Open Access Article
Open Access ArticleDouble conjugated nanogels for selective intracellular drug delivery†
Emanuele Mauria,
Pietro Veglianeseb,
Simonetta Papab,
Alessandro Marianic,
Massimiliano De Paolac,
Riccardo Rigamontia,
Giulia M. F. Chincarinia,
Irma Vismarab,
Stefano Rimondoa,
Alessandro Sacchetti‡
*a and
Filippo Rossi‡ *a
*a
aDipartimento di Chimica, Materiali e Ingegneria Chimica “Giulio Natta”, Politecnico di Milano, via Mancinelli 7, 20131 Milan, Italy. E-mail: alessandro.sacchetti@polimi.it; filippo.rossi@polimi.it; Fax: +39 0223993180; Tel: +39 0223993017 Tel: +39 0223993145
bDipartimento di Neuroscienze, IRCCS Istituto di Ricerche Farmacologiche “Mario Negri”, via La Masa 19, 20156 Milan, Italy
cDipartimento di Ambiente e Salute, IRCCS Istituto di Ricerche Farmacologiche “Mario Negri”, via La Masa 19, 20156 Milan, Italy
First published on 12th June 2017
Abstract
One of the most important drawbacks of nanomedicine is related to the unwanted rapid diffusion of drugs loaded within nanocarriers towards the external biological environment, according to the high clearance of body fluids. Therefore, colloids can carry only a small amount of their initial content in the target district, limiting their pharmacological activity and then the therapy. To overcome this limitation, we synthesized double conjugated nanogels: the first click strategy (1,3 Huisgen cycloaddition) guarantees the traceability of nanogels while the second one (disulfide bond) links drug molecules to polymeric chains. In this study, we proposed the above-mentioned double strategy and we validated the synthesized colloids and the selective release kinetics in microglia cells, dramatically involved in several diseases of the central nervous system. Cleavable linked drugs prove to be a promising tool for the selective administration of pharmacological compounds in microglia cells and potentially in many others counteracting some relevant events.
Introduction
Recent advances in polymer science have provided many innovations, underlining an increasing importance of polymeric nanocarriers as promising drug delivery tools.1–3 Indeed, thanks to their flexibility in terms of size, hydrophilic/lipophilic characteristics and surface functionalization, they offer relevant advantages in drug delivery by targeting molecules in specific cells and controlling drug release over time.4–7 In particular, one of the greatest advantages of nanocarriers resides in the fact that they can overcome some of the major limitations of conventional delivery systems, like drawbacks of both under- and over-dosing maintaining drug levels within a desired range for a long period.8–10 Moreover, several studies showed the intrinsic capacity of specific cells to internalize foreign nanobodies, which can be exploited for targeting drug delivery using nanodevices.11,12 In order to improve the selectivity of nanocarriers, specific targeting agents have been designed in recent years.13–15 These state-of-the-art systems are able to transport the loaded compounds specifically to the target cells and release their payload once there, following controlled or sustained kinetics.16–18 This approach, firstly used in cancer treatment, has now been extended to several other districts and diseases.19–22 Among them, in the last years a lot of attention was dedicated to nanosystems able to selectively deliver drugs in the damaged central nervous system using the high phagocytic activity of microglia.23,24Along these lines, Papa and coworkers developed, for example, poly-ε-caprolactone based nanoparticles able to efficiently modulate resident microglial cells reducing their pro-inflammatory response, maintaining a pro-regenerative milieu and ameliorating the behavioral outcome in mice up to 63 days post spinal cord injury.25 In addition, this novel pharmacological delivery by nanocarriers presents important improvements, namely a selective drug delivery and a maximized pharmacological activity (anti-inflammatory effect) compared to other administration routes such as direct parenchymal or intraperitoneal injection.
Cerqueira and coworkers proved that the intracellular internalization of high concentration of methylprednisolone-loaded CMCht/PAMAM dendrimer nanoparticles modulates the metabolic activity of microglia cells in vitro and in vivo.26 These strategies promised to be very efficient, but the main limitation resides in the fact that the drug loaded could be released during the transition to the target and the quantity of drug that can be delivered in the site is generally very low.27–29
To overcome these drawbacks, a promising strategy is represented by the use of cleavable bonds between polymer and drug molecules.30–32 Following this strategy, drug release is not driven by Fickian diffusion but it is delayed by the stability of the bond between drugs and polymer: the higher the stability, the slower the release rates.33,34 The most common strategy in order to functionalize polymers with drugs is represented by polymer post-polymerization modification also known as click strategies.30,35,36 Indeed, the presence of functional groups in polymerization reaction may completely prevent polymerization, modify polymer properties (molecular weight, polydispersity, crystallinity, etc.) or lead to side effects.
In this work we decided to functionalize nanogels (NGs) composed by polyethylene glycol (PEG) and polyethyleneimine (PEI) interconnections with a commonly used mimetic drug rhodamine B (RhB)37,38 (click 1) using disulfide bond that can be disrupted by glutathione present within cell cytosol.39,40 In order to be traceable, the nanogels were also functionalized with Cy5 dye (click 2) using Huisgen 1,3-cycloaddition. The synthesis, characterization and size of nanocarriers were evaluated together with their ability to release mimetic drug within microglia cells. This study represents a good proof of concept of the possibility to optimize dye and drug-conjunctions to polymers to guarantee traceability on one side and improve drug delivery performances on the other.
Experimental
Materials
Polyethyleneimine linear (PEI, MW = 2500 Da) was purchased from Polysciences Inc. (Warrington, USA), whereas polyethylene glycol 8000 (PEG, MW = 8000 Da), rhodamine, Cy5 dye and all other used chemicals were purchased from Sigma-Aldrich (Sigma Aldrich Chemie GmbH, Deisenhofen, Germany). The materials were used as received. Solvents were of analytical grade. Rhodamine derivatives were stored at 4 °C in dark. Synthetized products with Cy5 moiety were stored at −20 °C.Characterization and analyses
The NMR experiments were carried out on a Bruker AC (400 MHz) spectrometer using chloroform (CDCl3) or deuterium oxide (D2O) as solvent, and chemical shifts were reported as δ values in parts per million with respect to TMS as internal standard. Final sizes and ζ-potentials of nanogels were determined by dynamic light scattering (DLS) analysis using a Zetasizer Nano ZS from Malvern Instruments and the reported data are an average value of three measurements of the same sample, dissolved in distilled water. Atomic force microscopy (AFM) analysis was performed for the characterization of nanogel latexes using a NT-MDT Solver Pro instrument operating in non-contact mode with silicon tips. Samples were prepared by dropping latexes onto silicon substrate and drying. AFM images on 1 × 1 μm2 areas were recorded for the preliminary morphologic evaluation; 500 × 500 nm2 image were then cropped and height line profile performed for single gel. Surface morphology was evaluated by flattening of the images (first order) using NT-MDT software.Nanogel synthesis
Found: C, 74.08; H, 7.83; N, 11.52. Calc. for C30H38N4O2: C, 74.04; H, 7.87; N, 11.51%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate 9
ethyl acetate 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) resulting in a pale-yellow oil (60% yield), which was characterized by NMR analysis. 1H NMR (400 MHz, CDCl3): δ 4.20 (d, J = 2.0 Hz, 2H), 3.93–3.86 (t, 2H), 3.83–3.74 (t, 2H), 2.93 (t, J = 6.4 Hz, 2H), 2.89 (t, J = 5.9 Hz, 2H), 2.47 (t, J = 2.4 Hz, 1H), 2.04 (s, 1H).
1 v/v) resulting in a pale-yellow oil (60% yield), which was characterized by NMR analysis. 1H NMR (400 MHz, CDCl3): δ 4.20 (d, J = 2.0 Hz, 2H), 3.93–3.86 (t, 2H), 3.83–3.74 (t, 2H), 2.93 (t, J = 6.4 Hz, 2H), 2.89 (t, J = 5.9 Hz, 2H), 2.47 (t, J = 2.4 Hz, 1H), 2.04 (s, 1H).Found: C, 43.68; H, 6.30. Calc. for C7H12O2S2: C, 43.72; H, 6.29.
Found: C, 64.77; H, 6.89; N, 7.92. Calc. for C38H48N4O5S2: C, 64.74; H, 6.86; N, 7.95.
The final product was obtained as white solid dissolving the residue in excess of diethyl ether and collecting the formed precipitate by filtration under vacuum (97% yield). The epoxide moiety linked to the polymeric chain was evaluated by 1H-NMR analysis. 1H NMR (400 MHz, CDCl3) δ: 3.57 (s, 720H), 3.12–3.04 (m, 2H), 2.72 (dd, J = 5.1, 4.1 Hz, 2H), 2.54 (dd, J = 5.0, 2.7 Hz, 2H).
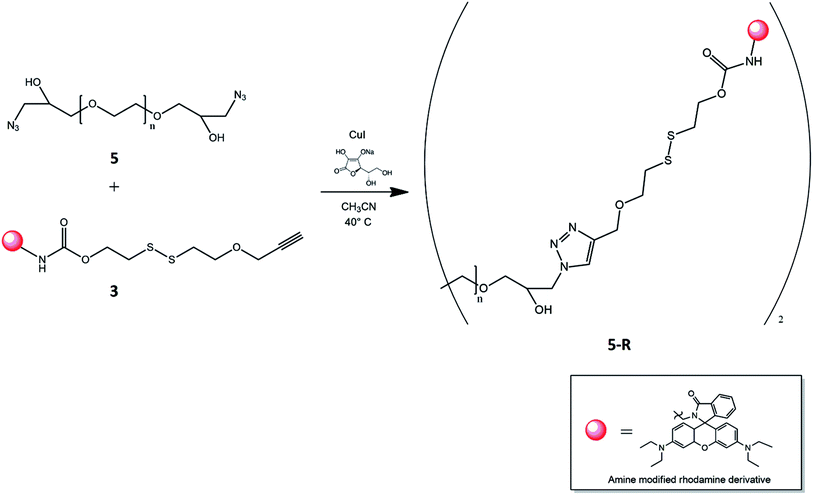
The resulting system was lyophilized, giving a purple solid. This intermediate was labelled as compound 5-R. Finally, the latter (30 mg, 4 μmol) was dissolved in acetonitrile (5 ml) and CDI (0.97 mg, 6 μmol) was added. The reaction mixture was left stirring at 40 °C for 70 h. Successively, the solvent was evaporated under vacuum and the residue was precipitated into diethyl ether. The formed solid was collected and dried by vacuum filtration and characterized by NMR analysis and gel permeation chromatography (ESI†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) propargyl bromide 1
propargyl bromide 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. The formation of triazole using click strategy was performed as follows: functionalized PEI (10 mg, 4 μmol) was dissolved in distilled water (2 ml) and a solution (200 μl, 0.4 μmol) of Cy5-azide in DMSO (2 mg ml−1) was added to the system. Copper iodide (0.02 mg, 0.2 μmol) and sodium ascorbate (0.02 mg, 0.2 μmol) were added to the mixture and the latter was heated to 50 °C for 24 h, under stirring. Then, the resulting system was dialyzed against distilled water (2 l), using dialysis membrane with Mw cutoff = 500 Da, for 2 days, with daily replacement of the aqueous media. The polymer was collected after lyophilization and 1H-NMR spectrum confirmed the polymer functionalization.
5. The formation of triazole using click strategy was performed as follows: functionalized PEI (10 mg, 4 μmol) was dissolved in distilled water (2 ml) and a solution (200 μl, 0.4 μmol) of Cy5-azide in DMSO (2 mg ml−1) was added to the system. Copper iodide (0.02 mg, 0.2 μmol) and sodium ascorbate (0.02 mg, 0.2 μmol) were added to the mixture and the latter was heated to 50 °C for 24 h, under stirring. Then, the resulting system was dialyzed against distilled water (2 l), using dialysis membrane with Mw cutoff = 500 Da, for 2 days, with daily replacement of the aqueous media. The polymer was collected after lyophilization and 1H-NMR spectrum confirmed the polymer functionalization.Nanogel formation
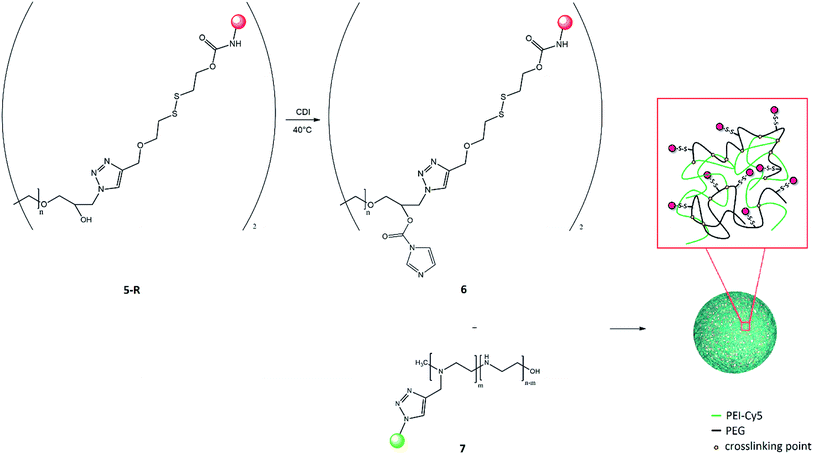
Nanogels were performed referring to the modified emulsification-evaporation method.46 Activated PEG-rhodamine 6 (28 mg, 2.88 μmol) was dissolved in CH2Cl2 (1 ml), whereas modified PEI-Cy5 7 (5 mg, 1.92 μmol) was added to distilled water (2 ml) until complete dissolution.The organic solution was added dropwise, under vigorous stirring, to the aqueous one and the resulting system was sonicated for 30 min. CH2Cl2 was evaporated and the mixture left stirring for 17 h at room temperature. Formed nanogels were purified by dialysis (membrane Mw cutoff = 3.5 kDa) against distilled water (2 l) and lyophilized. They were characterized by 1H-NMR and FT-IR spectra. Moreover, the diameter and the shape of these nanostructures were evaluated by DLS and AFM analyses.
Rhodamine release from nanogels
Drug release mechanism was investigated in two different release environments: the first one was a phosphate buffer saline solution (PBS) at pH 7.4, whereas the second one was a glutathione (GSH) solution 5 mM. Obtained nanogels (5 mg) were separately placed in excess of PBS and GSH solution (1 ml) and stored at 37 °C. At defined interval of time, for each sample, aliquots (3 × 100 μl) of the released media were collected for UV-vis measurements and substituted, respectively, with the same volume of fresh PBS or GSH solution, in order to avoid mass-transfer equilibrium with the surrounding release environment. The released amount of mimetic drug was spectrophotometrically measured at 570 nm and determined referring to the standard calibration curve for amine-rhodamine 1 (ESI†). The percentage of the released rhodamine was defined as the ratio of the released absolute amount in the aqueous media to the sum of the total amounts of the released and unreleased mimetic drug. All these experiments were repeated 3 times and the results were averaged.Biological evaluation: cell-mediated drug release
Cells obtained at this step were a mixed neuron/glia population and underwent centrifugation through a 6% iodixanol (OptiPrep™) cushion to separate large neurons from glial cells. The glial fraction was plated at a density of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 into 75 cm2 flasks, previously pre-coated with poly-L-lysine. Isolated microglia were obtained from flasks containing confluent mixed glial cultures after overnight shaking at 275 rpm in incubators. The supernatants (containing microglia) were collected and seeded at a density of 40
000 cells per cm2 into 75 cm2 flasks, previously pre-coated with poly-L-lysine. Isolated microglia were obtained from flasks containing confluent mixed glial cultures after overnight shaking at 275 rpm in incubators. The supernatants (containing microglia) were collected and seeded at a density of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 into 24-well plates. NGs (0.05% weight/volume) were then added to cell cultures up to 3 days.
000 cells per cm2 into 24-well plates. NGs (0.05% weight/volume) were then added to cell cultures up to 3 days.
Results and discussion
Chemical characterization
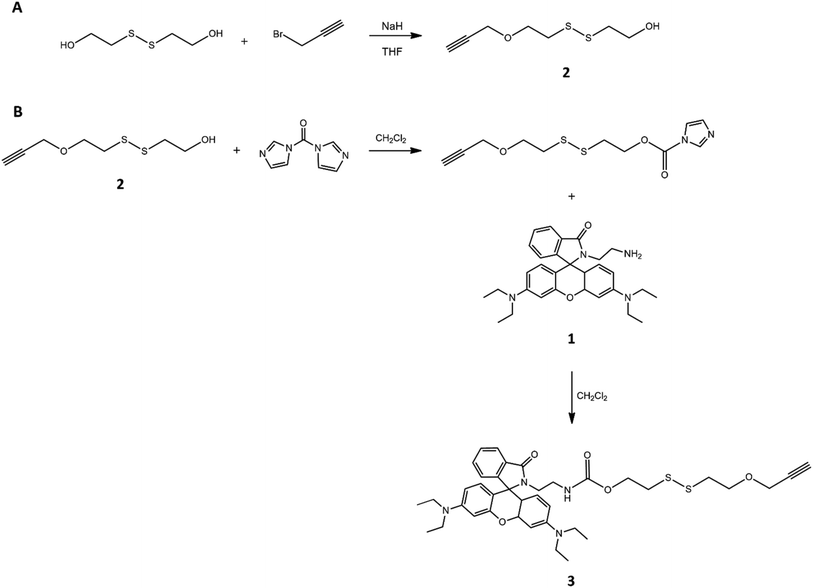 | ||
| Fig. 1 Steps of synthesis of disulfide derivative grafting on one side the terminal alkyne group (A) and on the other side rhodamine (B). | ||
In this way, the mimetic drug was linked to the S–S bridge structure, which in turn was grafted to a triple bond, needed for the formation of a stable and non-cleavable linkage with PEG. Obtaining the compound 3 necessarily involved the purification of the compound 2 (NMR spectrum visible in Fig. S1†) by chromatographic technique in order to separate the correct product carrying only one alkyne functionality and one –OH group.
Indeed, the hydroxyl groups of 2-hydroxyethyl disulfide showed the same reactivity, due to its molecular symmetry, and the foresight to use a molar ratio disulfide: propargyl between the reagents equal to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
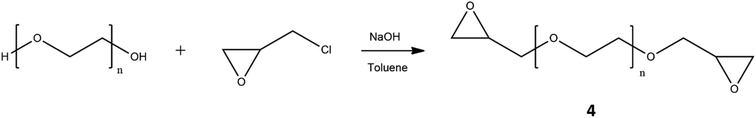
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, in order to potentially involve only one –OH group of each molecule in the reaction with the alkyne derivative, was not enough to produce the desired product because the formation of a mixture of mono-, bis-, or no functionalized system was obtained. The integral values of the characteristic signals of 2 in its 1H-NMR spectrum, confirmed the collection of the mono-functionalized product (ESI†). The introduction of triple bond was justified by the need to graft the sensitive drug-disulfide linker system to the polymeric chain using copper-catalyzed azide alkyne cycloaddition (CuAAC). The formed triazole ring possesses a number of desirable features reported in the medicinal chemistry and, in particular, is stable under standard physiological conditions, which suits well for gene and drug delivery system. The use of click chemistry required the functionalization of PEG with azide moiety, that was performed in two steps. The first one, as illustrated in Fig. 2, involved the preparation of the polymer with epoxide end groups, through reaction with epichlorohydrin. PEG functionalization was verified by 1H-NMR analysis (Fig. S2†), that showed the specific peaks of the epoxy ring at 3.12 ppm, 2.72 ppm and 2.54 ppm, and then estimated according to the literature procedure:43 resulting polymer was characterized by all hydroxyl terminal groups substituted by epoxide groups.
1, in order to potentially involve only one –OH group of each molecule in the reaction with the alkyne derivative, was not enough to produce the desired product because the formation of a mixture of mono-, bis-, or no functionalized system was obtained. The integral values of the characteristic signals of 2 in its 1H-NMR spectrum, confirmed the collection of the mono-functionalized product (ESI†). The introduction of triple bond was justified by the need to graft the sensitive drug-disulfide linker system to the polymeric chain using copper-catalyzed azide alkyne cycloaddition (CuAAC). The formed triazole ring possesses a number of desirable features reported in the medicinal chemistry and, in particular, is stable under standard physiological conditions, which suits well for gene and drug delivery system. The use of click chemistry required the functionalization of PEG with azide moiety, that was performed in two steps. The first one, as illustrated in Fig. 2, involved the preparation of the polymer with epoxide end groups, through reaction with epichlorohydrin. PEG functionalization was verified by 1H-NMR analysis (Fig. S2†), that showed the specific peaks of the epoxy ring at 3.12 ppm, 2.72 ppm and 2.54 ppm, and then estimated according to the literature procedure:43 resulting polymer was characterized by all hydroxyl terminal groups substituted by epoxide groups.
The second step, reported in Fig. 3, consisted in the ring-opening reaction of di-epoxide PEG using sodium azide and ammonium chloride as a coordinating salt;52 in this case, 1H-NMR analysis revealed new signals related to the methylene group adjacent to azide group at 3.28 ppm, while the epoxide proton resonances disappeared (Fig. S3†). In this way, we introduced –N3 moiety and restored –OH groups at both PEG sides. The importance of the presence of hydroxyl groups was related to their chemical modification with imidazole, essential for PEG-PEI nanogel synthesis.
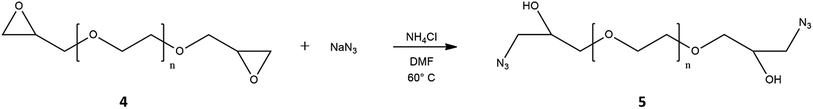 | ||
| Fig. 3 Functionalization of PEG with azide groups. The reaction allowed restoring the hydroxyl groups of the polymer. | ||
Finally, the click reaction between PEG–OH–N3, 5, and disulfide bridge with rhodamine on one side and alkyne group on the other end, 3, was carried out with copper iodide as the catalyst, sodium ascorbate as reducing agent and a mixture of acetonitrile-water as the solvent (Fig. 4). According to the presence of two azide groups for each PEG chain, the used molar ratio between 5 and 3 was equal to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, in order to ensure quantitatively the reaction. The disulfide functionalized PEG was purified by dialysis against distilled water to remove unreacted chemicals and salts. The formation of triazole was confirmed by its characteristic signal in 1H-NMR spectrum (Fig. S4†). Moreover the GPC chromatogram visible in Fig. S6† reveals the presence of functonalized polymer.
2, in order to ensure quantitatively the reaction. The disulfide functionalized PEG was purified by dialysis against distilled water to remove unreacted chemicals and salts. The formation of triazole was confirmed by its characteristic signal in 1H-NMR spectrum (Fig. S4†). Moreover the GPC chromatogram visible in Fig. S6† reveals the presence of functonalized polymer.
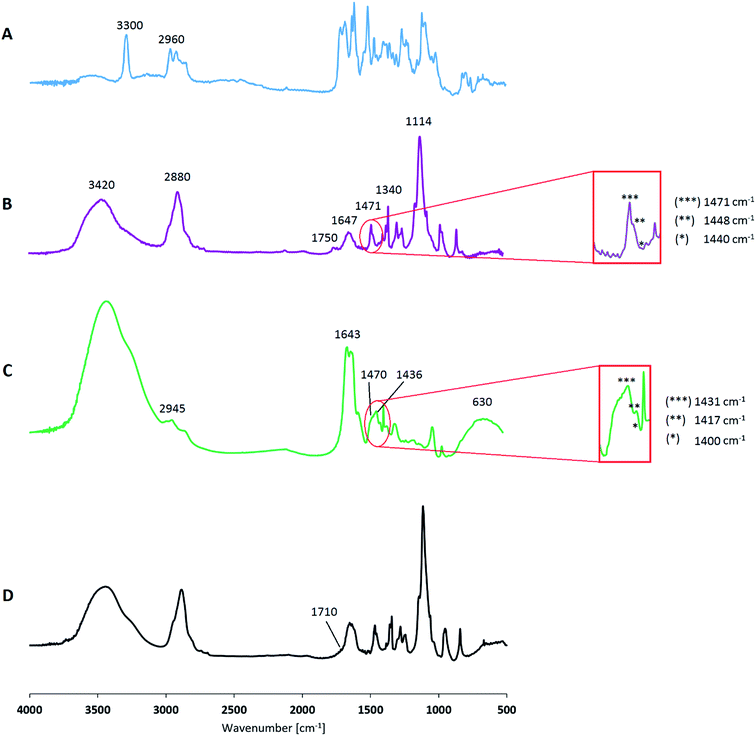
Fig. 5B shows the FT-IR spectrum of the modified PEG, where the characteristic signals of triazole can be recognized at 1471 cm−1, 1448 cm−1, 1440 cm−1, as outlined in the figure. The broad peak around 3420 cm−1 corresponding to the stretching vibration of terminal –OH bonds, whereas the signals at 2880 cm−1 and at 1114 cm−1 are respectively due to the C–H and C–O–C stretch of the polymer chain. Other characteristic PEG signals, related to the C–H bending, are found in the wavenumbers range around 1470 ÷ 800 cm−1. In addition, in the range 1750 ÷ 700 cm−1 peaks related to rhodamine are detectable according to the signals of the FT-IR spectrum of compound 3 (Fig. 5A): this confirms that PEG is linked in a stable manner to the modified mimetic drug and it is still reflected in the absence of the alkyne signal at 3300 cm−1, which is, otherwise, clearly visible in the spectrum of compound 3. The latter also shows the characteristic stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 1715 cm−1, represented also in the spectrum of the functionalized PEG.
O at 1715 cm−1, represented also in the spectrum of the functionalized PEG.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
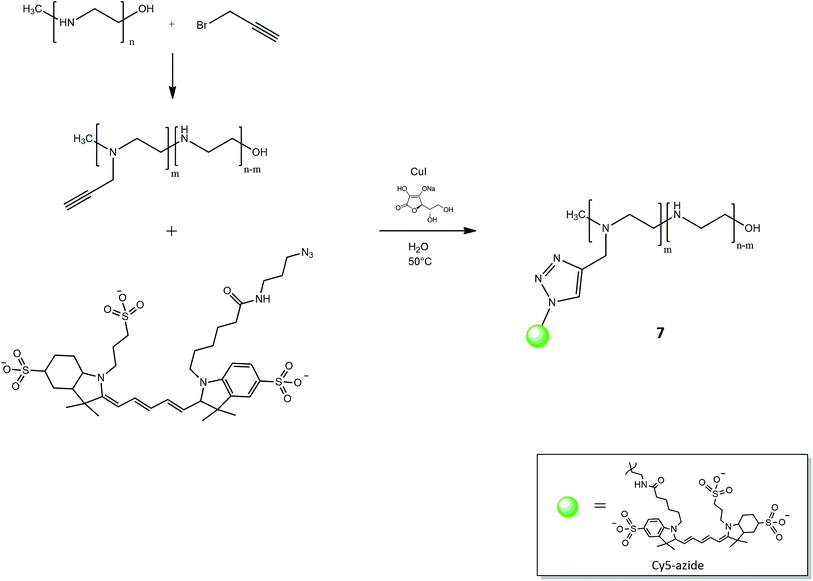
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and the excess amount of the alkyne groups in comparison to polymer was used to ensure the reaction conversion. The percentage of linked propargyl moiety was evaluated referring to 1H-NMR spectrum: as illustrated in our previous work,45 the ratio among the integral value of the signal at 3.55 ppm, related to monomer propargyl derivative, and the peaks corresponding to PEI-block in the range 3.50–2.90 ppm, suggested that the functionalization degree of the polymer was 10%. The signal corresponding to the H of alkyne was covered by the peak of the polymer chains. The resulting product reacted with Cy5-N3 using click strategy CuAAC (Fig. 6), in a similar manner as described in the previous section about the formation of triazole between PEG and disulfide derivatives.
5 and the excess amount of the alkyne groups in comparison to polymer was used to ensure the reaction conversion. The percentage of linked propargyl moiety was evaluated referring to 1H-NMR spectrum: as illustrated in our previous work,45 the ratio among the integral value of the signal at 3.55 ppm, related to monomer propargyl derivative, and the peaks corresponding to PEI-block in the range 3.50–2.90 ppm, suggested that the functionalization degree of the polymer was 10%. The signal corresponding to the H of alkyne was covered by the peak of the polymer chains. The resulting product reacted with Cy5-N3 using click strategy CuAAC (Fig. 6), in a similar manner as described in the previous section about the formation of triazole between PEG and disulfide derivatives.
PEI linked to Cy5 was collected after purification by dialysis to remove unreacted chemicals and avoid the separation of product as PEI complexes with copper. 1H-NMR spectrum showed the characteristic signal of triazole at 8.10 ppm and FT-IR spectrum sets out the corresponding peaks around 1470 cm−1. Fig. 5C also shows PEI peculiar signals: at 3240 cm−1 due to N–H stretching, in the range 2945 ÷ 2820 cm−1 of C–H stretching, at 1643 cm−1 representative of N–H bending, at 1463 cm−1 related to PEI C–H bending and in the wavenumber range 1400 ÷ 900 cm−1 the peaks are due to C–N stretching. According to the emission/absorbance of Cy5 dye,53 its corresponding signal is reported around 630 cm−1. PEI functionalization could occur at higher percentage of linked alkyne group and then of grafted Cy5, just increasing the amount of propargyl bromide involved in the corresponding reaction. However, our study suggests that a dye functionalization degree of 10% is satisfactory for ensuring the traceability of the final nanogel systems within cellular media. In addition, lower chemical modification of PEI chain allows maintaining its peculiar properties without any potential alterations, in particular the availability of amine moieties for the successive nanogel synthesis. Cy5 was chosen due to its functional detection in biological application; cyanine derivatives are characterized by the fact that absorbance and fluorescence wavelength can be controlled by polymethine bridge length: longer cyanines possess higher absorbance and emission wavelengths up to near infrared region. Cy5-N3 is fluorescent in the red region (∼650 excitation, 670 nm emission) and its signal is detectable where many charge-coupled devices detectors have better sensitivity, and biological compounds have lower background. In this way, it is possible to mark the used polymer and also investigate its fate in the in vitro experiments.
Nanogel composition
Synthesis of biocompatible PEG-PEI nanogels was achieved by coupling of modified PEG 6 and PEI 7 using a modified emulsification–evaporation method.46 According to this procedure, the solution of activated PEG (grafting the disulfide bridge-rhodamine system) in dichloromethane was emulsified in the aqueous solution of PEI (grafting Cy5) by sonication. After the completion of the cross-linking reaction, the organic solvent was removed under vacuum, and nanogel was allowed to form in the aqueous medium (Fig. 7). The growing stage completed the rearrangement of linked polymers, according to the chain entanglements in the forming nanogel networks. The resulting suspension was subjected to purification via dialysis to remove by-products and unreacted polymers.The effective synthesis of nanogels was verified by 1H-NMR data, where the signals at 4.12 ppm and at 3.38 ppm correspond, respectively, to the two methylene protons of PEG and PEI monomer forming the carbamate bond (Fig. S5†). This confirmed the cross-linking between the two polymer chains. The composition of this nanostructures was estimated in terms of molar ratio between the polymers, considering the integral values of the PEG (range 3.80–3.40 ppm) and PEI (range 2.85–0.80 ppm) chains signals and it turned out to be PEG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEI 1.5
PEI 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. FT-IR spectrum reported in Fig. 5D shows the distinctive signals of the polymers and their corresponding functionalizations according to the FT-IR spectra of modified PEG (Fig. 5B) and PEI (Fig. 5C); in particular, the presence of signal at about 1710 cm−1 is representative of C
1. FT-IR spectrum reported in Fig. 5D shows the distinctive signals of the polymers and their corresponding functionalizations according to the FT-IR spectra of modified PEG (Fig. 5B) and PEI (Fig. 5C); in particular, the presence of signal at about 1710 cm−1 is representative of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching related to the carbamate linkage.
O stretching related to the carbamate linkage.
Physical characterization
One of the most important issues in the field of nanoscience is the size effect on the biodistribution, kinetic of release, and cellular uptake of nanoparticles.54 Therefore, the investigation of nanogel dimensions is necessary to design system able to be internalized through active or passive uptake. Dynamic light scattering (DLS) is a widely used technique for nanoparticle size and ζ-potential determination in suspension. The measured hydrodynamic diameter represents the dimension of the polymeric sample together with layer of surface-bound solvent. Recorded DLS data provide nanogel average diameter equal to 462 nm (polydispersity index, PDI = 0.066) and a surface charge of +21.8 mV. The positive value of ζ-potential indicates that nanogels are characterized by a positive surface charge, probably due to the ability of PEI, although alkyne functionalization, to preserve part of its positive charge in the final polymeric network. Size distribution may be quantified by using atomic force microscopy (AFM) analysis, obtaining additional information about the morphology. In this case, the obtained size is 250 nm (data not shown). The size of the nanogels observed by AFM was less than those measured using DLS.This variation between AFM and DLS measurements is justifiable considering the difference in the sample processing: in DLS, the slight increase in size is due to the nanogel swelling behaviour in aqueous solution. This problem was not present with hydrophobic polymeric nanoparticles synthesized by our group and presented in previous work.55 In that case, the complete accordance between AFM and DLS was due to the fact that in that system swelling phenomenon was negligible. Therefore, obtained results suggested that nanogels presented particle diameter and surface charge within the range useful to promote cellular internalization, according to the guideline in the rational design of drug nanocarriers with maximized therapeutic efficacy.25
Drug release mechanism
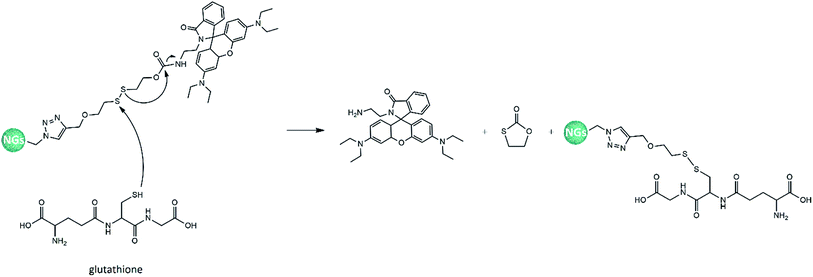
The efficacy of synthetized nanogels as selective drug delivery systems was investigated performing the rhodamine release experiments in two different media, at 37 °C. The first one was GSH solution 5 mM, prepared in a pH 7.4 phosphate buffer and representative of the average intracellular glutathione concentration in human body;49 whereas, the second one was PBS solution, chosen as a reference, in order to evaluate the potential GSH-responsive effect of our nanosystems. The aim was the evaluation of the interactions between a disulfide bond and a thiol compound: the first one is slightly degradable under physiological conditions but can be quickly cleaved in a highly reductive environment within the cells, achieving controlled intracellular rapid drug release. A schematic representation of the rhodamine release mechanism that could occur within cellular membrane is reported in Fig. 8: the S–S reduction could be performed by GSH or cysteine cell component (both present –SH group and interact in a similar manner).56,57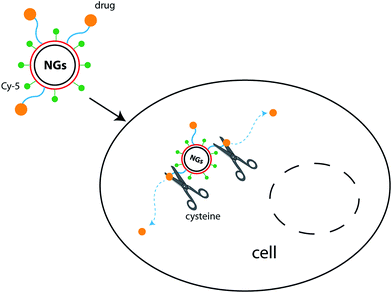 | ||
| Fig. 8 Scheme of rhodamine release within a cell, due to the reduction of S–S bond by thiol compound. | ||
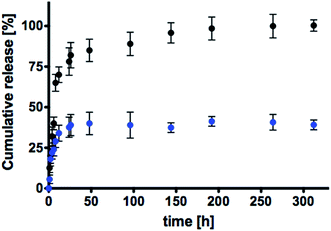
About thiol-sensing in biological samples, GSH has a leading role because it is the most abundant thiol in cells.51,58 The percentage of rhodamine released from the nanogels was defined as the ratio between the release amount in the aqueous media and the total amount linked to the polymeric network. The spectrophotometrical evaluation of escaped rhodamine was performed referring the calibration curve of the mimetic drug as amine modified rhodamine 1: the disulfide cleavage in the redox-responsive environment allowed the release of 1, as showed in Fig. 9, without any other linked functionalizations (which, otherwise, are needed to the synthesis of the final sensitive nanogels), that could distort the UV-vis measurements.
Fig. 10 shows the release profiles in GSH (in black) and in PBS (in blue) solutions. It is well visible that rhodamine release is faster in media containing the thiol compound and it is completed within 150 h. Release profile associated to the physiological PBS media is prolonged for a much longer period, underling that the key difference between the two release mechanisms is related to the presence or absence of GSH. In particular, it is evident that the synthetized nanogels are thiol-sensitive and the disulfide bridge plays a leading role in the development of redox-responsive nanogels. In these systems, the release of rhodamine occurred due to the selective reduction of S–S moiety caused by GSH that is the driving force influencing the release kinetic. In addition, considering that used GSH concentration is representative of the thiol amount detectable within cells, produced nanostructures seemed to be a promising tool to achieve a sensitive drug release caused only by common intracellular compounds, without any other external stimuli. In physiological environment without GSH, released rhodamine occurred with lower delivery rate, maintaining 70% of loaded mimetic drug still linked to the nanogel network at 14 days. In this case, the release was not caused by chemical interaction with GSH, but due to the progressive degradation of the disulphide bond in physiological conditions, considering its moderate instability towards the electrophilic and nucleophilic nature of polar solvents.59
The obtained results allow to state that the design of a disulfide linker make it possible to release a drug within intracellular environment due to the simple interactions with cell components, whereas an extracellular media (such as a PBS solution) decreases its susceptibility to reduction. In this way, the developed nanostructures could be considered useful for further investigations of intracellular drug release such as hydrophilic analgesic and anti-inflammatory drugs that are characterized by carboxyl terminal group, steric hindrance and resemblance in a similar manner to rhodamine structure (examples are: fenoprofen, diflunisal, mefenamic acid or ketorolac).
In vitro cell-selective drug release
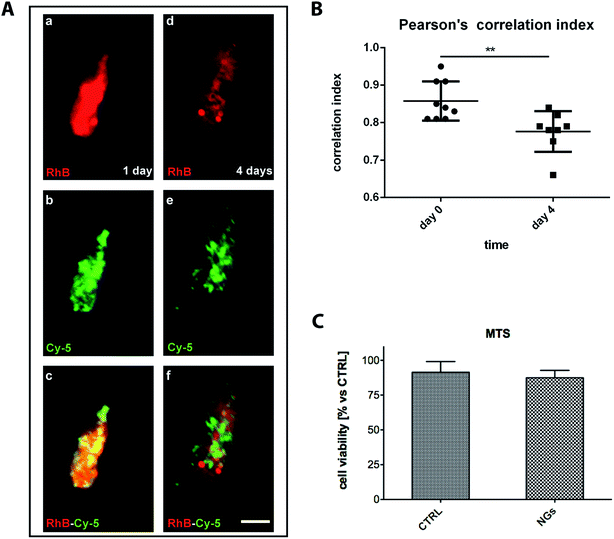
To further understand whether polymeric NGs were able to deliver linked compounds into the cytosol of microglia we observed release profile of RhB (as drug mimetic) from Cy5 decorated nanogels.The colocalization of single fluorescent Cy5-positive NGs (pseudocolor green) loaded with RhB (pseudocolor red) showed that the mimetic drug was efficiently encapsulated (Fig. 11A). To evaluate the rhodamine delivery by NGs in vitro the nanodevices were incubated for 24 h with microglial cells. At this time of analysis, microglia showed clusterized NGs in the cytosol with a colocalized Cy5 and RhB signals (Fig. 11Ac). After 4 days of incubation, a markedly diffused signal of RhB was revealed in the cytosol of the microglial cells and it was only partially colocalized with Cy5 (Fig. 11Ad and e).
This quantitatively evaluated by Pearson's correlation index, where rhodamine signal was more widespread diffused within the cytosol, compared to Cy5-positive NGs (Fig. 11B). Furthermore, we evaluated the biocompatibility of internalized NGs in microglial cell colture by MTS assay. Nanogel internalization did not affect microglia cell viability up to 4 days compared to microglia not exposed to the polymeric systems (Fig. 11C). To understand whether internalized NGs affected the integrity of cellular membranes we measured the level of lactate dehydrogenase (LDH) released (Fig. S7†) in the culture medium. No detectable amount of LDH release was revealed underlining the biocompatibility of this colloidal system.
Conclusions
In this work, we investigated the possibility to carry hydrophilic molecules within nanogels towards specific target and without losing the drugs content in body fluids. Indeed, the nanogel traceability was successfully performed through a one-step CuAAC click chemistry involving PEI and Cy5 dye. Then, to improve drug delivery performances, we introduced a sensitive cleavable bond (disulfide) between polymeric chains and drug molecules in order to create a selective drug delivery vehicle. NGs were internalized by microglia, without detectable toxic effect. In vitro studies showed a selective drug release within the cell cytosol of activated microglia confirming the efficacy of this tool. The incorporation of these types of linkers reduced the drug escape to the surrounding environment compared to systems where solute's release is only driven by a pure Fickian diffusion mechanism. In conclusion, the approach proposed in this work is a potential system to provide benefit in different human neurologic pathologies opening new perspectives to the inflammatory treatment in the central nervous system, through the modulated drug uptake and release of activated microglia.References
- L. Florez, C. Herrmann, J. M. Cramer, C. P. Hauser, K. Koynov, K. Landfester, D. Crespy and V. Mailänder, Small, 2012, 8, 2222–2230 CrossRef CAS PubMed.
- J. Nicolas, S. Mura, D. Brambilla, N. Mackiewicz and P. Couvreur, Chem. Soc. Rev., 2013, 42, 1147–1235 RSC.
- W. Liu, W. Zhang, X. Yu, G. Zhang and Z. Su, Polym. Chem., 2016, 7, 5749–5762 RSC.
- I. Elliott Donaghue and M. S. Shoichet, Acta Biomater., 2015, 25, 35–42 CrossRef CAS PubMed.
- F. Rossi, G. Perale, S. Papa, G. Forloni and P. Veglianese, Expert Opin. Drug Delivery, 2013, 10, 385–396 CrossRef CAS PubMed.
- Y.-C. Kuo and H.-F. Ko, Biomaterials, 2013, 34, 4818–4830 CrossRef CAS PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- M. Durán-Lobato, B. Carrillo-Conde, Y. Khairandish and N. A. Peppas, Biomacromolecules, 2014, 15, 2725–2734 CrossRef PubMed.
- L. A. Sharpe, A. M. Daily, S. D. Horava and N. A. Peppas, Expert Opin. Drug Delivery, 2014, 11, 901–915 CrossRef CAS PubMed.
- T. Ramasamy, Z. S. Haidar, T. H. Tran, J. Y. Choi, J. H. Jeong, B. S. Shin, H. G. Choi, C. S. Yong and J. O. Kim, Acta Biomater., 2014, 10, 5116–5127 CrossRef CAS PubMed.
- S. K. Mann, A. Dufour, J. J. Glass, R. De Rose, S. J. Kent, G. K. Such and A. P. R. Johnston, Polym. Chem., 2016, 7, 6015–6024 RSC.
- V. Mailänder and K. Landfester, Biomacromolecules, 2009, 10, 2379–2400 CrossRef PubMed.
- G. Villaverde, A. Baeza, G. J. Melen, A. Alfranca, M. Ramirez and M. Vallet-Regi, J. Mater. Chem. B, 2015, 3, 4831–4842 RSC.
- T. C. Johnstone, K. Suntharalingam and S. J. Lippard, Chem. Rev., 2016, 116, 3436–3486 CrossRef CAS PubMed.
- B. Karagoz, L. Esser, H. T. Duong, J. S. Basuki, C. Boyer and T. P. Davis, Polym. Chem., 2014, 5, 350–355 RSC.
- A. E. Arnold, P. Czupiel and M. Shoichet, J. Controlled Release, 2017 DOI:10.1016/j.jconrel.2017.02.019.
- W. L. Tung, S. H. Hu and D. M. Liu, Acta Biomater., 2011, 7, 2873–2882 CrossRef CAS PubMed.
- Y. Tian, S. Bian and W. Yang, Polym. Chem., 2016, 7, 1913–1921 RSC.
- S. Papa, F. Rossi, R. Ferrari, A. Mariani, M. De Paola, I. Caron, F. Fiordaliso, C. Bisighini, E. Sammali, C. Colombo, M. Gobbi, M. Canovi, J. Lucchetti, M. Peviani, M. Morbidelli, G. Forloni, G. Perale, D. Moscatelli and P. Veglianese, ACS Nano, 2013, 7, 9881–9895 CrossRef CAS PubMed.
- S. Papa, I. Caron, E. Erba, N. Panini, M. De Paola, A. Mariani, C. Colombo, R. Ferrari, D. Pozzer, E. R. Zanier, F. Pischiutta, J. Lucchetti, A. Bassi, G. Valentini, G. Simonutti, F. Rossi, D. Moscatelli, G. Forloni and P. Veglianese, Biomaterials, 2016, 75, 13–24 CrossRef CAS PubMed.
- J. Meng, V. Agrahari and I. Youm, Journal of Neuroimmune Pharmacology, 2017, 12, 84–98 CrossRef PubMed.
- M. Yu, X. Jie, L. Xu, C. Chen, W. Shen, Y. Cao, G. Lian and R. Qi, Biomacromolecules, 2015, 16, 2588–2598 CrossRef CAS PubMed.
- S. S. Minami, B. Sun, K. Popat, T. Kauppinen, M. Pleiss, Y. Zhou, M. E. Ward, P. Floreancig, L. Mucke, T. Desai and L. Gan, J. Neuroinflammation, 2012, 9, 22 CAS.
- E. Nance, F. Zhang, M. K. Mishra, Z. Zhang, S. P. Kambhampati, R. M. Kannan and S. Kannan, Biomaterials, 2016, 101, 96–107 CrossRef CAS PubMed.
- C. He, Y. Hu, L. Yin, C. Tang and C. Yin, Biomaterials, 2010, 31, 3657–3666 CrossRef CAS PubMed.
- S. R. Cerqueira, J. M. Oliveira, N. A. Silva, H. Leite-Almeida, S. Ribeiro-Samy, A. Almeida, J. F. Mano, N. Sousa, A. J. Salgado and R. L. Reis, Small, 2013, 9, 738–749 CrossRef CAS PubMed.
- S. Kabu, Y. Gao, B. K. Kwon and V. Labhasetwar, J. Controlled Release, 2015, 219, 141–154 CrossRef CAS PubMed.
- H. Zou, Z. Wang and M. Feng, J. Controlled Release, 2015, 214, 121–133 CrossRef CAS PubMed.
- J. McMasters and A. Panitch, Acta Biomater., 2017, 49, 78–88 CrossRef CAS PubMed.
- E. Mauri, F. Rossi and A. Sacchetti, Mater. Sci. Eng., C, 2016, 61, 851–857 CrossRef CAS PubMed.
- C. Porsch, Y. Zhang, M. I. Montañez, J. M. Malho, M. A. Kostiainen, A. M. Nyström and E. Malmström, Biomacromolecules, 2015, 16, 2872–2883 CrossRef CAS PubMed.
- M. Li, Z. Tang, H. Sun, J. Ding, W. Song and X. Chen, Polym. Chem., 2013, 4, 1199–1207 RSC.
- E. L. Schneider, J. Henise, R. Reid, G. W. Ashley and D. V. Santi, Bioconjugate Chem., 2016, 27, 1210–1215 CrossRef CAS PubMed.
- K. Ulbrich, K. Holá, V. Šubr, A. Bakandritsos, J. Tuček and R. Zbořil, Chem. Rev., 2016, 116, 5338–5431 CrossRef CAS PubMed.
- D. Ossipov, S. Kootala, Z. Yi, X. Yang and J. Hilborn, Macromolecules, 2013, 46, 4105–4113 CrossRef CAS.
- K. Vulic and M. S. Shoichet, J. Am. Chem. Soc., 2012, 134, 882–885 CrossRef CAS PubMed.
- C. L. Modery-Pawlowski, A. M. Master, V. Pan, G. P. Howard and A. Sen Gupta, Biomacromolecules, 2013, 14, 910–919 CrossRef CAS PubMed.
- G. Reina, S. Orlanducci, C. Cairone, E. Tamburri, S. Lenti, I. Cianchetta, M. Rossi and M. L. Terranova, J. Nanosci. Nanotechnol., 2015, 15, 1022–1029 CrossRef CAS PubMed.
- J. F. Quinn, M. R. Whittaker and T. P. Davis, Polym. Chem., 2017, 8, 97–126 RSC.
- M. Bilzer and B. H. Lauterburg, Eur. J. Clin. Invest., 1991, 21, 316–322 CrossRef CAS PubMed.
- J. Alam, T. H. Keller and T. P. Loh, Chem. Commun., 2011, 47, 9066–9068 RSC.
- L. Li, X. Wang, J. Yang, X. Ye and C. Wu, Macromolecules, 2014, 47, 650–658 CrossRef CAS.
- M. Teodorescu, B. Cursaru, P. Stanescu, C. Draghici, N. D. Stanciu and D. M. Vuluga, Polym. Adv. Technol., 2009, 20, 907–915 CrossRef CAS.
- B. Zhang, H. Zhang, R. Elupula, A. M. Alb and S. M. Grayson, Macromol. Rapid Commun., 2014, 35, 146–151 CrossRef CAS PubMed.
- E. Mauri, I. Moroni, L. Magagnin, M. Masi, A. Sacchetti and F. Rossi, React. Funct. Polym., 2016, 105, 35–44 CrossRef CAS.
- S. Vinogradov, E. Batrakova and A. Kabanov, Colloids Surf., B, 1999, 16, 291–304 CrossRef CAS.
- M. De Paola, A. Mariani, P. Bigini, M. Peviani, G. Ferrara, M. Molteni, S. Gemma, P. Veglianese, V. Castellaneta, V. Boldrin, C. Rossetti, C. Chiabrando, G. Forloni, T. Mennini and R. Fanelli, Mol. Med., 2012, 18, 971–981 CAS.
- S. Papa, R. Ferrari, M. De Paola, F. Rossi, A. Mariani, I. Caron, E. Sammali, M. Peviani, V. Dell'Oro, C. Colombo, M. Morbidelli, G. Forloni, G. Perale, D. Moscatelli and P. Veglianese, J. Controlled Release, 2014, 174, 15–26 CrossRef CAS PubMed.
- D. Yang, W. Chen and J. Hu, J. Phys. Chem. B, 2014, 118, 12311–12317 CrossRef CAS PubMed.
- D. Jung, S. Maiti, J. H. Lee, J. H. Lee and J. S. Kim, Chem. Commun., 2014, 50, 3044–3047 RSC.
- M. H. Lee, J. H. Han, J.-H. Lee, H. G. Choi, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2012, 134, 17314–17319 CrossRef CAS PubMed.
- N. V. Tsarevsky, S. A. Bencherif and K. Matyjaszewski, Macromolecules, 2007, 40, 4439–4445 CrossRef CAS.
- S. Luo, E. Zhang, Y. Su, T. Cheng and C. Shi, Biomaterials, 2011, 32, 7127–7138 CrossRef CAS PubMed.
- L. Shang, K. Nienhaus and G. U. Nienhaus, J. Nanobiotechnol., 2014, 12, 5 CrossRef PubMed.
- C. Colombo, L. Galletti, M. Lepri, I. Caron, L. Magagnin, P. Veglianese, F. Rossi and D. Moscatelli, Eur. Polym. J., 2015, 68, 216–225 CrossRef CAS.
- R. Dringen, Prog. Neurobiol., 2000, 62, 649–671 CrossRef CAS PubMed.
- K. Aquilano, S. Baldelli and M. R. Ciriolo, Front. Pharmacol., 2014, 5, 196 Search PubMed.
- C. Hwang, A. J. Sinskey and H. F. Lodish, Science, 1992, 257, 1496–1502 CAS.
- G. Leriche, L. Chisholm and A. Wagner, Bioorg. Med. Chem., 2012, 20, 571–582 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04584k |
| ‡ These authors share senior authorship. |
| This journal is © The Royal Society of Chemistry 2017 |