Open Access Article
Open Access ArticleLuminescent detection of the lipopolysaccharide endotoxin and rapid discrimination of bacterial pathogens using cationic platinum(II) complexes†
Yiwen Zhuabc,
Chen Xuabc,
Yu Wangabc,
Yaqing Chend,
Xiaokang Ding *abc and
Bingran Yu*abc
*abc and
Bingran Yu*abc
aState Key Laboratory of Chemical Resource Engineering, Beijing University of Chemical Technology, Beijing 100029, China. E-mail: yubr@mail.buct.edu.cn; dingxk@mail.buct.edu.cn; Tel: +86-10-64421243
bKey Laboratory of Carbon Fiber and Functional Polymers, Beijing University of Chemical Technology, Ministry of Education, Beijing 100029, China
cBeijing Laboratory of Biomedical Materials, Beijing Advanced Innovation Center for Soft Matter Science and Engineering, Beijing University of Chemical Technology, Beijing 100029, China
dAffiliated Hospital of Hebei University, Baoding 071000, China
First published on 27th June 2017
Abstract
A luminescence probe based on chloroplatinum(II) complexes of 2,6-bis(benzimidazol-2′-yl)pyridine with hexaethylene glycol methyl ether groups ([Pt(N^N^N)Cl]+) was reported for sensing of the lipopolysaccharide (LPS) endotoxin and rapid discrimination of Gram-negative and Gram-positive bacterial pathogens. [Pt(N^N^N)Cl]+ can be dissolved in aqueous solution with minimal luminescence emission. In the presence of LPS, [Pt(N^N^N)Cl]+ binds to negatively charged LPS to form LPS–Pt(II) aggregates. The formation of LPS–Pt(II) aggregates enhances the intermolecular Pt⋯Pt and π–π stacking interactions, and consequently leads to luminescence emission centered at 650 nm due to triplet metal-to-metal-to-ligand charge-transfer (3MMLCT). The limit of detection (LOD) of LPS is 5.7 nM. We also demonstrate a proof-of-concept application of [Pt(N^N^N)Cl]+ for rapid and washing-free discrimination of Gram-negative Escherichia coli and Gram-positive Staphylococcus aureus within 5 min. The above features of [Pt(N^N^N)Cl]+ make it a promising sensor for clinical applications in the detection of endotoxins and discrimination of bacterial pathogens.
1. Introduction
Lipopolysaccharide (LPS) is a common endotoxin which can be found in the outer membrane of Gram-negative bacteria.1 As an exogenous pyrogen, LPS strongly binds to the membrane receptor proteins such as CD14, TLR4, and MD2, and triggers strong immune responses in animals.2,3 As a result, the detection of the lipopolysaccharide endotoxin is important in the manufacturing of pharmaceutical products, medical devices and the food industry.4 Currently, the most prevalent LPS test relies on the limulus amebocyte lysate (LAL) assay, where the LAL is extracted from the blood of horseshoe crabs. However, the LAL assay is expensive, and the result can be either inhibited5 or enhanced6 especially in complicated biological samples. Besides, the population of horseshoe crabs is endangered because of over-fishing and environmental pollution. To overcome the above limitations, several analytical methods have been developed, such as gas chromatography-mass spectrometry (GC-MS),7 the enzyme-linked immunosorbent assay (ELISA),8 and piezoelectric biosensors.9 More recently, several turn-on fluorescent probes have been reported to detect LPS in serum or in urine samples. For example, Lei and coworkers employed a pair of anionic and cationic fluorescent dyes to detect LPS in aqueous solutions.10 In the absence of LPS, the anionic fluorophore of 9,10-anthracenediyl-bis(methylene)dimalonic acid (AMDA) and cationic fluorophore of coralyne formed complex via electrostatic attraction and π–π interaction, and the fluorescence of AMDA is selectively quenched due to ground charge transfer, excited state electron transfer, and energy transfer. Upon addition of LPS, the negatively charged LPS tended to facilitate the aggregation of positively charged coralyne molecules, and meanwhile dissociated the AMDA/coralyne complex. Therefore, the fluorescence of coralyne was significantly quenched and the fluorescence of AMDA restored, resulting in detectable changes in their fluorescent profiles. Alternatively, the magnetic nanoparticles were functionalized with perylene diimide (PDI) conjugated peptide which selectively binds to LPS.11 At first, the fluorescence of PDI was self-quenched due to the strong π–π stacking interaction. In the presence of LPS, the binding of LPS would cause conformational changes in the peptide, which subsequently resulted in the disruption of the π–π stacking of PDI molecules and thus their fluorescence was recovered. Although the above studies exhibited adequate sensitivity for detection of LPS, their abilities for detection and discrimination of bacteria were not shown.Bacteria are common pathogens which would cause infectious diseases and foodborne illnesses especially in developing countries where the medical resources and public hygiene are limited.12 As a result, rapid detection and preliminary discrimination of bacterial pathogens are important in the treatment of infectious disease and food safety inspection.13 The conventional methods for pathogen detection include culture and colony counting,14 polymerase chain reaction (PCR),15 and immunoassay-based methods.16,17 However, the above methods are either time-consuming or require tedious operations. For example, the colony counting methods usually take several days to grow the bacteria culture.18 In the recent years, much efforts have been made to achieve rapid detection and discrimination of bacterial pathogens. For example, Jiang et al. synthesized a tetraphenylethylene based fluorescent probe (TPEPyE) containing a cationic pyridinium group.19 The LPS in the outer membrane of Gram-negative bacteria triggers the aggregation of TPEPyE and thus generates fluorescence due to aggregation induced emission. In contrast, the fluorescence cannot be observed in Gram-positive bacteria because of their different envelop structures.
The chloroplatinum(II) complexes of 2,6-bis(benzimidazol-2′-yl)pyridine with hexaethylene glycol methyl ether groups exhibit strong luminescence in the aggregation state due to the Pt⋯Pt and π–π stacking interactions, while the luminescence is minimal in dilute aqueous solution.20 These complexes have attracted much attention due to their luminescence properties and their potential applications in chemical sensing and biosensing.21–24 For example, an alkynylplatinum(II) terpyridine complex has been reported to detect human serum albumin (HSA).22 Herein, we explored employing the chloroplatinum(II) complex ([Pt(N^N^N)Cl]+) as probes for detection of LPS endotoxin and discrimination of bacterial pathogens, as shown in Scheme 1. The positively charged [Pt(N^N^N)Cl]+ tended to complex with negatively charged LPS via electrostatic interactions, forming LPS–Pt(II) aggregates, and led to enhancement in luminescence emission. Afterwards, the [Pt(N^N^N)Cl]+ was employed in a “proof-of-concept” for the discrimination of Gram-negative and Gram-positive bacteria. Since the LPS is a component in the outer membrane of Gram-negative bacteria, they can be easily stained by the formation of LPS–Pt(II) aggregates, and the Gram-negative bacteria can be observed under laser scanning confocal microscope (LSCM). In contrast, the outer cell membrane of Gram-positive bacteria contains a thick layer of peptidoglycan, and the [Pt(N^N^N)Cl]+ cannot form aggregates, and the luminescence emission is minimal.
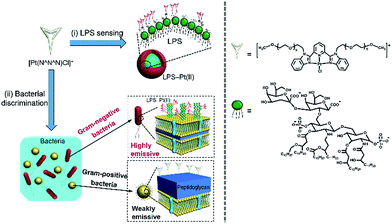 | ||
| Scheme 1 Schematic illustration of the chloroplatinum(II) complex [Pt(N^N^N)Cl]+ for (i) detection of LPS endotoxin, and (ii) discrimination of bacterial pathogens. | ||
2. Experimental
2.1 Materials and instrumentation
The LPS (from Escherichia coli O55:B5, MW 29.5 kDa) was purchased from Sigma-Aldrich. The sodium chloride saline solution (0.9%) was purchased from Shijiazhuang no. 4 Pharmaceutical Co., Ltd. The strains of Gram-negative Escherichia coli (E. coli, JM 109), Gram-positive Staphylococcus aureus (S. aureus, FRF 1169), and Bacillus subtilis (B. subtilis, ATCC 6051) were purchased from Promega (Madison, USA). The chloroplatinum(II) complex ([Pt(N^N^N)Cl]+PF6−) was obtained from Prof. Bu's group (Lanzhou University).20 To increase the solubility in aqueous solution, the counter anions of PF6− in the chloroplatinum(II) complex was exchanged to Cl− by titrating saturated solution of LiCl in acetone. The precipitates of ([Pt(N^N^N)Cl]+Cl−) were thoroughly rinsed with copious of acetone, and collected after drying in vacuum desiccator. Deionized water (18 MΩ cm) was obtained from a Millipore filtration system.2.2 Characterization of LPS–Pt(II) aggregates
The particle size distributions of [Pt(N^N^N)Cl]+ and LPS–Pt(II) aggregates were measured using Zetasizer Nano ZS (Malvern Instruments, Southborough, MA). 1 mL of aqueous solution containing [Pt(N^N^N)Cl]+ (70.1 μM) or a mixture of [Pt(N^N^N)Cl]+ (70.1 μM) and LPS (3.0 μM) was transferred into a cuvette, and the particle size was measured at room temperature. For the atomic force microscope (AFM) imaging, the above solution was spun-coated onto a piece of clean silicon wafer to disperse the LPS–Pt(II) aggregates. After the samples were dried under vacuum, the images were captured using an AFM system (Bruker Dimension Icon, Bruker). For transmission electron microscope (TEM) imaging, the same solution was deposited onto a copper grid (75 mesh), and the images were captured using a TEM system (FEI Company, Hillsboro, OR, 200 kV).2.3 Titration of LPS
The LPS (0.1 mg mL−1 in deionized water) was gradually added into an aqueous solution containing 70.1 μM of [Pt(N^N^N)Cl]+, to achieve an LPS final concentrations ranging from 0 to 6.65 μM. The fluorescence emission spectra of the LPS–Pt(II) aggregates were obtained from a fluorescence spectrophotometer (Hitachi F-7000), with an excitation wavelength of 450 nm.2.4 Discrimination of Gram-negative and Gram-positive bacteria
The sodium chloride saline solution containing Gram-negative E. coli (107 cfu mL−1) or Gram-positive S. aureus (107 cfu mL−1), B. subtilis (107 cfu mL−1) was mixed with [Pt(N^N^N)Cl]+ to achieve a final concentration of 350 μM. After incubating for 5 min, the above solution was transferred into a plastic Petri dish for laser scanning confocal microscope (CLSM, SP8, Leica, Germany) imaging. The excitation wavelength is 448 nm.3. Results and discussion
First, the size distributions of [Pt(N^N^N)Cl]+ before and after forming aggregation with LPS were determined by dynamic light scattering (DLS). Fig. 1a shows that the hydrodynamic diameters of [Pt(N^N^N)Cl]+ complex are 2.1 nm and 233 nm before and after forming aggregates with LPS. The aggregation is caused by the electrostatic interaction between negatively charged LPS and positively charged [Pt(N^N^N)Cl]+. This result is in accordance with the reported value in literature.20 Fig. 1b and c shows the AFM and TEM images of the LPS–Pt(II) aggregates, respectively. The average particle size of the LPS–Pt(II) aggregates obtained from the AFM and TEM images are 123 nm and 137 nm, which are significantly smaller than the hydrodynamic value obtained from DLS. This hydrodynamic value is both contributed by the swelling of the hexaethylene glycol substitutes in the aqueous solution and their abilities to drag a large number of water molecules with it as the particles move through the solution.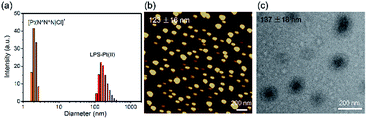 | ||
| Fig. 1 (a) Size distribution of the [Pt(N^N^N)Cl]+ and LPS–Pt(II) aggregates. (b) AFM and (c) TEM images of LPS–Pt(II) aggregates. | ||
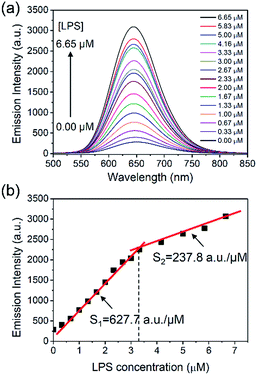
Next, the luminescence emission of the LPS–Pt(II) aggregates was characterized by titration of LPS into a solution containing 70.1 μM of [Pt(N^N^N)Cl]+. Fig. 2a shows the emission spectra of [Pt(N^N^N)Cl]+ after gradual addition of LPS. Before addition of LPS, the [Pt(N^N^N)Cl]+ is weakly emissive in aqueous solution due to the radiationless decay via a low-lying triplet ligand field state.25 However, the emission intensity of the LPS–Pt(II) aggregates increases with increasing concentration of LPS. This emission band at 650 nm is attributed to a triplet metal-to-metal-to-ligand charge-transfer (3MMLCT), which originates from the intermolecular Pt⋯Pt and π–π stacking interactions due to the formation of LPS–Pt(II) aggregates.20 As a blank control, the fluorescence emission of an aqueous solution containing 6.65 μM of LPS is minimal at 650 nm (see the ESI†). Fig. 2b shows the plotting of emission intensity at 650 nm with different LPS concentrations ranging from 0 to 6.65 μM. Intriguingly, we notice a junction point in the diagram at the point of 3.33 μM of LPS. When the concentration of LPS is 3.33 μM or below, the emission intensity increases in a linear relationship with a slope (S1) of 627.7 a.u. μM−1. When the concentration of LPS is 3.33 μM or above, the slope (S2) is 237.8 a.u. μM−1. Although the mechanism of the LPS–Pt(II) coordination is still unknown, this result may imply the binding of [Pt(N^N^N)Cl]+ to LPS is more complicated than other anionic ligands such as sodium dodecyl sulfonate,25 and acrylate-based block polymers.26 The limit of detection (LOD) of LPS is determined to be 5.7 nM (LOD = 3SD/S1, where SD is the standard deviation of 10 measurements of blank samples).4,19 This LOD is significantly lower than the earlier results reported in the literatures.27,28
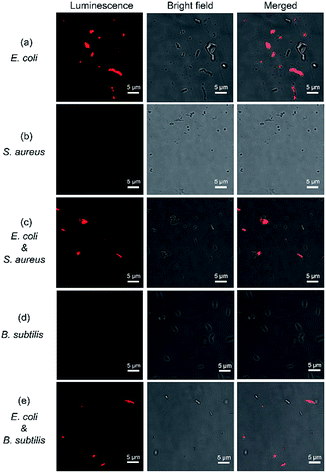
Rapid discrimination of bacterial pathogens is important in clinical diagnosis and establishing therapeutic strategies. Conventionally, the bacteria are divided into two groups by Gram staining based on the different permeability of the dyes against the bacterial cell walls. However, the Gram staining is tedious and time-consuming as it requires multiple washing steps to remove redundant dyes. LPS is abundant only in the outer membrane of Gram-negative bacteria, thus it is an ideal marker to discriminate Gram-negative and Gram-positive bacteria. To explore the potential application of [Pt(N^N^N)Cl]+ for discrimination of bacterial pathogens, the freshly prepared bacteria were mixed with [Pt(N^N^N)Cl]+ and incubated for 5 min. Fig. 3 shows the luminescent, bright field, and merged images of bacteria staining with [Pt(N^N^N)Cl]+. Fig. 3a shows that most of the Gram-negative E. coli have been successfully stained due to the formation of LPS–Pt(II) aggregates on the surface of the cell wall of E. coli. In contrast, the Gram-positive S. aureus were not stained, due to the lack of LPS component on the cell wall (Fig. 3b). It is notable that this method is washing-free and only requires 5 min to obtain final result. Moreover, the Gram-negative E. coli and Gram-positive S. aureus were mixed together, and stained with [Pt(N^N^N)Cl]+. Fig. 3c shows that the bacillus Gram-negative E. coli cells were successfully stained, while the cocci Gram-positive S. aureus cells remain unstained. Next, we explored the feasibility of [Pt(N^N^N)Cl]+ for the discrimination of Gram-negative and Gram-positive bacteria with similar cell morphology. For example, E. coli and B. subtilis are both bacillus bacteria, which cannot be discriminated from their cell morphology. Fig. 3d shows that the [Pt(N^N^N)Cl]+ cannot stain Gram-positive B. subtilis, due to the lack of LPS on the cell wall. When the [Pt(N^N^N)Cl]+ is incubated with a mixture of E. coli and B. subtilis, only the Gram-negative E. coli is stained. These results, when combined, show that the [Pt(N^N^N)Cl]+ can be used for rapid bacterial discrimination.
4. Conclusions
In conclusion, a chloroplatinum(II) complex [Pt(N^N^N)Cl]+ was employed for detection of LPS endotoxin and discrimination of bacterial pathogens. The [Pt(N^N^N)Cl]+ binds to negatively charged LPS to form LPS–Pt(II) aggregates, and consequently leads to luminescence emission due to 3MMLCT. The LOD for LPS is 5.7 nM. As a proof-of-concept, we also demonstrate that the [Pt(N^N^N)Cl]+ can be used to discriminate the Gram-negative and Gram-positive bacteria. This study provides novel approaches for detection of LPS and washing-free discrimination of bacterial pathogens, making the [Pt(N^N^N)Cl]+ a promising probe for clinical applications.Acknowledgements
The authors gratefully acknowledge Dr Jianjun Liang from Lanzhou University for generous providing chloroplatinum(II) complexes of hexaethylene glycol methyl ether substituted 2,6-bis-(benzimidazol-2′-yl)pyridine. This work was partially supported by National Natural Science Foundation of China (NNSFC, grant numbers 21504006, 51573014, and 51503012) and the Fundamental Research Funds for the Central Universities (PYBZ1708, PY201603).Notes and references
- H. Bai, H. Chen, R. Hu, M. Li, F. Lv, L. Liu and S. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 31550–31557 CAS.
- T. Ohnishi, M. Muroi and K.-i. Tanamoto, FEMS Immunol. Med. Microbiol., 2007, 51, 84–91 CrossRef CAS PubMed.
- R. J. Elin, S. M. Wolff, K. P. W. J. McAdam, L. Chedid, F. Audibert, C. Bernard and F. Oberling, J. Infect. Dis., 1981, 144, 329–336 CrossRef CAS PubMed.
- M. Lan, J. Wu, W. Liu, W. Zhang, J. Ge, H. Zhang, J. Sun, W. Zhao and P. Wang, J. Am. Chem. Soc., 2012, 134, 6685–6694 CrossRef CAS PubMed.
- A. Hollander, D. Heederik, P. Versloot and J. Douwes, Am. Ind. Hyg. Assoc. J., 1993, 54, 647–653 CrossRef CAS PubMed.
- S. Tanaka, J. Aketagawa, S. Takahashi, Y. Shibata, Y. Tsumuraya and Y. Hashimoto, Carbohydr. Res., 1993, 244, 115–127 CrossRef CAS PubMed.
- N. Binding, S. Jaschinski, S. Werlich, S. Bletz and U. Witting, J. Environ. Monit., 2004, 6, 65–70 RSC.
- G. H. Zhang, L. Baek and C. Koch, J. Clin. Microbiol., 1988, 26, 1464–1470 CAS.
- H. Muramatsu, M. Suda, T. Ataka, A. Seki, E. Tamiya and I. Karube, Sens. Actuators, A, 1990, 21, 362–368 CrossRef CAS.
- W. Lei, G. Jiang, Q. Zhou, Y. Hou, B. Zhang, X. Cheng and X. Wang, Sens. Actuators, B, 2012, 166–167, 853–858 CrossRef CAS.
- F. Liu, J. Mu, X. Wu, S. Bhattacharjya, E. K. L. Yeow and B. Xing, Chem. Commun., 2014, 50, 6200–6203 RSC.
- P. Pashazadeh, A. Mokhtarzadeh, M. Hasanzadeh, M. Hejazi, M. Hashemi and M. de la Guardia, Biosens. Bioelectron., 2017, 87, 1050–1064 CrossRef CAS PubMed.
- O. Lazcka, F. J. D. Campo and F. X. Muñoz, Biosens. Bioelectron., 2007, 22, 1205–1217 CrossRef CAS PubMed.
- P. M. Fratamico, Mol. Cell. Probes, 2003, 17, 215–221 CrossRef CAS PubMed.
- D. Rodriguez-Lazaro, M. Hernandez, T. Esteve, J. Hoorfar and M. Pla, J. Microbiol. Methods, 2003, 54, 381–390 CrossRef CAS PubMed.
- G. Garvey, D. Shakarisaz, F. Ruiz-Ruiz, A. E. V. Hagström, B. Raja, C. Pascente, A. Kar, K. Kourentzi, M. Rito-Palomares, P. Ruchhoeft and R. C. Willson, Anal. Chem., 2014, 86, 9029–9035 CrossRef CAS PubMed.
- H. J. Pandya, M. K. Kanakasabapathy, S. Verma, M. K. Chug, A. Memic, M. Gadjeva and H. Shafiee, Biosens. Bioelectron., 2017, 91, 32–39 CrossRef CAS PubMed.
- K.-M. Lee, M. Runyon, T. J. Herrman, R. Phillips and J. Hsieh, Food Control, 2015, 47, 264–276 CrossRef.
- G. Jiang, J. Wang, Y. Yang, G. Zhang, Y. Liu, H. Lin, G. Zhang, Y. Li and X. Fan, Biosens. Bioelectron., 2016, 85, 62–67 CrossRef CAS PubMed.
- J. Liang, X. Zheng, L. He, H. Huang and W. Bu, Dalton Trans., 2014, 43, 13174–13177 RSC.
- T. J. Wadas, Q.-M. Wang, Y.-j. Kim, C. Flaschenreim, T. N. Blanton and R. Eisenberg, J. Am. Chem. Soc., 2004, 126, 16841–16849 CrossRef CAS PubMed.
- C. Y.-S. Chung and V. W.-W. Yam, J. Am. Chem. Soc., 2011, 133, 18775–18784 CrossRef CAS PubMed.
- S. D. Cummings, Coord. Chem. Rev., 2009, 253, 449–478 CrossRef CAS.
- I. Eryazici, C. N. Moorefield and G. R. Newkome, Chem. Rev., 2008, 108, 1834–1895 CrossRef CAS PubMed.
- N. Liu, B. Wang, W. Liu and W. Bu, J. Mater. Chem. C, 2013, 1, 1130–1136 RSC.
- N. Liu, B. Wang, W. Liu and W. Bu, Chem. Commun., 2011, 47, 9336–9338 RSC.
- V. Ganesh, K. Bodewits, S. J. Bartholdson, D. Natale, D. J. Campopiano and J. C. Mareque-Rivas, Angew. Chem., Int. Ed., 2009, 48, 356–360 CrossRef CAS PubMed.
- S. Voss, R. Fischer, G. Jung, K.-H. Wiesmüller and R. Brock, J. Am. Chem. Soc., 2007, 129, 554–561 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra03312e |
| This journal is © The Royal Society of Chemistry 2017 |