Open Access Article
Open Access ArticleBiotechnological production of the mussel byssus derived collagen preColD†
Adrian V. Golsera and
Thomas Scheibel *ab
*ab
aLehrstuhl Biomaterialien, Fakultät für Ingenieurwissenschaften, Universität Bayreuth, Universitätsstraße 30, 95440 Bayreuth, Germany
bBayreuther Zentrum für Kolloide und Grenzflächen (BZKG), Bayerisches Polymerinstitut (BPI), Bayreuther Zentrum für Molekulare Biowissenschaften (BZMB), Bayreuther Materialzentrum (BayMAT), Universität Bayreuth, Universitätsstraße 30, 95440 Bayreuth, Germany. E-mail: thomas.scheibel@uni-bayreuth.de
First published on 3rd August 2017
Abstract
Marine mussels adhere to substrates within the intertidal zone by means of a bundle of threads called the mussel byssus which contains several collagen-like proteins responsible for the mechanical properties of the fibers. Here, we demonstrate the biotechnological production of one of three identified byssus collagens, preColD, from Mytilus galloprovincialis using Pichia pastoris as the expression host. Previously detected structural features of natural preColD, such as collagen triple helix formation, could also be detected in the recombinant preColD, even in the absence of hydroxylation of proline or tyrosine residues.
Introduction
Several marine species produce a variety of distinct materials for underwater attachment with mechanical properties ranging from those of cement1 to silk2 to softer and more elastic materials.3 These holdfasts have the ability to dissipate mechanical stress without losing their structural integrity. One of these remarkable devices is the byssus of Mytilidae,4 e.g. Mytilus galloprovincialis, which consists of several distinct threads each comprising a mechanical gradient along the fiber axis.5 This gradient of the material's Young's modulus allows the soft muscle (the mussel foot) to seamlessly connect to hard substrates without causing radial stress within the material.6The byssus contains a variety of different proteins, including collagen-like preCols,7 which assemble into fibrils being the byssus' main load-bearing structure located in the fiber core.8 preCols (Fig. 1), are a class of structural proteins comprising a collagen core domain flanked by specific domains which strongly contribute to the overall mechanical properties of the byssus threads:9 the elastic, proximal portion of a thread is rich in preColP10 comprising elastin-like flanking domains, whilst the tougher, more rigid portion of the thread consists mostly of preColD11 which incorporates crystalline silk-like flanking domains.
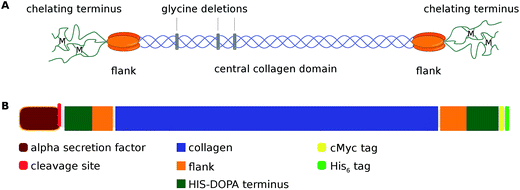 | ||
| Fig. 1 (A) Schematic representation of a preColD triple helix. The central collagen sequence is based on the collagen-typical (GXY)n-repeat which is interrupted by glycine deletions. The flanking regions of preColD contain poly-alanine stretches similar to those of silks and putatively form nanocrystalline beta sheets upon higher order assembly of the thread. The chelating termini of preCols contain both histidine and DOPA residues and thereby can promote a reversible, metal dependent end-to-end arrangement between fibrils that become increasingly chemically crosslinked during the maturation of the thread.9 (B) Block schematic of the domain structure of monomeric, recombinant preColD (domain size drawn to scale). | ||
The influence of the preCol-composition on the thread's mechanics is significant: even though the preCols are embedded in a complex array of matrix proteins,12–14 the mechanical properties of the byssus thread have been largely reproduced by spinning a fiber from matrix-free preCols extracted from the mussel foot.15,16
In addition to mechanical stability, all known preCols mediate various intermolecular interactions with other preCols, matrix proteins and substrates. The amino- and carboxyterminal domains of the natural preCols contain DOPA moieties for adhesion, metal binding and crosslinking, as well as histidine-based metal chelate complexes, which are likely responsible for previously detected self-healing properties of byssus threads,15,17 since the latter are fully reversible. Like other collagens, proline residues within the natural preCols are often hydroxylated at the Y-position in the collagen triplet (GXY) by a mussel specific hydroxylase; it is known, however, that most types of collagen will readily form triple-helices even when the content of hydroxyproline is low, albeit at lower melting points.
In this context, the lack of DOPA-residues will also reduce the crosslinking, but it is not expected to influence the protein's folding. Therefore, recombinant production of preColD was chosen as one possible strategy to investigate individual mussel byssus collagens and their properties in greater detail.
Experimental section
Molecular cloning and microbiology
The general methodology for the handling of P. pastoris, as well as the molecular biology methods utilized for obtaining the preColD expression plasmid are described in the pPICZ user manual18 and other relevant literature.19Fermentation of Pichia pastoris
Fermentation was carried out in a Minifors fermenter (Infors, Basel Switzerland) according to established protocols23,24 with slight modifications. Cells were grown at 30 °C in BMGY medium (13.4 g L−1 YNB, 1% (v/v) glycerol, 1% (w/v) yeast extract, 2% (w/v) peptone, 100 mM potassium phosphate, pH 6.0) until the initial carbon sources were depleted and subsequently fed with 86% glycerol, 1.2% PTM1 trace salts25 at 10 mL h−1 L−1 starting volume until the wet cell mass reached 200 g L−1. Production of recombinant protein was induced with methanol (final concentration of 0.5% (v/v)). The concentration was kept stable with an automated feeding procedure controlled by a methanol sensor. The pH was held constant at 6.0, and pO2 was kept at 40%.Purification of recombinant preColD
Cells were harvested by centrifugation (6000 xG) and washed once with phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). The pellet was resuspended in an equal volume of buffer A (4 M guanidinium hydrochloride in PBS) and disrupted by 5 passes through a Microfluidics M-110S homogenizer at 80 psi inlet pressure. After centrifugation (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 xG), the supernatant of the extract was loaded onto a high-performance Ni-NTA IMAC column (approx. 200 mL bed volume), washed extensively with 8 M urea in PBS and eluted with 200 mL buffer A + 500 mM imidazole. The eluate was dialyzed against 10 mM ammonium bicarbonate and subsequently dried via lyophilization (see ESI Fig. S2† for SDS-PAGE).
000 xG), the supernatant of the extract was loaded onto a high-performance Ni-NTA IMAC column (approx. 200 mL bed volume), washed extensively with 8 M urea in PBS and eluted with 200 mL buffer A + 500 mM imidazole. The eluate was dialyzed against 10 mM ammonium bicarbonate and subsequently dried via lyophilization (see ESI Fig. S2† for SDS-PAGE).
Transmission electron microscopy
200 μL of preColD were desalted using a 5 mL HiTrap desalting column equilibrated with 50 mM sodium acetate, pH 5.5 and concentrated to approx. 25 mg mL−1 using a centricon centrifugation tube with a MW cutoff of 30 kDa.5 μL of 50 mM TRIS/HCl pH 8.0 were applied to TEM-grids, then adding 5, 1 or 0.1 μL of preColD solution, respectively. After incubating for 1 min, the grids were washed twice with 10 μL TRIS-buffer and negatively stained with uranyl acetate. The samples were visualized using a Zeiss CEM 902 microscope at 80 kV.
CD and FTIR spectroscopy
CD spectra were recorded with a Jasco J-815 spectrometer at 20 °C (scanning speed: 50 nm min−1; band width: 1.7 nm; D.I.T.: 2 s; accumulations: 5). Films were poured onto glass slides from a 5 mg mL−1 solution in formic acid and measured directly after the evaporation of the solvent. preColD in solution was measured in a 1 mm quartz cuvette after dialyzing the same solution against PBS and PBS containing 4 M urea at 4 °C. FTIR spectra were recorded with a Bruker Tensor 27 spectrometer, using a PIKE Miracle Ga-ATR crystal. The films were prepared directly on the freshly cleaned crystal by allowing the solvent to evaporate from 10 mg mL−1 solutions of protein (preColD, gelatin, spider silk protein eADF4(C16)) in formic acid. The spider silk protein, after measuring the freshly prepared film, was additionally treated with methanol to induce beta-sheet formation. The obtained spectra were smoothed, baseline-adjusted and corrected for atmospheric moisture.Results and discussion
Production and purification of preColD
P. pastoris can be a suitable host for the production of secreted proteins, which simplifies downstream processing steps during recombinant protein production. Therefore, preColD was cloned with an α-secretion factor inducing translocation into the endoplasmic reticulum (ER) and subsequent secretion via the Golgi apparatus. Accordingly, the selection for the highest producing P. pastoris expression strain was performed by screening for both intracellular as well as secreted transgenic cMyc-Antigen that is present at the carboxyterminus of recombinant preColD (Fig. 1B). The screening was performed in shake flask culture, and secreted protein levels corresponded well with intracellular retention (see ESI Fig. S1†).Nevertheless, apparently no secretion past the yeast cell wall took place during fermentation, since no recombinant protein could be detected in the medium. Therefore, the recombinant protein was purified from the cell pellet. A single band at the expected molecular weight could be detected by SDS-PAGE (ESI Fig. S2†), suggesting that the recombinant preColD was successfully translocated into the endoplasmic reticulum (ER), had its secretion factor cleaved as expected, but could not be secreted due to unknown reasons. The most likely explanation therefore is a difference in relative expression levels due to the scale-up, since the induction level is dependent on the methanol concentration, which is kept constant during fermentation but isn't monitored in shake flasks.
The yield of pure recombinant preColD from a 1 L fermentation up to 350 g L−1 wet cell mass was 252 mg when purified according to the protocol in the experimental section. While this is low compared to industrial processes employing P. pastoris,26 this number is adequate for lab scale production using a non-optimized system.
Determination of secondary structures
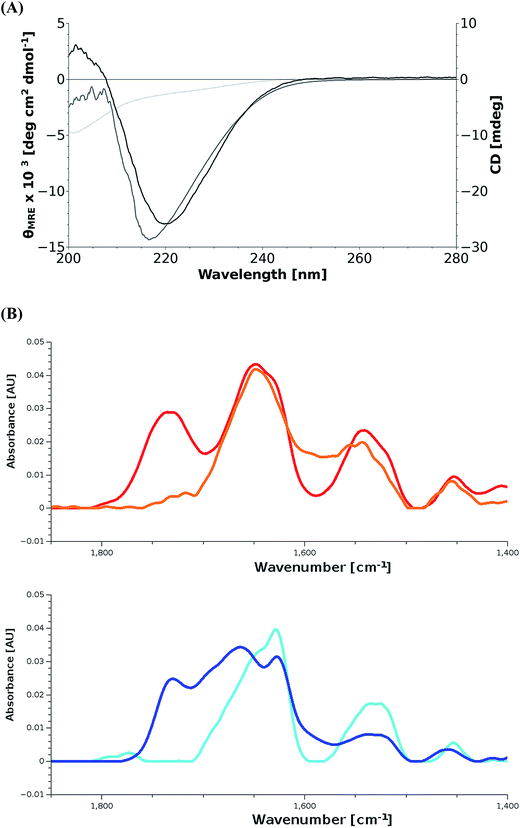
The β-sheet signal observed in the CD spectra (Fig. 2A) suggest a strong influence of the silk-like flanking domains on the overall structure of preColD, as they are present even under denaturing conditions (4 M urea) and when dried from formic acid, conditions that would usually favor random coil structures.Under physiological conditions, the signal intensity was reduced, suggesting that the negative β-sheet-peak (218 nm) is partially canceled out by the positive peak from triple-helical collagen (220 nm). The emergence of a negative peak around 200 nm, which is also expected for folded collagen, supports this hypothesis. It is known that denatured collagen mostly loses its CD-active properties (loss of positive peak at 220 nm, reduction of mean residual ellipticity at 200 nm from −30 to −5 × 103 deg cm2 dmol−1), while non-collagenous proteins would instead show a random-coil structure, which would interfere with the β-sheet-signal also under denaturing conditions.27,28 The spectra of preColD in solution at 4 M and 0 M urea have been compared to reference spectra using the web-based tool K2D3,29 showing no similarity to standard proteins present in the database, which supports the hypothesis of having mixed signals (ESI Fig. S3†).
Additional experiments suggesting the presence of β-sheets as well as unstructured collagen have been obtained for films of preColD via FTIR (Fig. 2B). The recombinant spider silk protein eADF4(C16) was used as a control to visualize the peak shift towards lower wavenumbers that occurs when beta sheet formation is induced in the previously unstructured film30,31 which, in this measurement, manifests itself as a shift of the amide I peak from 1645 cm−1 to 1630 cm−1. Commercial gelatin was used as a control to determine the signal obtained by an unstructured collagen-like protein, and corresponds to that of a random coil protein at 1645 cm−1. preColD has two peaks within the amide I band, one suggesting the presence of beta-sheets (1645 cm−1), the other suggesting the presence of random coil structures or helices (1660 cm−1).
While no Fourier self deconvolution was performed, it is evident that a part of preColD contains structural features that can be compared to those of silk proteins under the given conditions, while another portion of the protein seems to contain structures that correspond to unstructured or alpha-helical proteins, similar to those obtained by unstructured gelatin.
We hypothesize that the silk-like flanking domains of preColD form a stable, β-sheet-rich structure under virtually all conditions, being largely responsible for the stiffness of the distal portion of the natural mussel byssus. The collagen core appears to be stable at room temperature even when not hydroxylated, but loses its folding under denaturing conditions. Additionally, the formation of the collagen triple-helix seems to be kinetically hindered,32 since it does not appear when the denaturing solvent is removed quickly, such as during the preparation of films.
Fibrillization of preColD
Natural preCols extracted from the foot of M. edulis undergo fibrillization when the pH is increased above the pKa of histidine, as this enables the chelation of metal ions by the HIS/DOPA-rich termini of the proteins which thereby promote intermolecular interactions.17,33 Here, we analyzed the assembly of recombinant preColD under similar conditions. Preliminary experiments haven shown that the dilution of a highly concentrated solution of preColD in formic acid rapidly results in unspecific aggregation (ESI Fig. S4†), while dilute solutions of preColD undergo fibrillization within the same timescale after shifting the pH from 5.5 to 8.0 (Fig. 3). This has also been described for the natural extract of mixed preCols in ref. 33. Our recombinant fibrils also show similar properties as the natural extract in regard to size and intermolecular arrangement and, as expected, lack any amyloid-like features such as Thioflavin T binding. The presence of globular structures, as seen here, has not been described for the natural extract and seems to be depending on the preparation of the samples. However, since these structures are only observable in the vicinity of high densities of fibrils, they likely result from higher order assembly and/or aggregation of preColD trimers, which might likely be the result of artifacts due to the sample preparation.Conclusions
The mussel byssus collagen preColD of M. galloprovincialis could be recombinantly produced in P. pastoris and successfully purified under denaturing conditions using IMAC. It was shown that, even when no posttranslational modifications are present, preColD still has the ability to form nanofibrils under native conditions, which appeared to be similar to those obtained when subjecing natural preCols extracted from mussel feet to the same conditions.33The secondary structure of soluble preColD and films made thereof showed β-sheet structures even under mild denaturing conditions, which suggests that the silk-like flanking domains are involved in early-on structure formation of preColD. Further, they might play a central role in providing physical stability and chemical resilience in the natural byssus thread, since they have been shown in silk to withstand even stronger denaturing conditions.34–36 Furthermore, the data obtained by CD and FTIR spectroscopy suggest the presence of correctly folded collagen triplehelices. Since the formation of collagen triplehelices is kinetically hindered due to the very slow proline cis/trans isomerization, it is believed that the unique structure of the flanking regions15,37,38 is involved in premature assembly similar to the function of pro-domains in vertebrate collagens39 and certain foldons in bacterial collagens.40,41 This stabilizing effect seems to be sufficient to compensate for the decrease in stability due to the absence of hydroxyprolines.
Conclusively, the deeper understanding of byssus collagens can serve as an inspiration for the development of biotechnologically produced, protein-based, highly resilient biopolymers with adjustable functions.
Acknowledgements
The authors thank Dr A. Hagenau for providing the mussel foot cDNA library as well as K. Schacht (Biomaterials, University of Bayreuth), R. Grotjahn and PD Dr S. Geimer (Electron Microscopy, University of Bayreuth) for assistance with TEM. Fig. 1B was created using DomainDraw.42 This work was funded by DFG Grant SCHE603/14-1. A. Golser (MSc) received a scholarship from the Bayerisches Eliteförderungsgesetz (BayEFG).References
- R. R. Despain, K. L. De Vries, R. D. Luntz and M. L. Williams, J. Dent. Res., 1973, 52, 674–679 CrossRef CAS PubMed.
- K. Kronenberger, C. Dicko and F. Vollrath, Naturwissenschaften, 2012, 99, 3–10 CrossRef CAS PubMed.
- M. E. Callow and J. E. Callow, Biologist, 2002, 49, 10–14 Search PubMed.
- J. P. Pujol, Nature, 1967, 214, 204–205 CrossRef.
- J. E. Smeathers and J. F. V. Vincent, J. Mollus. Stud., 1979, 45, 219–230 CrossRef.
- J. H. Waite, E. Vaccaro, C. Sun and J. M. Lucas, Philos. Trans. R. Soc. London, Ser. B, 2002, 357, 143–153 CrossRef CAS PubMed.
- A. Hagenau, M. H. Suhre and T. Scheibel, Prog. Polym. Sci., 2014, 39, 1564–1583 CrossRef CAS.
- X. Qin and J. H. Waite, J. Exp. Biol., 1995, 198, 633–644 CAS.
- J. H. Waite, X. X. Qin and K. J. Coyne, Matrix Biol., 1998, 17, 93–106 CrossRef CAS PubMed.
- K. J. Coyne, X. X. Qin and J. H. Waite, Science, 1997, 277, 1830–1832 CrossRef CAS PubMed.
- X. X. Qin, K. J. Coyne and J. H. Waite, J. Biol. Chem., 1997, 272, 32623–32627 CrossRef CAS PubMed.
- M. H. Suhre and T. Scheibel, J. Struct. Biol., 2014, 186, 75–85 CrossRef CAS PubMed.
- M. H. Suhre, M. Gertz, C. Steegborn and T. Scheibel, Nat. Commun., 2014, 5, 3392 Search PubMed.
- M. H. Suhre, T. Scheibel, C. Steegborn and M. Gertz, Acta Crystallogr., Sect. F: Struct. Biol. Commun., 2014, 70, 769–772 CrossRef CAS PubMed.
- A. Hagenau, P. Papadopoulos, F. Kremer and T. Scheibel, J. Struct. Biol., 2011, 175, 339–347 CrossRef CAS PubMed.
- T. Priemel, E. Degtyar, M. N. Dean and M. J. Harrington, Nat. Commun., 2017, 8, 14539 CrossRef PubMed.
- M. J. Harrington, H. S. Gupta, P. Fratzl and J. H. Waite, J. Struct. Biol., 2009, 167, 47–54 CrossRef CAS PubMed.
- ppiczalpha_man.pdf, https://tools.thermofisher.com/content/sfs/manuals/ppiczalpha_man.pdf, accessed June 28, 2017.
- D. R. Higgins and J. Cregg, Pichia Protocols, Humana Press, 2008 Search PubMed.
- J. M. Lucas, E. Vaccaro and J. H. Waite, J. Exp. Biol., 2002, 205, 1807–1817 CAS.
- C. A. Scorer, J. J. Clare, W. R. McCombie, M. A. Romanos and K. Sreekrishna, Biotechnology, 1994, 12, 181–184 CAS.
- J. M. Cregg and K. A. Russell, Methods Mol. Biol., 1998, 103, 27–39 CAS.
- K. Sreekrishna, R. H. Potenz, J. A. Cruze, W. R. McCombie, K. A. Parker, L. Nelles, P. K. Mazzaferro, K. A. Holden, R. G. Harrison and P. J. Wood, J. Basic Microbiol., 1988, 28, 265–278 CrossRef CAS PubMed.
- G. P. L. Cereghino, J. L. Cereghino, C. Ilgen and J. M. Cregg, Curr. Opin. Biotechnol., 2002, 13, 329–332 CrossRef PubMed.
- J. Stratton, V. Chiruvolu and M. Meagher, Methods Mol. Biol., 1998, 103, 107–120 CAS.
- B. Brodsky and J. A. M. Ramshaw, in Fibrous Proteins: Structures and Mechanisms, ed. D. A. D. Parry and J. M. Squire, Springer International Publishing, cham, 2017, vol. 82, pp. 601–629 Search PubMed.
- N. J. Greenfield, Nat. Protoc., 2007, 1, 2527–2535 CrossRef PubMed.
- T. Hyashi, S. Curran-Patel and D. J. Prockop, Biochemistry, 1979, 18, 4182–4187 CrossRef CAS PubMed.
- C. Louis-Jeune, M. A. Andrade-Navarro and C. Perez-Iratxeta, Proteins: Struct., Funct., Bioinf., 2012, 80, 374–381 CrossRef CAS PubMed.
- K. Spiess, A. Lammel and T. Scheibel, Macromol. Biosci., 2010, 10, 998–1007 CrossRef CAS PubMed.
- U. Slotta, S. Hess, K. Spieß, T. Stromer, L. Serpell and T. Scheibel, Macromol. Biosci., 2007, 7, 183–188 CrossRef CAS PubMed.
- S. Boudko, S. Frank, R. A. Kammerer, J. Stetefeld, T. Schulthess, R. Landwehr, A. Lustig, H. P. Bächinger and J. Engel, J. Mol. Biol., 2002, 317, 459–470 CrossRef CAS PubMed.
- M. J. Harrington and J. H. Waite, Biomacromolecules, 2008, 9, 1480–1486 CrossRef CAS PubMed.
- F. Bauer and T. Scheibel, Angew. Chem., Int. Ed. Engl., 2012, 51, 6521–6524 CrossRef CAS PubMed.
- G. Lang, H. Herold and T. Scheibel, Subcell. Biochem., 2017, 82, 527–573 Search PubMed.
- D. Huemmerich, T. Scheibel, F. Vollrath, S. Cohen, U. Gat and S. Ittah, Curr. Biol., 2004, 14, 2070–2074 CrossRef CAS PubMed.
- A. Hagenau and T. Scheibel, J. Adhes., 2010, 86, 10–24 CrossRef CAS.
- A. Hagenau, H. A. Scheidt, L. Serpell, D. Huster and T. Scheibel, Macromol. Biosci., 2009, 9, 162–168 CrossRef CAS PubMed.
- J. P. Malone, K. Alvares and A. Veis, Biochemistry, 2005, 44, 15269–15279 CrossRef CAS PubMed.
- Y. Xu, D. R. Keene, J. M. Bujnicki, M. Höök and S. Lukomski, J. Biol. Chem., 2002, 277, 27312–27318 CrossRef CAS PubMed.
- Z. Yu, O. Mirochnitchenko, C. Xu, A. Yoshizumi, B. Brodsky and M. Inouye, Protein Sci., 2010, 19, 775–785 CrossRef CAS PubMed.
- J. L. Fink and N. Hamilton, In Silico Biol., 2007, 7, 145–150 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra04515h |
| This journal is © The Royal Society of Chemistry 2017 |