483c |
| X4 × X6 | 0.948 | 1 | 28.77 | 28.77 | 7.41 | 0.012b |
| X5 × X6 | −0.707 | 1 | 15.99 | 15.99 | 4.12 | 0.053c |
| Residual error | 25 | 97.01 | 3.88 | |||
| Lack of fit | 17 | 78.52 | 4.62 | 2.00 | 0.161c | |
| Pure error | 8 | 18.49 | 2.31 | |||
| Total error | 53 | 7657.14 | ||||
Effects of the factors with linear, square, and interactive coefficients are shown in Table 2. The pH, inoculum size, sucrose, and atrazine concentrations all showed significant linear effects. The inoculum size and atrazine concentration had higher coefficient values, indicating a higher significant effect on the degradation of atrazine compared with the other factors. Inoculum size showed a positive effect on atrazine degradation, and atrazine concentration had a negative effect. When square terms were considered, only temperature and atrazine concentration were significant.
By applying multiple regression analysis, a predictive quadratic model was fitted with experimental results, and the following regression equation was obtained when only significant terms for atrazine degradation were included (eqn (2)):
| Y = 48.442 + 2.811X2 + 8.255X3 + 2.235X4 − 9.964X6 − 3.21X12 + 3.94X62 − 1.020X1X3 − 1.427X1X4 + 1.137X1X6 − 1.307X2X5 − 0.781X3X4 − 0.829X3X5 + 4.923X3X6 + 0.948X4X6 | (2) |
3.5 Effects of interactive factors
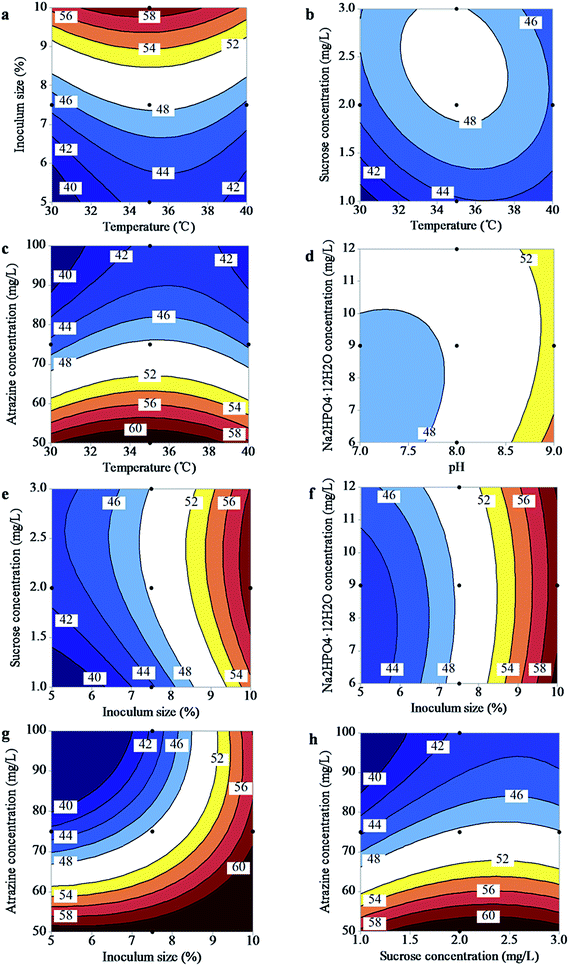
The RSM explained the effects of the different factors in a pronounced way as well as described the interactive effects between factors on atrazine degradation. With atrazine degradation as the response, the two dimensional response plots of significant mutual effects (see Table 2) with four factors kept at their central levels and the other two varying within the experimental ranges are shown in Fig. 2.The interactive effects between temperature and inoculum size, temperature and sucrose, temperature and atrazine concentration (Fig. 2a–c) indicated that the atrazine degradation was highly affected by temperature as compared to the other three factors. Similar contour plots were observed in Fig. 2e–g, where the pair-wise interaction of the two-dimensional contours showed significant interactive effects on atrazine degradation between inoculum size and sucrose concentration, inoculum size and Na2HPO4·12H2O concentration, and inoculum size and atrazine concentration.
From Fig. 2a, the increased inoculum size caused a considerable increase in atrazine-degrading efficiency. For instance, an atrazine degradation value of 56.43% was obtained at 30 °C and 10% (v/v) inoculum size, compared with that of 37.75% at 30 °C and 5% (v/v). Within the range tested, inoculum size could play a stimulative role in the consideration of atrazine degradation by ZXY-2. However, Mohana et al.24 reported that pollutant removal rate increased with inoculum size to its low condition. Hence, different inoculum size levels result in different degradation efficiency.
The elliptical contour plot in Fig. 2b indicates that the atrazine degradation rate increased with rising temperature and sucrose concentration to an optimal point, and then decreased with further increase in temperature and sucrose concentration. The maximum atrazine removal was observed in the range of temperature from 32.02 to 37.68 °C while sucrose concentration from 1.82 g L−1 to 3 g L−1. In Fig. 2d, Na2HPO4·12H2O concentration reached a plateau level response above the mid-range in its interaction with pH, indicating that the Na2HPO4·12H2O concentration had already reached an asymptotic value for atrazine degradation.
Fig. 2g showed that atrazine degradation rates were similar at all ranges of inoculum size at low atrazine concentration. However, the degradation rate increased with increasing inoculum size when the atrazine concentration was high, indicating that high inoculum size might have positive effects on atrazine removal. Furthermore, it must be highlighted that atrazine degradation changed from 31.98% at 100 mg L−1 atrazine concentration to 61.35% at 50 mg L−1 atrazine concentration when inoculum size was 5% (v/v). Higher atrazine concentrations led to longer lag phases on the microbial biodegradation.25 Therefore, higher atrazine concentration resulted in lower atrazine degradation at the fixed time of 3 h.
The interactive effects of atrazine concentration with temperature and sucrose concentration had a positive effect on the degradation of atrazine as shown in Fig. 2c and h. The contour plots show that at any given atrazine concentration the atrazine removal efficiency changed only slightly between low and high temperature and sucrose concentrations. However, the response value changed sharply (from 37.61% to 62.34% and 36.72% to 62.29%, respectively) as the atrazine concentration decreased. This indicated that atrazine concentration had a greater effect on the model output than either temperature or sucrose concentration in the current experimental ranges. Similarly, the atrazine concentration (X6) coefficient value was 4.5-fold higher than the sucrose concentration term (X4) and 20.8-fold higher than the temperature term (X1) (Table 2). These results support the suggestion that initial atrazine concentration may be one of the most critical factors in the biodegradation process of atrazine.1
3.6 Optimal atrazine removal and validation experiments
Based on the results of the RSM, the optimal culturing conditions of 34.04 °C, pH 9.0, 10% (v/v) inoculum size, 2.212 g L−1 sucrose, 6 g L−1 Na2HPO4·12H2O, and 50 mg L−1 atrazine predicted a maximum atrazine degradation of 74.65% after 3 h cultivation (12.73 mg L−1 h−1). Three replicate runs at optimal conditions were carried out to verify the accuracy of the quadratic model. Under optimal conditions, the observed removal value was 76.36% within 3 hours. This coincided with the predicted value, and the model was validated.3.7 Evaluation of the modified culturing conditions
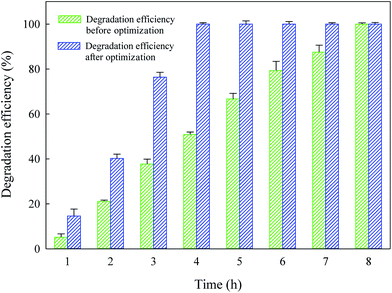
This study introduced modified culturing conditions to significantly enhance the biodegradation efficiency of atrazine under optimal conditions. Compared to MSM and other commonly used media with at least ten components, the optimized culture medium could generate a higher atrazine-degrading efficiency of 12.73 mg L−1 h−1 with only two components (sucrose and Na2HPO4·12H2O).4,15,26,27 Additionally, the optimal conditions established in this study permitted a high cell density of 2 × 108 CFU mL−1 to be reached within 3 h, a 28.6-fold increase as compared to that of the basal culturing conditions (see Section 2.1 Isolation and enrichment). To our knowledge, the cell density of 2 × 108 CFU mL−1 is the highest reported to date among the atrazine-degrading strains.8,14The degradation performance by strain ZXY-2 was compared before and after optimization of conditions (Fig. 3). In the optimized medium, the degradation efficiency reached 100% within 4 h, while it was 50.9% in the original mineral salts medium. Results revealed that strain ZXY-2 had an increased ability to biodegrade atrazine in a shorter period of time under the optimal conditions. As a consequence, the modified culture conditions will greatly improve atrazine biodegradation by strain ZXY-2.
4. Conclusions
This study isolated and identified a new atrazine-degrading Arthrobacter sp. ZXY-2 with high atrazine removal efficiency and high salinity tolerance. PB design and RSM were used to determine the significant factors influencing atrazine degradation in order to achieve maximum atrazine degradation. The most favorable conditions for biodegradation were 34.04 °C, pH 9.0, inoculum size of 10% (v/v), 2.212 g L−1 sucrose, 6 g L−1 Na2HPO4·12H2O, and 50 mg L−1 atrazine, providing a maximum atrazine degradation efficiency of 12.73 mg L−1 h−1. The optimal conditions were further validated and showed significant improvement in atrazine degradation. A more thorough investigation will focus on the genomics and transcriptomics in order to understand the highly-efficient and salt-tolerant mechanisms of ZXY-2. We propose that the present study be used as an application framework for atrazine bioremediation.Conflict of interest
None declared.Acknowledgements
This work was supported by the Major Science and Technology Program for Water Pollution Control and Treatment (2012ZX07201003).References
- E. Silva, A. M. Fialho, I. Sa-Correia, R. G. Burns and L. J. Shaw, Environ. Sci. Technol., 2004, 38, 632–637 CrossRef CAS PubMed.
- Fazlurrahman, M. Batra, J. Pandey, C. R. Suri and R. K. Jain, Lett. Appl. Microbiol., 2009, 49, 721–729 CrossRef CAS PubMed.
- F. Govantes, O. Porrua, V. Garcia-Gonzalez and E. Santero, Microb. Biotechnol., 2009, 2, 178–185 CrossRef CAS PubMed.
- J. Wang, L. Zhu, Q. Wang, J. Wang and H. Xie, PLoS One, 2014, 9, e107270 Search PubMed.
- M. S. B. M. L. Wackett, Appl. Microbiol. Biotechnol., 2002, 58, 39–45 CrossRef PubMed.
- M. A. Shir, M. Dehghani and M. R. Samaei, J. Health Sci. Surveillance Sys., 2016, 4, 121–128 Search PubMed.
- P. Singh, C. R. Suri and S. S. Cameotra, Biochem. Biophys. Res. Commun., 2004, 317, 697–702 CrossRef CAS PubMed.
- T. El Sebaï, M. Devers-Lamrani, F. Changey, N. Rouard and F. Martin-Laurent, Int. Biodeterior. Biodegrad., 2011, 65, 1249–1255 CrossRef.
- R. T. Mandelbaum, D. L. Allan and L. P. Wackett, Appl. Environ. Microbiol., 1995, 61, 1451–1457 CAS.
- X. Zhao, L. Wang, F. Ma, S. Bai, J. Yang and S. Qi, J. Environ. Sci., 2017, 54, 152–159 CrossRef PubMed.
- N. Shapir, R. T. Mandelbaum and H. Gottlieb, J. Ind. Microbiol. Biotechnol., 1998, 20, 153–159 CrossRef CAS.
- S. Xie, R. Wan, Z. Wang and Q. Wang, Environ. Sci. Pollut. Res., 2013, 20, 4078–4084 CrossRef CAS PubMed.
- Q. Wang and S. Xie, Int. Biodeterior. Biodegrad., 2012, 71, 61–65 CrossRef CAS.
- X. Zhou, Q. Wang, Z. Wang and S. Xie, Environ. Sci. Pollut. Res., 2013, 20, 2484–2491 CrossRef CAS PubMed.
- J. Wang, L. Zhu, A. Liu, T. Ma, Q. Wang, H. Xie, J. Wang, T. Jiang and R. Zhao, Environ. Geochem. Health, 2011, 33, 259–266 CrossRef CAS PubMed.
- L. Du, Y. Yang, G. Li, S. Wang, X. Jia and Y. Zhao, Int. Biodeterior. Biodegrad., 2010, 64, 566–573 CrossRef CAS.
- Y. Kong, P. Zou, L. Miao, J. Qi, L. Song, L. Zhu and X. Xu, Environ. Sci. Pollut. Res., 2014, 21, 5983–5990 CrossRef CAS PubMed.
- Y. Xiao, S. Chen, Y. Gao, W. Hu, M. Hu and G. Zhong, Appl. Microbiol. Biotechnol., 2015, 99, 2849–2859 CrossRef CAS PubMed.
- I. Amalraj Appavoo, J. Hu, Y. Huang, S. F. Li and S. L. Ong, Water Res., 2014, 57, 270–279 CrossRef CAS PubMed.
- C. A. Velis, C. Franco-Salinas, C. O'Sullivan, J. Najorka, A. R. Boccaccini and C. R. Cheeseman, Environ. Sci. Technol., 2014, 48, 7527–7535 CrossRef CAS PubMed.
- C. Zhang, L. Jia, S. Wang, J. Qu, K. Li, L. Xu, Y. Shi and Y. Yan, Bioresour. Technol., 2010, 101, 3423–3429 CrossRef CAS PubMed.
- Y. Yus Azila, M. D. Mashitah and S. Bhatia, Bioresour. Technol., 2008, 99, 8549–8552 CrossRef CAS PubMed.
- S. I. Mulla, H. Wang, Q. Sun, A. Hu and C. P. Yu, Sci. Rep., 2016, 6, 21965 CrossRef CAS PubMed.
- S. Mohana, S. Shrivastava, J. Divecha and D. Madamwar, Bioresour. Technol., 2008, 99, 562–569 CrossRef CAS PubMed.
- X. Peng and X. Jia, Bioresour. Technol., 2013, 148, 386–393 CrossRef CAS PubMed.
- Y. Zhang, Z. Jiang, B. Cao, M. Hu, Z. Wang and X. Dong, Int. Biodeterior. Biodegrad., 2011, 65, 1140–1144 CrossRef CAS.
- Q. Li, Y. Li, X. Zhu and B. Cai, J. Environ. Sci., 2008, 20, 1226–1230 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04661h |
| This journal is © The Royal Society of Chemistry 2017 |