Open Access Article
Open Access ArticleTwo novel Co(II) and Ni(II) complexes of tebuconazole with enhanced antifungal activities†
Jie Lia,
Yao Zhanga,
Mingyan Yangb and
Haixia Ma *a
*a
aSchool of Chemical Engineering, Northwest University, Xi'an, Shaanxi 710069, China. E-mail: mahx@nwu.edu.cn; Fax: +86-029-88307755; Tel: +86-029-88307755
bDepartment of Environmental Science and Engineering, Chang'an University, Xi'an, Shaanxi 710054, China
First published on 30th June 2017
Abstract
Two transition metal complexes, [CoL4Cl2]·4MeOH 1 and [NiL4Cl2]·4MeOH 2 (L = (RS)-1-(4-chloro-phenyl)-4,4-dimethyl-3-(1,2,4-triazole-1-ylmethyl)pentane-3-ol), tebuconazole were synthesized and their structures were determined using single crystal X-ray diffraction (XRD). Crystal structural analysis shows that complexes 1 and 2 have similar structures, both with the metal cation lying on a crystallographic inversion center and coordinated with four triazole groups and two chloride anions. The antifungal activities of L and its complexes against four selected plant pathogenic fungi were evaluated. The results show that both complexes have stronger bioactivities than the ligand L and that complex 2 has slightly higher bioactivities than complex 1. To elucidate the mechanisms behind the increased antifungal activities of the title complexes in comparison with L, cumulative release studies in static water and theoretical investigations of the complexes were carried out. The results indicate that there are three factors contributing to the enhanced bioactivities: attractive controlled release properties, synergic interaction between metal cations and L, and improved penetration into the lipid membranes.
1 Introduction
Although complexes containing transition metals and multidentate ligands have been extensively investigated over the past decades due to their unique structures1–3 and potential applications in different fields,4–8 studies on the complexation of commercial 1,2,4-triazole fungicides with transition metals are still rare. Evans reported the formation and structures of CuAc2 and ZnAc2 complexes with tebuconazole and propiconazole.9 Yang reported the synthesis, controlled release properties and antifungal activities of a series of metal complexes with cis and trans propiconazole.10 Fujii prepared difenoconazole copper complexes and used them as wood preservatives.11 Our group reported the structures and antifungal activities of two Cu(II) complexes with triadimefon12 and four Cu(II) complexes with tebuconazole.13 In the literature, it has been shown that the pharmacological and toxicological properties of many drugs are modified when they form metallic complexes.10–15 Complexation of pesticides with metals has potential advantages including the enhancement of persistence, longer shelf life, reduction of mammalian toxicity and conversion of pesticides from non-systemic to systemic.16–18 Moreover, the pesticides coordinated with transition metals could be utilized for controlled release formulations with the capacity of alleviating the toxicity and decreasing the pesticide residue.10,19Tebuconazole (C16H22ClN3O, CASRN: 107534-96-3), chemically named (RS)-1-(4-chloro-phenyl)-4,4-dimethyl-3-(1,2,4-triazole-1-ylmethyl)pentane-3-ol, is an important 1,2,4-triazole fungicide with broad bactericidal spectrum, high antibacterial efficiency and low toxicity, which has been widely used to control mildews and rusts of cereal grains, fruits, vegetables and ornamentals by inhibiting the synthesis of ergosterol to prevent fungal mycelium development.20–22 However, the intensive use of tebuconazole and their single active site have led to substantial environmental contamination and a rapid selection of resistance strains.23–25 Given the limitations of tebuconazole and with the aim of possibly improving its properties and potential applications, a series of metal complexes with tebuconazole have been studied in our lab.
In an earlier study, we reported four Cu(II) complexes of tebuconazole13 and found that the obtained copper complexes exhibited potent antifungal activity toward the selected fungi Gibberella nicotiancola, Botryosphaeria berengriana, Botryosphaeria ribis, Alternaria solani. To continue our work on the complexes with tebuconazole and study the impact of different metal cations (Co2+ and Ni2+) on the crystal structures and the bioactivities, in this work, we have conducted experimental and theoretical investigations on the molecular and electronic structures of [CoL4Cl2]·4MeOH 1 and [NiL4Cl2]·4MeOH 2. Meanwhile, the antifungal activities of these two complexes against four selected plant pathogenic fungi and the possible reasons for the increased bioactivities after complexation are also discussed.
2 Results and discussion
2.1 Synthesis and IR spectroscopy
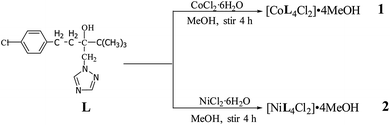
As shown in Scheme 1, reactions of tebuconazole with the inorganic metal salts CoCl2·6H2O and NiCl2·6H2O under the same reaction conditions led to similar mononuclear 1,2,4-triazole complexes with four ligands, two chloride anions and four free methanol molecules, namely, [CoL4Cl2]·4MeOH 1 and [NiL4Cl2]·4MeOH 2.The IR spectra of the complexes 1 and 2 show that both the complexes display characteristic bands of alcoholic hydroxyl groups and the alcoholic hydroxyl ν(O–H) stretching vibrations in the IR spectrums are present at 3288 cm−1 and 3291 cm−1, respectively. The strong absorption bands at 1491 cm−1 and 1492 cm−1 in the IR spectra of 1 and 2, respectively attribute to the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) stretching vibration. The bands observed in the 3134–2876 cm−1 region are assigned to the C–H stretching vibrations.
C) stretching vibration. The bands observed in the 3134–2876 cm−1 region are assigned to the C–H stretching vibrations.
2.2 Description of structures
As shown in Fig. 1b, there is one kind of intramolecular hydrogen bond O4–H4⋯O2 in the crystal structure of complex 1, which is generated by the O4 atom of the free methanol molecule as hydrogen bond donor to O2 of the ligand. And there are three kinds of intermolecular hydrogen bonds: O1 and O3 atoms act as hydrogen bond donors to Cl3 atom from the adjacent molecule, generating O1–H1⋯Cl3ii and O3–H3C⋯Cl3ii (symmetry code: ii −x, −y, 1 − z). Meanwhile, O2 atom also as hydrogen bond donor takes part in the hydrogen bonds of O2–H2⋯O3iii (symmetry code: iii 1 + x, y, z). The intermolecular hydrogen bonds contribute to the formation of a one-dimensional chain along the b-axis in the crystal structure of complex 1. Furthermore, the 1D chains are connected by van der Waals forces to give rise to the 3D framework along the a-axis, as shown in Fig. 1c.
Unlike complex 1, complex 2 only has intermolecular hydrogen bonds: O2 and O3 atoms act as hydrogen bond donors to Cl3 atom from the adjacent molecule, generating O2–H2⋯Cl3ii and O3–H3⋯Cl3ii (symmetry code: ii 1 − x, 1 − y, 1 − z). O1 atom as hydrogen bond donor takes part in hydrogen bond of O1–H1⋯O3iii (symmetry code: iii −1 + x, y, z). Meanwhile, O1 atom acts as hydrogen bond acceptor to O4 atom from the other adjacent molecule, generating O4–H4⋯O1iv (symmetry code: iv 1 + x, y, z). Each structural unit of [NiL4Cl2]·4MeOH is bound to two adjacent structural units by the intermolecular hydrogen bonds resulting in the formation of a one-dimensional chain along the b-axis (Fig. 2b). The 1D chains are also connected by van der Waals forces to form the 3D framework along the a-axis, as shown in Fig. 2c.
Structure refinement details of both the complexes are summarized in Table 1. Selected bond lengths and angles are listed in Table 2, and hydrogen bonds are shown in Table 3.
| Parameter | 1 | 2 |
|---|---|---|
| Empirical formula | C68H104Cl6CoN12O8 | C68H104Cl6NiN12O8 |
| Formula weight | 1489.26 | 1489.04 |
| Crystal size (mm) | 0.31 × 0.27 × 0.24 | 0.32 × 0.25 × 0.21 |
| Crystal system | Triclinic | Triclinic |
| Space group | ![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) 1 1 |
![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) 1 1 |
| a/(Å) | 9.427(3) | 9.350(3) |
| b/(Å) | 14.221(5) | 14.154(4) |
| c/(Å) | 15.414(5) | 15.270(4) |
| α/(°) | 92.275(6) | 92.110(6) |
| β/(°) | 95.615(6) | 95.644(6) |
| γ/(°) | 101.926(6) | 101.787(5) |
| V/(Å3) | 2008.3(12) | 1965.3(10) |
| Z | 1 | 1 |
| Dc (g cm−3) | 1.231 | 1.258 |
| μ | 0.469 | 0.508 |
| F(000) | 789 | 790 |
| Reflns. collected | 9940 | 9918 |
| Unique/observed | 7009/3305 | 6914/2733 |
| R1(I ≥ 2σ(I)) | 0.0753 | 0.0830 |
| wR2 (I ≥ 2σ(I)) | 0.1934 | 0.1820 |
| Goodness-of-fit on F2 | 0.957 | 0.933 |
| 1 | 2 | ||
|---|---|---|---|
| a Symmetry codes: complex 1 (i) 1 − x, −y, 1 − z. Complex 2 (i) −x, 1 − y, 1 − z. | |||
| Co–N1 | 2.153(4) | Ni–N1 | 2.080(6) |
| Co–N1i | 2.153(4) | Ni–N1i | 2.080(6) |
| Co–N4 | 2.141(5) | Ni–N4 | 2.094(5) |
| Co–N4i | 2.141(5) | Ni–N4i | 2.094(5) |
| Co–Cl3 | 2.5508(16) | Ni–Cl3 | 2.4979(18) |
| Co–Cl3i | 2.5508(16) | Ni–Cl3i | 2.4979(18) |
| N1–Co–N1i | 180.000(1) | N1–Ni–N1i | 180.000(1) |
| N4–Co–N4i | 180.000(1) | N4–Ni–N4i | 180.0(3) |
| Cl3–Co–Cl3i | 180.0 | Cl3–Ni–Cl3i | 180.0 |
| N1–Co–N4 | 90.38(17) | N1–Ni–N4 | 90.5(2) |
| N1–Co–Cl3 | 89.85(13) | N1–Ni–Cl3 | 89.32(16) |
| D–H⋯A | D–H | H⋯A | D⋯A | ∠D–H⋯A |
|---|---|---|---|---|
| a Symmetry code: complex 1 (ii) −x, −y, 1 − z; (iii) 1 + x, y, z; complex 2 (ii) 1 − x, 1 − y, 1 − z; (iii) −1 + x, y, z; (iv) 1 + x, y, z. | ||||
| Complex 1 | ||||
| O4–H4⋯O2 | 0.820 | 2.407 | 2.848 | 114.67 |
| O2–H2⋯O3ii | 0.820 | 1.933 | 2.735 | 165.71 |
| O1–H1⋯Cl3iii | 0.820 | 2.496 | 3.290 | 163.45 |
| O3–H3C⋯Cliii | 0.820 | 2.328 | 3.115 | 161.21 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Complex 2 | ||||
| O2–H2⋯Cl3ii | 0.820 | 2.477 | 3.266 | 161.76 |
| O3–H3⋯Cl3ii | 0.820 | 2.283 | 3.086 | 166.07 |
| O1–H1⋯O3iii | 0.820 | 1.915 | 2.725 | 169.16 |
| O4–H4⋯O1iv | 0.820 | 2.279 | 2.837 | 125.67 |
2.3 Biological activities
| Compound | G. nicotiancola | B. berengriana | B. ribis | A. solani |
|---|---|---|---|---|
| 1 | 1.65 | 2.87 | 1.01 | 1.33 |
| 2 | 1.22 | 2.60 | 0.72 | 1.02 |
| L | 2.50 | 11.9 | 1.75 | 3.54 |
| CoCl2·6H2O | 45.9 | 39.9 | 34.5 | 73.4 |
| NiCl2·6H2O | 41.5 | 39.5 | 31.8 | 76.3 |
It is noteworthy that the interaction levels of complex 2 were higher than those of complex 1 for the same tested fungus. It indicates that the synergy levels for the molecular-level mixture of Ni2+ and tebuconazole were better than those for the molecular-level mixture of Co2+ and tebuconazole. This also leads to slightly higher bioactivities for complex 2 than complex 1 against the selected fungi. We can also obtain from Fig. 4 that both the synthesized complexes show different synergy levels to the different tested fungi. The synergy levels concerning the tested fungi is in the sequence of B. berengriana > A. solani > B. ribis > G. Nicotiancola, which implies that the antifungal activities against B. berengriana are improved to the greatest extent, and against G. Nicotiancola to the smallest extent after complexation.
2.4 Controlled release properties
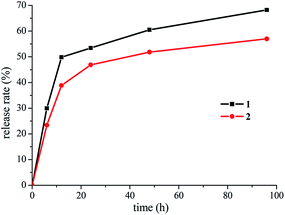
The title complexes can gradually release L in the static water as shown in Fig. 5, suggesting that they were unstable in their coordinating mode in the aqueous solution. The releases of L from complexes 1 and 2 showed different rates yet similar profiles. The release rate pattern indicated that L from complex 1 was released more quickly than from complex 2. For example, after 24 and 96 h in static water, complex 1 released 53.4 and 68.2% of L, respectively, while complex 2 released 46.9, and 57.0% of L, respectively.The reason for the above difference could be related to the different stabilities of the complexes in water: the coordination bonds in complex 1 are weaker (the bond lengths of Co–N and Co–Cl in complex 1 are longer than those of Ni–N and Ni–Cl in complex 2), which led to complex 1 being more unstable in water. The release rate of L from both the complexes decreased dramatically after about 20 h. This result suggests the possibility to develop the title complexes as new medicament for slower controlled release.
2.5 DFT calculations
The plots of HOMO, LUMO and the energy gaps (ΔE = ELUMO − EHOMO) of complexes 1 and 2 are shown in Fig. 6. The modes in each HOMO and LUMO are symmetrically placed as illustrated in Fig. 6. The HOMO of complex 1 is localized predominantly on Co cation, two Cl anions and two coordinated triazole rings, and the LUMO of complex 1 is localized predominantly on Co cation and two Cl anions. The HOMO and LUMO of complex 2 are very similar and localized predominantly on Ni cation, two Cl anions and four coordinated triazole rings. These results imply that the metal cations, triazole rings and coordinated Cl anions may be the reactive sites of the two complexes. The energy gaps in complex 1 and complex 2 are found to be much less (2.357 eV and 1.875 eV, respectively) than that of L (4.326 eV), which indicates the complexes have lower chemical activity than the ligand, and complex 2 is more reactive than complex 1.
| Atoms | Atomic charges (e) | f+k | f−k | f0k | |
|---|---|---|---|---|---|
| 1 | Co | 0.722 | 0.263 | 0.134 | 0.199 |
| N1 | −0.374 | 0.012 | 0.008 | 0.010 | |
| C2 | 0.191 | 0.007 | 0.005 | 0.006 | |
| N3 | −0.234 | 0.016 | 0.014 | 0.015 | |
| N2 | −0.132 | 0.025 | 0.023 | 0.024 | |
| C1 | 0.230 | 0.011 | 0.007 | 0.009 | |
| Cl3 | −0.631 | 0.106 | 0.161 | 0.133 | |
| 2 | Ni | 0.759 | 0.187 | 0.093 | 0.140 |
| N1 | −0.378 | 0.023 | 0.027 | 0.025 | |
| C2 | 0.178 | 0.005 | 0.006 | 0.006 | |
| N2 | −0.131 | 0.010 | 0.010 | 0.010 | |
| N3 | −0.228 | 0.009 | 0.0100 | 0.009 | |
| C1 | 0.223 | 0.008 | 0.008 | 0.008 | |
| Cl3 | −0.656 | 0.119 | 0.173 | 0.146 | |
| L | N1 | −0.344 | 0.010 | 0.006 | 0.008 |
| C2 | 0.170 | 0.007 | 0.003 | 0.005 | |
| N2 | −0.150 | 0.003 | 0.000 | 0.001 | |
| N3 | −0.227 | 0.007 | 0.003 | 0.005 | |
| C1 | 0.200 | 0.009 | 0.002 | 0.006 |
Based on the analysis above, it can be observed that the results of HOMO–LUMO, Fukui analysis and Mulliken atomic charge calculations are consistent, which indicate that the metal cations, coordinated anions and triazole rings should be the active sites of the complexes. However, previous bioactivity results show that the antifungal activities have poor correlation with the coordinated anions,12,13 thus it may be concluded that the Co2+ or Ni2+ cations and the triazole rings originating from the ligand should be the main active sites inhibiting the growth of fungi.
2.6 Discussion of reasons for enhanced antifungal activities
As a result of the controlled release properties, the complexes release L step by step. The complexes can have a positive effect against the plant pathogenic fungi in a longer residual period and show different bioactivities from their ligand, which can affect the antifungal activities of the complexes. It can be further proved by the fact that complex 2 shows higher bioactivities with slower controlled release rate in comparison with complex 1. Thereby, the controlled release properties should be one of the reasons for the enhanced bioactivities of metal complexes.The results of DFT calculations and antifungal activities indicate that the metal cations and the triazole ring are the main active sites of the complexes against the plant fungi. While it is well known that only the triazole ring is the active site of the ligand L, which is also responsible for a rapid selection of resistance strains of L. Thus we can infer that the increased active site of the metal cations and the synergic interactions between the metal cations and L should be another reason for the increased bioactivities of the complexes.
Tebuconazole belongs to the ergosterol biosynthesis inhibitors, which can impair the pathogenic ability of plant fungi by destroying the structure of the cell membrane. As a result of Mulliken atomic charge calculations, the polarity of the atoms in the complexes is reduced, which is helpful for the enhanced penetration of the metal complexes into the lipid membranes and consequently enhance the bioactivities. The result is in accord with chelation theory.10,41,42
Therefore, the controlled release properties, the synergetic interactions between the metal cations and L, as well as the enhanced penetration of the metal complexes into the lipid membranes are three main reasons for the enhanced antifungal activities after complexation.
3 Experimental section
3.1 Materials and general methods
Tebuconazole was purchased from commercial source and recrystallized by isopropyl alcohol solvent. IR (KBr, σ/cm−1): 3300 (m), 3140 (w), 2973 (s), 2876 (w), 1510 (vs), 1490 (vs), 1009 (s), 735 (m). All other reagents were of reagent grade and used as received. And the crystal samples of the complexes were used for all the tests. Elemental analysis was performed on a Vario EL III elemental analyzer. IR spectrum was carried out on EQUINX 55 with KBr presser bit. X-ray diffraction data were collected on a Bruker SMART APEX CCD diffractometer. Release properties were measured by high performance liquid chromatography HPLC type shimadzu LC-20A, infusion pump type LC-20ATvp, detector type SPD-20Avp, the mobile phase was methanol and water with 80% (V/V).3.2 Procedures and synthesis of the complexes 1 and 2
3.3 Crystal structure determination
Single-crystal X-ray diffraction (XRD) measurements of 1 and 2 were carried out on a Bruker SMART APEX CCD diffractometer equipped with a graphite monochromator using Mo Kα radiation (0.71073 Å) at 296(2) K in the F–ω scan mode. Unit cell dimensions were obtained with least-squares refinements and semi-empirical absorption corrections were applied using the SADABS program.45 The structures were solved by direct methods and refined by full-matrix least squares techniques based on F2 with the SHELXTL program.46 All non-hydrogen atoms were obtained from the difference Fourier map and refined with atomic anisotropic thermal parameters. The hydrogen atoms were added according to the theoretical models. Crystallographic data for structures 1 and 2 have been deposited at the Cambridge Crystallographic Data Centre as supplementary publication CCDC 934445 and 934446.†3.4 Biological activities tests
Four important phytopathogens (Gibberella nicotiancola, Botryosphaeria berengriana, Botryosphaeria ribis and Alternaria solani) were provided by Shaanxi Microbiology Institute, China and selected for antifungal activity studies. The antifungal activities of the ligand L, the title complexes and the metal salts CoCl2·6H2O, NiCl2·6H2O were diluted by starch and ground into dust, then added to potato dextrose agar (PDA) medium, respectively, to obtain a range of concentrations (1, 2, 4 and 8 mg L−1) before pouring into the Petri dishes (7.5 cm in diameter).10,47,48Each concentration was tested in triplicate. Parallel controls were maintained with starch mixed with PDA medium. The diameter of fungal colonies on PDA plates was measured after 72 h. Percentage inhibition of mycelial growth was calculated using the formula (1). Because the synthesized complexes can be regarded as molecular-level mixtures of L and inorganic salts, synergy ratios (SR) were calculated to investigate the extent of the interactions between L and inorganic salts according to Wadley approach32 using the formulas (2) and (3).
 | (1) |
| EC50 (expected) = (a + b)/(a/ECA50 + b/ECB50) | (2) |
| SR = EC50 (expected)/EC50 (observed) | (3) |
3.5 Release assay
The release properties were carried out according to the ref. 10. Both the complexes (25 mg each sample) were placed, respectively, in 25 mL of distilled water in a beaker with film to minimize loss of water by evaporation and then allowed to stand at 28 °C. After the desired time had elapsed (6, 12, 24, 48 and 96 h), L released in each beaker was extracted with ether (30 mL). After ether was completely volatilized, the extract was diluted to 25 mL methanol in a 25 mL volumetric flask. The extracts were analyzed for L content by HPLC using calibration curves obtained from standard samples.3.6 Computational method
The entire calculations were carried out with the DMol3 software,49 which is based on the density functional theory (DFT). Symmetry operations were applied for all structures. The generalized gradient approximation (GGA) of Perdew, Burke, and Ernzerhof (PBE) exchange–correlation functional50 was used. Double numerical basis sets including polarization functions (DNP)51,52 were performed to describe the valence orbitals of all the atoms in our calculations. To describe the cores, all-electron relativistic calculations were used.4 Conclusion
Two new complexes of the fungicide tebuconazole, [CoL4Cl2]·4MeOH (1) and [NiL4Cl2]·4MeOH (2), have been synthesized and characterized by elemental analysis, IR spectroscopy and single crystal XRD. The two complexes have similar structural geometry and both form distorted octahedron around the metal atoms. And the one-dimension chains of the complexes are formed by different kinds of hydrogen bonds. The bioactivities tests reveal that the obtained complexes are more efficient to inhibit the four selected fungi relative to the ligand tebuconazole, which indicates the potential applications of these complexes in the fields as antifungal agents. To elucidate the possible mechanism behind the increased antifungal activities, the controlled release properties and the synergistic interactions between Co2+, Ni2+ and tebuconazole, were studied, in conjunction with theoretical investigations of the electronic structures of the complexes. The results show that the good controlled release properties, the synergic interactions between the metal cations and tebuconazole, as well as the greater penetration through the cell membrane of microorganisms all contribute to the enhanced biocidal properties. However, because of the complexity of biological systems and the limited condition of our laboratory, it is rather difficult to stipulate the detailed mechanism for the antifungal activity using the tebuconazole complexes, which may be conducted in future work.Acknowledgements
This work is funded by the Provincial Natural Science Foundation of Shaanxi, China (Grant No. 2016JZ003). The authors would like to thank Prof. J.Z. Zhang (Department of Chemistry and Biochemistry, University of California, Santa Cruz, USA) for help in revising the manuscript.Notes and references
- S. S. Han, L. L. Shi, K. Li, S. Zhao, B. L. Li and B. Wu, RSC Adv., 2015, 5, 107166–107178 RSC.
- M. H. Klingele, A. Noble, P. D. W. Boyd and S. Brooker, Polyhedron, 2007, 26, 479–485 CrossRef CAS.
- X. R. Meng, Y. L. Song, H. W. Hou, H. Y. Han and B. Xiao, Inorg. Chem., 2004, 43, 3528–3536 CrossRef CAS PubMed.
- P. P. Xiong, J. Li, H. Y. Bu, Q. Wei, R. L. Zhang and S. P. Chen, J. Solid State Chem., 2014, 215, 421–435 CrossRef.
- L. Habalaad, C. Bartelb, G. Giesterc, M. A. Jakupecb, B. K. Kepplerb and A. Rompela, J. Inorg. Biochem., 2015, 147, 147–152 CrossRef PubMed.
- B. Q. Zou, X. Lu, Q. P. Qin, Y. X. Bai, Y. Zhang, M. Wang, Y. C. Liu, Z. F. Chen and H. Liang, RSC Adv., 2017, 7, 17923–17933 RSC.
- N. Boechat, V. F. Ferreira, S. B. Ferreira and M. L. G. Ferreira, J. Med. Chem., 2011, 54, 5988–5999 CrossRef CAS PubMed.
- J. M. Liu, L. Wang, K. Yu, Z. H. Su, C. X. Wang, C. M. Wang and B. B. Zhou, New J. Chem., 2015, 39, 1139–1147 RSC.
- P. D. Evans, K. J. Schmalzl, C. M. Forsyth, G. D. Fallon, S. Schmid, B. Bendixen and S. Heimdal, J. Wood Chem. Technol., 2007, 27, 243–256 CrossRef CAS.
- X. Chen and C. L. Yang, J. Agric. Food Chem., 2009, 57, 2441–2446 CrossRef CAS PubMed.
- K. Fujii, K. Hatano and Y. Kanetsuki, Preparation of difenoconazole copper salts and their use as wood preservatives, JP 2006273776A, 2006-10-12.
- J. Li, T. Xi, B. Yan, M. Y. Yang, J. R. Song and H. X. Ma, New J. Chem., 2015, 39, 6997–7003 RSC.
- J. Li, T. Xi, B. Yan, Y. L. Guan, M. Y. Yang, J. R. Song and H. X. Ma, New J. Chem., 2015, 39, 9550–9556 RSC.
- Z. H. Chohan, S. H. Sumrra, M. H. Youssoufi and T. B. Hadda, Eur. J. Med. Chem., 2010, 45, 2739–2747 CrossRef CAS PubMed.
- A. K. Sadana, Y. Mirza, K. R. Aneja and O. Prakash, Eur. J. Med. Chem., 2003, 38, 533–536 CrossRef CAS PubMed.
- Y. H. Wang, D. Y. Yu, P. Xu, B. Y. Guo, Y. F. Zhang, J. Z. Li and H. L. Wang, Ecotoxicol. Environ. Saf., 2014, 107, 276–283 CrossRef CAS PubMed.
- M. Kamiya and K. Kameyama, Chemosphere, 2001, 45, 231–235 CrossRef CAS PubMed.
- E. Morillo, T. Undabeytia, C. Maqueda and A. Rams, Chemosphere, 2002, 47, 747–752 CrossRef CAS PubMed.
- P. Z. Zhang, Q. Y. Fu, R. X. Chi, C. X. Yang and J. G. Xu, J. Zhejiang Univ. Sci. Technol., 2003, 15, 142–145 Search PubMed , (in Chinese).
- Z. Kang, L. Huang, U. Krieg, A. Maulermachnik and H. Buchenauer, Pest Manage. Sci., 2001, 57, 491–500 CrossRef CAS PubMed.
- J. P. Zubrod, M. Bundschuh, A. Feckler, D. Englert and R. Schulz, Environ. Toxicol. Chem., 2011, 30, 2718–2724 CrossRef CAS PubMed.
- C. Wang, F. Wang, Q. Zhang and W. Liang, Eur. J. Soil Biol., 2016, 72, 6–13 CrossRef CAS.
- R. Akallal, D. Debieu, C. Lanen, M. J. Daboussi, R. Fritz, C. Malosse, J. Bach and P. Leroux, Pestic. Biochem. Physiol., 1998, 60, 147–166 CrossRef CAS.
- R. D. Horsley, J. D. Pederson, P. B. Schwarz, K. McKay, M. R. Hochhalter and M. P. McMullen, Agron. J., 2006, 98, 194–197 CrossRef CAS.
- L. Lucini and G. P. Molinari, Pest Manage. Sci., 2009, 65, 440–443 CrossRef CAS PubMed.
- I. D. Brown and D. Altermatt, Acta Crystallogr., Sect. B: Struct. Sci., 1985, 41, 244–247 CrossRef.
- S. Gao, X. F. Zhang, L. H. Huo, Z. Z. Lu, H. Zhao and J. G. Zhao, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2004, 60, 1128–1130 Search PubMed.
- J. Matsumoto, T. Suzuki, Y. Kajita and H. Masuda, Dalton Trans., 2012, 41, 4107–4117 RSC.
- F. D. Salvo, F. Teixidor, C. Viñas, J. G. Planas, M. E. Light, M. B. Hursthouse and N. Aliaga-Alcalde, Cryst. Growth Des., 2012, 12, 5720–5736 Search PubMed.
- Y. Q. Huang, B. Ding, H. L. Gao, P. Cheng, D. Z. Liao, S. P. Yan and Z. H. Jiang, J. Mol. Struct., 2005, 743, 201–207 CrossRef CAS.
- S. L. Gao, Q. Yang, J. L. Li, G. Fan, X. X. Li, L. P. Chen, X. T. Dang and Z. Y. Wu, Periodic Table of Chemical Element, Science Press, Beijing, 3rd edn, 2012, (in Chinese) Search PubMed.
- F. M. Wadley, Experimental Statistics in Entomology, Graduate School Press, Washington State University, Pullman, 1976 Search PubMed.
- J. X. Mu, Z. W. Zhai, M. Y. Yang, Z. H. Sun, H. K. Wu and X. H. Liu, Crystals, 2016, 6, 4 CrossRef.
- M. Larif, A. Adad, R. Hmammouchi, A. I. Taghki, A. Soulaymani, A. Eimidaoui, M. Bouachrine and T. Lakhlifi, Arabian J. Chem., 2017, 10, S946–S955 CrossRef CAS.
- J. X. Mu, Y. X. Shi, M. Y. Yan, Z. H. Sun and X. H. Liu, Molecules, 2016, 21, 68 CrossRef PubMed.
- W. Boufas, N. Dupont, M. Berredjem, K. Berrezag, I. Becheker, H. Berredjem and N. E. Aouf, J. Mol. Struct., 2014, 1074, 180–185 CrossRef CAS.
- J. Aihara, J. Phys. Chem. A, 1999, 103, 7487–7495 CrossRef CAS.
- P. Govindasamy, S. Gunasekaran and S. Srinivasan, Spectrochim. Acta, Part A, 2014, 130, 329–336 CrossRef CAS PubMed.
- K. C. Zheng, J. P. Wang, W. L. Peng, Y. Shen and F. C. Yun, Inorg. Chim. Acta, 2002, 328, 247–253 CrossRef CAS.
- A. Chaudhary, N. Bansal, A. Gajraj and R. V. Singh, J. Inorg. Biochem., 2003, 96, 393–400 CrossRef CAS PubMed.
- C. R. A. María, C. L. S. Estefania, S. Jesús, P. Pelagatti and F. Zani, J. Inorg. Biochem., 2005, 99, 2231–2239 CrossRef PubMed.
- K. Fukui, T. Yonezzawa and H. Shingui, J. Chem. Phys., 1952, 20, 722–725 CrossRef CAS.
- C. J. Brala, I. Fabijanić, A. K. Marković and V. Pilepić, Comput. Theor. Chem., 2014, 1049, 1–6 CrossRef.
- S. Saravanan and V. Balachandran, Spectrochim. Acta, Part A, 2014, 130, 604–620 CrossRef CAS PubMed.
- G. M. Sheldrick, SADABS, University of Göttingen, Germany, 2000 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 CrossRef CAS PubMed.
- D. Saetae and W. Suntornsuk, J. Microbiol. Biotechnol., 2010, 20, 319–324 CAS.
- R. K. Devappaa, S. K. Rajeshb and V. Kumara, Ecotoxicol. Environ. Saf., 2012, 78, 57–62 CrossRef PubMed.
- B. Delley, Phys. Rev. B: Condens. Matter Mater. Phys., 2002, 65, 85403–85409 CrossRef.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865 CrossRef CAS PubMed.
- B. Delley, J. Phys. Chem., 1996, 100, 6107–6110 CrossRef CAS.
- W. J. Hehre, L. Radom, P. V. R. Schlyer and J. A. Pople, Ab Initio Molecular Orbital Theory, Wiley, New York, 1986 Search PubMed.
Footnote |
| † CCDC 934445 1 and 934446 2. For crystallographic data in CIF or other electronic format see DOI: 10.1039/c7ra03629a |
| This journal is © The Royal Society of Chemistry 2017 |