Open Access Article
Open Access ArticleProperties of a flavonol-based photoCORM in aqueous buffered solutions: influence of metal ions, surfactants and proteins on visible light-induced CO release†
Marina Popovaa,
Tatiana Sobolevaa,
Atta M. Arifb and
Lisa M. Berreau *a
*a
aDepartment of Chemistry & Biochemistry, Utah State University, 0300 Old Main Hill, Logan, UT 84322-0300, USA. E-mail: lisa.berreau@usu.edu
bDepartment of Chemistry, University of Utah, 315 S. 1400 E., Salt Lake City, UT 84112-0850, USA
First published on 19th April 2017
Abstract
The properties of the extended flavonol 3-hydroxy-2-phenyl-benzo[g]chromen-4-one (2a) in DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) aqueous buffer solutions at pH = 7.4, including in the presence of metal ions, surfactants and serum albumin proteins, have been examined. Absorption and emission spectral studies of 2a in 1
aqueous buffer solutions at pH = 7.4, including in the presence of metal ions, surfactants and serum albumin proteins, have been examined. Absorption and emission spectral studies of 2a in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS buffer (pH = 7.4) indicate that a mixture of neutral and anionic forms of the flavonol are present. Notably, in 1
PBS buffer (pH = 7.4) indicate that a mixture of neutral and anionic forms of the flavonol are present. Notably, in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TRIS buffer (pH = 7.4) only the neutral form of the flavonol is present. These results indicate that the nature of the buffer influences the acid/base equilibrium properties of 2a. Introduction of a Zn(II) complex of 2a− to a 1
TRIS buffer (pH = 7.4) only the neutral form of the flavonol is present. These results indicate that the nature of the buffer influences the acid/base equilibrium properties of 2a. Introduction of a Zn(II) complex of 2a− to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) aqueous buffer (TRIS or PBS, pH = 7.4) solution produces absorption and emission spectral features consistent with the presence of a mixture of neutral 2a along with Zn(II)-coordinated or free 2a−. The nature of the anionic species present depends on the buffer composition. PBS buffered solutions (pH = 7.4) containing the surfactants CTAB or SDS enable 2a to be solubilized at a much lower percentage of DMSO (3.3–4.0%). Solutions containing the cationic surfactant CTAB include a mixture of 2a and 2a− whereas only the neutral flavonol is present in SDS-containing buffered solution. Compound 2a is also solubilized in TRIS buffer solutions at low cocentrations of DMSO (3.3%, pH = 7.4) in the presence of serum albumin proteins. Stern–Volmer analysis of the quenching of the inherent protein fluorescence indicates static binding of 2a to the proteins. The binding constant for this interaction is lower than that found for naturally-occurring flavonols (quercetin or morin) or 3-hydroxyflavone. Compound 2a binds to Site I of bovine and human serum albumin proteins as indicated by competition studies with warfarin and ibuprofen, as well as by docking investigations. The quantum yield for CO release from 2a (λirr = 419 nm) under aqueous conditions ranges from 0.0006(3) when the compound is bound to bovine serum albumin to 0.017(1) when present as a zinc complex in a 1
aqueous buffer (TRIS or PBS, pH = 7.4) solution produces absorption and emission spectral features consistent with the presence of a mixture of neutral 2a along with Zn(II)-coordinated or free 2a−. The nature of the anionic species present depends on the buffer composition. PBS buffered solutions (pH = 7.4) containing the surfactants CTAB or SDS enable 2a to be solubilized at a much lower percentage of DMSO (3.3–4.0%). Solutions containing the cationic surfactant CTAB include a mixture of 2a and 2a− whereas only the neutral flavonol is present in SDS-containing buffered solution. Compound 2a is also solubilized in TRIS buffer solutions at low cocentrations of DMSO (3.3%, pH = 7.4) in the presence of serum albumin proteins. Stern–Volmer analysis of the quenching of the inherent protein fluorescence indicates static binding of 2a to the proteins. The binding constant for this interaction is lower than that found for naturally-occurring flavonols (quercetin or morin) or 3-hydroxyflavone. Compound 2a binds to Site I of bovine and human serum albumin proteins as indicated by competition studies with warfarin and ibuprofen, as well as by docking investigations. The quantum yield for CO release from 2a (λirr = 419 nm) under aqueous conditions ranges from 0.0006(3) when the compound is bound to bovine serum albumin to 0.017(1) when present as a zinc complex in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O solution. Overall, the results of these studies demonstrate that 2a is a predictable visible light-induced CO release compound under a variety of aqueous conditions, including in the presence of proteins.
H2O solution. Overall, the results of these studies demonstrate that 2a is a predictable visible light-induced CO release compound under a variety of aqueous conditions, including in the presence of proteins.
Introduction
Carbon monoxide (CO) is generated in humans via the oxidative degradation of heme, which is catalyzed by heme oxygenase enzymes.1 The discovery of beneficial health effects associated with the delivery of small amounts of CO, including anti-inflammatory, anti-apoptotic, and anti-proliferative effects, as well as the promotion of vasodilation and protection of tissues against reperfusion injury, has led to investigations of the possible use of CO as a therapeutic.2–8 Several clinical trials are currently in progress involving the use of inhaled carbon monoxide.9–14 A challenge in using inhaled CO as a therapeutic is that it does not enable the delivery of controlled amounts of CO to specific targets and diffusion into tissues is unpredictable. As an approach toward introducing controlled amounts of CO, several research labs have worked on the development of CO-releasing molecules (CORMs).15,16 The most widely investigated CORMs to date are metal carbonyl complexes,17 with CORM-1 and CORM-3 (Fig. 1(top)) having been employed in a variety of biological investigations. Metal carbonyl-based CORMs release CO either spontaneously via ligand exchange, or via triggering with light (photoCORMs),18–25 enzyme activity,26–29 or magnetic heating30–32 (Fig. 1). A drawback of the use of metal carbonyl complexes as CORMs is the possible (or perceived) toxicity of the residual low-valent metal fragment remaining following CO release. Additionally, little is currently known in terms of the reactivity of metal carbonyl-based CORMs with other biomolecules. It was only after CORM-3 had been used in a several biological studies that the active form for CO-release in vivo was determined to likely be a protein-bound Ru(CO)2 species.33,34 In general, although many metal carbonyl species have been demonstrated to be CORMs and to enable CO release both in vitro and in cellular environments, very few have been studied with regard to their interactions with biological molecules, such as proteins.35,36 These interactions could dramatically affect the CO-release reactivity.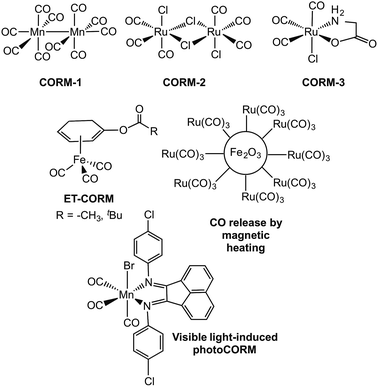 | ||
| Fig. 1 Examples of metal carbonyl-based CORMs.17,20,28,30 | ||
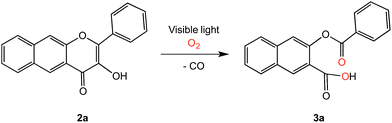
Metal-free CORMs37,38 and photoCORMs39–42 (Fig. 2) have recently emerged as alternatives to metal carbonyl-based CO-releasing molecules. Of the structural frameworks reported thus far for metal-free photoCORMs, the BODIPY and extended flavonol motifs (Fig. 2) are notable in that they can be tuned to enable the use of low energy visible light (>575 nm) to induce CO release, which is important for use in biological environments. The CO release unit in 1a/1b is a carboxyl moiety whereas the extended flavonol motif in 2a–d contains a pyrone ring that is similar to naturally occurring flavonols (e.g., quercetin), which are known to undergo enzyme-catalyzed CO release.43 The visible light-induced CO release reaction involving 1a or 1b proceeds in high yield (87–92%) only under anaerobic conditions, with the yield of CO under aerobic conditions dropping to 42–44%.39 Visible light-induced carbon monoxide release from 2a–c requires the presence of O2 for a quantitative, dioxygenase-type reaction to produce an O-benzoylsalicylic acid product (3a, Scheme 1). Complex 2d exhibits both aerobic and anaerobic visible light-induced quantitative CO release reactivity.42
 | ||
| Fig. 2 Metal-free CORMs and photoCORMs reported to date.36–41 | ||
Development of an understanding of the properties of metal-free CO-releasing photoCORM frameworks in aqueous buffered solutions and in the presence of biomolecules is essential toward the further development of these motifs as potential biological tools and therapeutics. Klán and coworkers have reported that at pH = 7.4 in PBS buffer, 1a and 1b exist as monoanions, with a deprotonated carboxyl group (pKa = 3.0 ± 0.2).39 Under anaerobic conditions in PBS (pH = 7.4) visible light-induced CO release occurs from 1a and 1b with quantum yields of 1.2(4) × 10−4 and 1.2(4) × 10−5, respectively. It is currently unknown how a biological environment, including the presence of proteins, will affect these values. The organic products in these CO release reactions also remain only partially characterized.
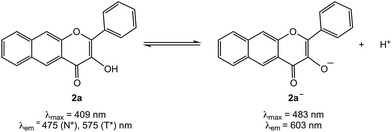
We have previously reported the visible light-induced CO release reactivity of 2a–d42 and zinc complexes of these flavonols in organic solvent (acetonitrile).44 All exhibit quantitative CO release under aerobic conditions. The quantum yields for these reactions are significantly affected by the presence of a neutral versus anionic form of the flavonol. Specifically, for 2a the quantum yield for CO release in CH3CN is 0.007(3) whereas for zinc complex of 2a− it is 0.651(2), a >90-fold increase.42,44 Experimental and computational studies of 3-hydroxyflavone (3-HflH) indicate that the pKa for this compound is ∼8.5 in aqueous solution.45 In this contribution, we examine the aqueous solution chemistry of 2a at pH = 7.4 in buffered solutions that include metal ions, surfactants or serum albumin proteins. Our results indicate that the components of the aqueous buffered environment affect the 2a/2a− equilibrium (Scheme 2) and modestly influence the quantum yield for CO release. Our studies reveal that 2a is a reliable CO release motif under a variety of aqueous buffered conditions. Combined with its straight-forward preparation, moderate toxicity, and the non-toxic nature of the O-benzoylsalicylate product (3a) remaining following CO release, 2a exhibits many features desirable in a photoCORM motif to be further developed for biomedical applications.
Experimental
Reagents
Compound 2a was prepared according to the literature procedure.42 All reagents were used as received unless otherwise noted. Cetrimonium bromide (CTAB) was purchased from TCI. Sodium dodecylsulfate (SDS) was purchased from Acros Organics. Bovine serum albumin (BSA, heat shock fraction) and human serum albumin (HAS, lyophilized powder, ≥97%) were purchased from Sigma Aldrich. Warfarin was purchased from Cayman Chemicals Company. Ibuprofen was purchased from Alfa Aesar. Double-distilled or distilled water was used in all experiments.Instrumentation
1H and 13C NMR spectra were collected using a Bruker Avance III HD Ascend-500 spectrometer. UV-vis spectra were recorded at ambient temperature using a Cary 50Bio or a Hewlett–Packard 8453A diode array spectrophotometer. Fluorescence emission spectra were recorded using a Shimadzu RF-530XPC spectrometer using 1.0 cm quartz cells. The excitation and emission slit widths were set at 3.0 nm. IR spectra were collected using a Shimadzu FTIR-8400 spectrometer. Mass spectral data were collected at the Mass Spectrometry Facility, University of California, Riverside. Elemental analyses were performed by Robertson Microlit Laboratory, Ledgewood, NJ. A Rayonet photoreactor equipped with RPR-419 lamps was used for all photochemical reactions. Quantum yields were determined as previously described using potassium ferrioxalate as a standard to measure photon flux.46–48[(bpy)Zn(2a−)]ClO4 (4)
Zn(ClO4)2·6H2O (50 mg, 0.13 mmol) dissolved in 1 ml of methanol was added to 2,2′-bipyridine (21 mg, 0.13 mmol) in 1 ml of methanol. The resulting mixture was added to a mixture of 2a (39 mg, 0.13 mmol) and Me4NOH·5H2O (24 mg, 0.13 mmol) in methanol (2 mL). The reaction mixture was stirred at room temperature for 3 hours. The resulting solution was divided into three parts and solvent was removed under reduced pressure. The remaining residue was dissolved in CH2Cl2, the solution was filtered through Celite plug, and the solvent was removed under vacuum. The product was recrystallized from ACN/Et2O, which resulted in the deposition of X-ray quality crystals (45%). IR (KBr, cm−1) 1100 (νClO4), 623 (νClO4); UV-vis (DMSO, nm) (ε, M−1 cm−1) 378 (3100), 479 (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100); ESI/APCI MS (positive ion): m/z calculated for C29H19N2O3Zn: 507.0722 [M–ClO4]+, found: 507.0699; elemental analysis calc. for C29H19N2O7ClZn·0.5H2O: C, 56.42%; H, 3.27%; N, 4.61%. Found: C 56.31; H, 2.85; N, 4.60.
100); ESI/APCI MS (positive ion): m/z calculated for C29H19N2O3Zn: 507.0722 [M–ClO4]+, found: 507.0699; elemental analysis calc. for C29H19N2O7ClZn·0.5H2O: C, 56.42%; H, 3.27%; N, 4.61%. Found: C 56.31; H, 2.85; N, 4.60.
X-ray crystallography
A translucent light orange plate-like crystal of 4 of approximate dimensions 0.140 mm × 0.168 mm × 0.392 mm was mounted using a viscous oil. Data were collected using a Nonius Kappa CCD diffractometer (Mo Kα, λ = 0.71073 Å). A total of 2079 frames were collected and subsequently integrated with the Bruker SAINT software package using a narrow-frame algorithm. The integration of the data using a monoclinic unit cell yielded a total of 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 427 reflections to a maximum θ angle of 30.03° (0.71 Å resolution), of which 7184 were independent (average redundancy 7.159, completeness = 99.8%, Rint = 2.75%, Rsig = 1.73%) and 6280 (87.42%) were greater than 2σ(F2). The final cell constants of a = 8.2518(2) Å, b = 14.8377(4) Å, c = 20.0844(6) Å, β = 92.219(2), volume = 2457.24(12) Å3, are based upon the refinement of the XYZ-centroids of reflections above 20 σ(I). Data were corrected for absorption effects using the multi-scan method (SADABS). The calculated minimum and maximum transmission coefficients (based on crystal size) are 0.6600 and 0.8540, respectively.
427 reflections to a maximum θ angle of 30.03° (0.71 Å resolution), of which 7184 were independent (average redundancy 7.159, completeness = 99.8%, Rint = 2.75%, Rsig = 1.73%) and 6280 (87.42%) were greater than 2σ(F2). The final cell constants of a = 8.2518(2) Å, b = 14.8377(4) Å, c = 20.0844(6) Å, β = 92.219(2), volume = 2457.24(12) Å3, are based upon the refinement of the XYZ-centroids of reflections above 20 σ(I). Data were corrected for absorption effects using the multi-scan method (SADABS). The calculated minimum and maximum transmission coefficients (based on crystal size) are 0.6600 and 0.8540, respectively.
The structure of 4 was solved and refined using the Bruker SHELXTL Software Package, using the space group P2![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) n with Z = 4 for the formula unit C29H19ClN2O7Zn. The final anisotropic full-matrix least-squares refinement on F2 with 370 variables converged at R1 = 3.18%, for the observed data and wR2 = 8.97%. The goodness-of-fit was 1.062. The largest peak in the final difference electron density was 1.263 e− Å−3 with an RMS deviation of 0.066 e− Å−3. On the basis of the final model, the calculated density was 1.644 g cm−3 and F(000), 1240 e−. The structure was deposited in the Cambridge Crystallographic Database (CCDC 1523782).
n with Z = 4 for the formula unit C29H19ClN2O7Zn. The final anisotropic full-matrix least-squares refinement on F2 with 370 variables converged at R1 = 3.18%, for the observed data and wR2 = 8.97%. The goodness-of-fit was 1.062. The largest peak in the final difference electron density was 1.263 e− Å−3 with an RMS deviation of 0.066 e− Å−3. On the basis of the final model, the calculated density was 1.644 g cm−3 and F(000), 1240 e−. The structure was deposited in the Cambridge Crystallographic Database (CCDC 1523782).
Buffer preparation
TRIS (0.05 M) at pH = 7.4 was prepared using a mixture of TRIS–HCl and TRIS-base. Sodium chloride (0.1 M) was added to maintain ionic strength. Phosphate buffered saline (PBS) was prepared containing 140 mM NaCl, 3 mM KCl, and 10 mM phosphate and was adjusted to pH = 7.4 using aqueous HCl.Solution studies of 2a in the presence of CTAB and SDS
Experiments were performed in PBS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (96.7
DMSO (96.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.3% v:v, pH = 7.4). The absorption and emission properties of 2a were evaluated in the presence of varying amounts of CTAB or SDS.
3.3% v:v, pH = 7.4). The absorption and emission properties of 2a were evaluated in the presence of varying amounts of CTAB or SDS.
Solution studies of 2a in the presence of bovine or human serum albumin
Emission spectra of solutions of 2a (1.4 × 10−6 M) in TRIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (96
DMSO (96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4% v:v, pH = 7.4) were collected from 410 to 800 nm in the presence of varying amounts of bovine or human serum albumin (1
4% v:v, pH = 7.4) were collected from 410 to 800 nm in the presence of varying amounts of bovine or human serum albumin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 to 1
0 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40). The excitation wavelength was 408 nm, which corresponds to the absorption maximum of 2a.
40). The excitation wavelength was 408 nm, which corresponds to the absorption maximum of 2a.
Serum albumin binding studies
Binding studies of 2a to bovine or human serum albumin were performed using tryptophan fluorescence quenching experiments in TRIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (96
DMSO (96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4% v:v, pH = 7.4). The excitation wavelengths used for BSA and HSA were 282 and 285 nm, respectively. Emission spectra were recorded from 300 to 600 nm. The maximum emission intensities at 340 nm were used to calculate binding constants and to determine the number of binding sites.
4% v:v, pH = 7.4). The excitation wavelengths used for BSA and HSA were 282 and 285 nm, respectively. Emission spectra were recorded from 300 to 600 nm. The maximum emission intensities at 340 nm were used to calculate binding constants and to determine the number of binding sites.
Molecular docking
Studies were performed using AutoDock Vina 1.1.2 software.49 The crystal structure of BSA was downloaded from the RSCB Protein Data Bank (PDB entry code 3V03). The X-ray crystallographically determined structure of 2a42 was converted to pdbqt format with MGLTools for use as an input file for AutoDock Vina. A structure of the O-benzoylsalicylic acid CO release product 3a was energy minimized using Chem 3D (MM2 force field) and saved in pdbqt format using MGLTools for input into AutoDock Vina. A semi-flexible docking procedure was performed wherein BSA was kept rigid and stationary and was assigned with polar hydrogen atoms. Compound 2a with two rotatable bonds and 3a with five rotatable bonds were allowed to be flexible as they were docked into the protein. A grid box with dimensions 60 × 68 × 68 points with the spacing of 1.0 Å was used to accommodate the ligands to move freely during each docking run. The lowest energy docked conformations identified for 2a and 3a with BSA were visualized using Pymol.Independent synthesis of 3-(benzoyloxy)-2-naphthoic acid (3a)
3-Hydroxy-2-naphthoic acid, (0.50 g, 2.7 mmol) in 3 ml dry diethyl ether was mixed with pyridine (0.58 ml, 7.2 mmol). The resulting mixture was cooled in an ice bath and a solution of benzoyl chloride (0.31 ml, 2.7 mmol) in diethyl ether (2 mL) was added. The reaction mixture was warmed to room temperature and then stirred for 1 hour. H2O (10 ml) was then added. The solution was acidified with 2 M HCl until pH ∼4 was obtained. The mixed aqueous/organic mixture was then extracted using chloroform (3 × 15 ml). The organic fractions were combined, dried over Na2SO4, the solution was filtered, and the solvent was removed from the filtrate under reduced pressure. The product was purified by column chromatography using hexanes![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (1
ethyl acetate (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as the eluent. 3-(Benzoyloxy)-2-naphthoic acid (3a) was obtained as colorless needles (0.20 g, 25%) following removal of solvent. The 1H NMR features of the product in CD3CN matched with those previously reported.42 1H NMR (500 MHz, CD3OD) δ ppm 8.66 (s, 1H), 8.23 (d, J = 9 Hz, 2H), 8.05 (d, J = 9.5 Hz, 1H), 7.93 (d, J = 9 Hz, 1H), 7.73 (s, 1H), 7.69 (t, J = 7.5 Hz, 1H), 7.65 (t, J = 7.5 Hz, 1H), 7.56 (m, 3H); 13C{1H} NMR (125 MHz, CD3OD) δ 169.4, 169.0, 149.9, 138.5, 136.2, 136.1, 133.6, 132.7, 132.6, 131.5, 131.4, 131.1, 129.8, 129.3, 125.9, 123.6 ppm (16 signals expected and observed); IR (KBr, cm−1) 1743 (νC
1) as the eluent. 3-(Benzoyloxy)-2-naphthoic acid (3a) was obtained as colorless needles (0.20 g, 25%) following removal of solvent. The 1H NMR features of the product in CD3CN matched with those previously reported.42 1H NMR (500 MHz, CD3OD) δ ppm 8.66 (s, 1H), 8.23 (d, J = 9 Hz, 2H), 8.05 (d, J = 9.5 Hz, 1H), 7.93 (d, J = 9 Hz, 1H), 7.73 (s, 1H), 7.69 (t, J = 7.5 Hz, 1H), 7.65 (t, J = 7.5 Hz, 1H), 7.56 (m, 3H); 13C{1H} NMR (125 MHz, CD3OD) δ 169.4, 169.0, 149.9, 138.5, 136.2, 136.1, 133.6, 132.7, 132.6, 131.5, 131.4, 131.1, 129.8, 129.3, 125.9, 123.6 ppm (16 signals expected and observed); IR (KBr, cm−1) 1743 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); ESI/APCI MS (negative ion): m/z calculated for C18H12O4: 291.0657, found: 291.0674. m.p. 173–175 °C.
O); ESI/APCI MS (negative ion): m/z calculated for C18H12O4: 291.0657, found: 291.0674. m.p. 173–175 °C.
Results
Properties of 2a in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) aqueous buffer at pH = 7.4
aqueous buffer at pH = 7.4
When dissolved in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Tris buffer at pH 7.4, compound 2a exhibits features consistent with the presence of the neutral flavonol (Fig. 3). Specifically, an absorption band with a maximum at approximately ∼409 nm is similar to that observed for the compound in CH3CN and 1
Tris buffer at pH 7.4, compound 2a exhibits features consistent with the presence of the neutral flavonol (Fig. 3). Specifically, an absorption band with a maximum at approximately ∼409 nm is similar to that observed for the compound in CH3CN and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O.42 When 2a is dissolved in 1
H2O.42 When 2a is dissolved in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS buffer at pH 7.4 (Fig. 3), an absorption feature at ∼480 nm is also present. This red-shifted band is consistent with the presence of the flavonolato anion (2a−) based on comparison to spectroscopic features of Zn(II) complexes of 2a−.44 The same 480 nm band is present when 2a is dissolved in 1
PBS buffer at pH 7.4 (Fig. 3), an absorption feature at ∼480 nm is also present. This red-shifted band is consistent with the presence of the flavonolato anion (2a−) based on comparison to spectroscopic features of Zn(II) complexes of 2a−.44 The same 480 nm band is present when 2a is dissolved in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O to which two equivalents of sodium hydroxide has been added (Fig. S1(a)†).
H2O to which two equivalents of sodium hydroxide has been added (Fig. S1(a)†).
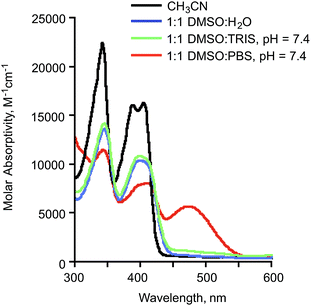 | ||
Fig. 3 Absorbance spectra of 2a (0.10 mM) in various solution environments. The ∼480 nm absorption feature present in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO 1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS at pH 7.4 is associated with the presence of 2a−. PBS at pH 7.4 is associated with the presence of 2a−. | ||
Fluorescence spectra (λex = 410 and 480 nm) of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer solutions (pH = 7.4) of 2a provide additional evidence for the presence of neutral and anionic forms of 2a under various conditions. When excited at 410 nm, solutions of 2a in 1
buffer solutions (pH = 7.4) of 2a provide additional evidence for the presence of neutral and anionic forms of 2a under various conditions. When excited at 410 nm, solutions of 2a in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, DMSO
H2O, DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TRIS and 1
TRIS and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
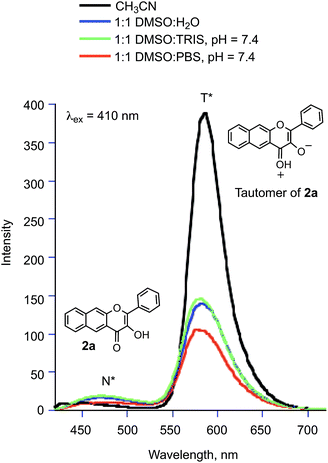
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS at pH = 7.4 exhibit two emission bands at ∼475 and ∼580 nm (Fig. 4), respectively. These emissions are from the normal (N*) and tautomeric (T*) excited state forms of 2a.50 The T* form is the result of excited state proton transfer (ESPT) within the flavonol and formation of the pyrillium ion (Fig. 4). The intensity of the T* emission is lower than that exhibited by 2a in organic solvent (CH3CN) due to the availability of additional non-radiative pathways in the hydrogen bonding aqueous solutions which results in quenching of the excited state.51 The emission maximum in aqueous environments is also slightly blue-shifted relative to the spectrum of 2a in CH3CN. Excitation of the same solutions at 480 nm produced an emission at ∼603 nm in the solution of 2a in 1
PBS at pH = 7.4 exhibit two emission bands at ∼475 and ∼580 nm (Fig. 4), respectively. These emissions are from the normal (N*) and tautomeric (T*) excited state forms of 2a.50 The T* form is the result of excited state proton transfer (ESPT) within the flavonol and formation of the pyrillium ion (Fig. 4). The intensity of the T* emission is lower than that exhibited by 2a in organic solvent (CH3CN) due to the availability of additional non-radiative pathways in the hydrogen bonding aqueous solutions which results in quenching of the excited state.51 The emission maximum in aqueous environments is also slightly blue-shifted relative to the spectrum of 2a in CH3CN. Excitation of the same solutions at 480 nm produced an emission at ∼603 nm in the solution of 2a in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS buffer. A 1
PBS buffer. A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O solution containing two equivalents of sodium hydroxide gives a similar emission feature (Fig. S1(c)†) providing evidence that this emission is associated with 2a−.
H2O solution containing two equivalents of sodium hydroxide gives a similar emission feature (Fig. S1(c)†) providing evidence that this emission is associated with 2a−.
Zinc complex
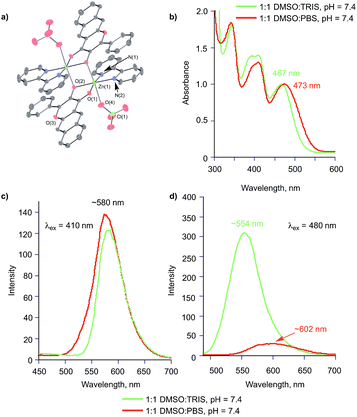
To gain insight into the properties of 2a− in aqueous buffer in the presence of a divalent metal ion, we have prepared and characterized [(bpy)Zn(2a−)]ClO4 (4) and performed spectroscopic studies of this complex. In the solid state, 4 exhibits a binuclear structure (Fig. 6(a)) with the deprotonated oxygen atom of each flavonolato ligand bridging between two zinc centers. This coordination motif is similar to that found in [(bpy)Zn(3-Hfl)]ClO4 (3-Hfl = 3-hydroxyflavonolato anion).52 ESI APCI mass spectral studies of 4 in wet CH3CN (Fig. S2†) indicate that the mononuclear [(bpy)Zn(2a−)]+ ion is present indicating breakup of the dimer in solution. Additionally, species resulting from ligand exchange (e.g., [(bpy)2Zn(ClO4)]+ and [(bpy)2Zn]2+) and ligand loss (e.g., [bpyH]+ and [2a] H+) are evident in the mass spectrum. The UV-vis spectrum of 4 in dry CH3CN, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TRIS and 1
TRIS and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS at pH = 7.4, are shown in Fig. 6(b). The DMSO
PBS at pH = 7.4, are shown in Fig. 6(b). The DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer solutions show features consistent with the presence of neutral and anionic forms of 2a. The emission spectra of 4 (λex = 410 nm; Fig. 6(c)) in 1
buffer solutions show features consistent with the presence of neutral and anionic forms of 2a. The emission spectra of 4 (λex = 410 nm; Fig. 6(c)) in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TRIS and 1
TRIS and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS at pH = 7.4 confirm the presence of the neutral flavonol. Excitation at 480 nm (Fig. 6(d)) produces an emission feature at ∼554 nm in 1
PBS at pH = 7.4 confirm the presence of the neutral flavonol. Excitation at 480 nm (Fig. 6(d)) produces an emission feature at ∼554 nm in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TRIS and ∼603 nm in 1
TRIS and ∼603 nm in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS. The differences in these spectra are indicative of the presence of zinc-coordinated 2a− and free 2a− anion, respectively. The attribution of the emission at ∼563 nm to Zn(II) coordinated 2a− is made on the basis of prior studies.44 These results indicate that the zinc ion stays associated with the 2a− anion in 1
PBS. The differences in these spectra are indicative of the presence of zinc-coordinated 2a− and free 2a− anion, respectively. The attribution of the emission at ∼563 nm to Zn(II) coordinated 2a− is made on the basis of prior studies.44 These results indicate that the zinc ion stays associated with the 2a− anion in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TRIS at pH = 7.4, but is displaced from the zinc center in 1
TRIS at pH = 7.4, but is displaced from the zinc center in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS at the same pH.
PBS at the same pH.
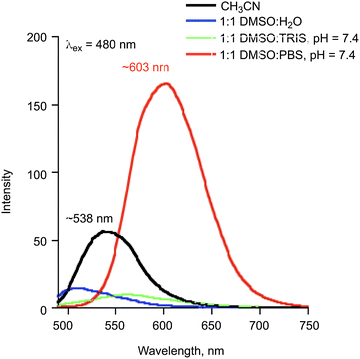 | ||
| Fig. 5 Emission spectra (λex = 480 nm) of solutions of 2a (0.10 mM). The emission feature for 2a at ∼538 nm in CH3CN is attributed to the neutral flavonol. | ||
Effects of surfactants
The flavonol 2a exhibits low solubility in PBS buffer in the presence of low amounts of DMSO (∼3%). This is evident in the lack of a flavonol emission feature of a solution of this composition and observed precipitation. However, in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer, 2a is soluble. In order to minimize the amount of DMSO present and gain insight into how the solubility of 2a may be impacted by interactions with other biomolecules or charged species, we have investigated the solution spectroscopic properties of 2a in the presence of surfactants. In the presence of the cationic surfactant CTAB (centrimonium bromide), compound 2a exhibits absorption and emission spectra consistent with the presence of a mixture of 2a and 2a− (Fig. 7). The emission features associated with these species increase in intensity with increasing CTAB concentration (Fig. 7(b) and (c)), which is similar to studies involving 3-hydroxyflavone.53
buffer, 2a is soluble. In order to minimize the amount of DMSO present and gain insight into how the solubility of 2a may be impacted by interactions with other biomolecules or charged species, we have investigated the solution spectroscopic properties of 2a in the presence of surfactants. In the presence of the cationic surfactant CTAB (centrimonium bromide), compound 2a exhibits absorption and emission spectra consistent with the presence of a mixture of 2a and 2a− (Fig. 7). The emission features associated with these species increase in intensity with increasing CTAB concentration (Fig. 7(b) and (c)), which is similar to studies involving 3-hydroxyflavone.53
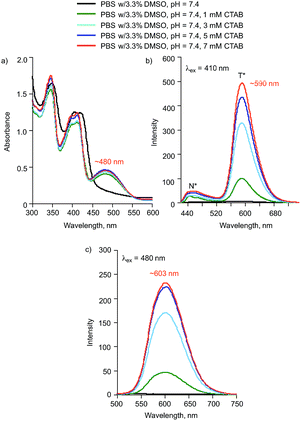 | ||
| Fig. 7 (a) Absorption spectra of 2a (0.10 mM) in the presence of various concentrations of CTAB. (b) Emission spectra produced with λex = 410 nm. (c) Emission spectra produced with λex = 480 nm. | ||
In the presence of sodium dodecyl sulphate (SDS), the absorption and emission features of 2a are consistent with the presence of only the neutral form of the flavonol (Fig. 8). This is particularly evident in the lack of an emission feature upon excitation at 480 nm.
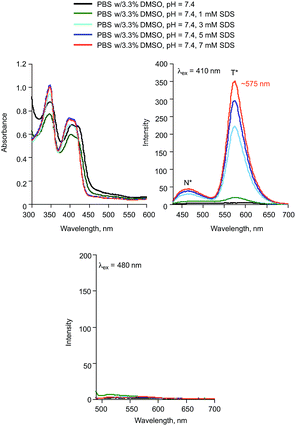 | ||
| Fig. 8 (a) Absorption spectra of 2a (0.10 mM) in the presence of various concentrations of SDS. (b) Emission spectra produced with λex = 410 nm. (c) Emission spectra produced with λex = 480 nm. | ||
Serum albumins
Several studies of the binding properties of flavonols to serum albumin proteins using fluorescence spectroscopy have been previously reported.54–62 In the case of 2a, we selectively examined the fluorescence properties of both the flavonol and the tryptophan residues of the protein to probe the 2a/albumin interaction. Addition of aliquots of BSA to the flavonol in TRIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (96
DMSO (96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4% v:v, pH = 7.4) results in a saturatable increase in the intensity of the flavonol T* emission band (Fig. 9). A bathochromic shift (∼30 nm) of this emission is observed. We note that a similar red shift of the tautomer fluorescence band was identified for 3-hydroxyflavone (3-HflH) upon binding to BSA.54 Notably, the emission maximum observed for 2a in the presence of BSA matches that found in a polar aprotic organic solvent (e.g., CH3CN (Fig. 4)) wherein the ESIPT process is efficient. This suggests a hydrophobic binding site for the flavonol within the protein environment. As shown in Fig. 10, addition of 2a to BSA in TRIS
4% v:v, pH = 7.4) results in a saturatable increase in the intensity of the flavonol T* emission band (Fig. 9). A bathochromic shift (∼30 nm) of this emission is observed. We note that a similar red shift of the tautomer fluorescence band was identified for 3-hydroxyflavone (3-HflH) upon binding to BSA.54 Notably, the emission maximum observed for 2a in the presence of BSA matches that found in a polar aprotic organic solvent (e.g., CH3CN (Fig. 4)) wherein the ESIPT process is efficient. This suggests a hydrophobic binding site for the flavonol within the protein environment. As shown in Fig. 10, addition of 2a to BSA in TRIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (96
DMSO (96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4% v:v, pH = 7.4) results in a decrease of the 340 nm intrinsic emission (λex = 282 nm) associated with the tryptophan residues of the protein. This quenching suggests that 2a binds in close proximity to the tryptophan residues (positions 134 and 213). Notably, similar quenching is seen upon titration of human serum albumin (HSA), which contains a single tryptophan residue at position 214 in subdomain IIA of the protein (Fig. S3†). For each protein, the Stern–Volmer equation (eqn (1)) was used to classify the type of fluorescence quenching induced by 2a as static or dynamic.54
4% v:v, pH = 7.4) results in a decrease of the 340 nm intrinsic emission (λex = 282 nm) associated with the tryptophan residues of the protein. This quenching suggests that 2a binds in close proximity to the tryptophan residues (positions 134 and 213). Notably, similar quenching is seen upon titration of human serum albumin (HSA), which contains a single tryptophan residue at position 214 in subdomain IIA of the protein (Fig. S3†). For each protein, the Stern–Volmer equation (eqn (1)) was used to classify the type of fluorescence quenching induced by 2a as static or dynamic.54
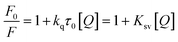 | (1) |
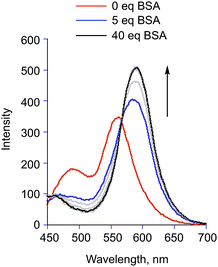 | ||
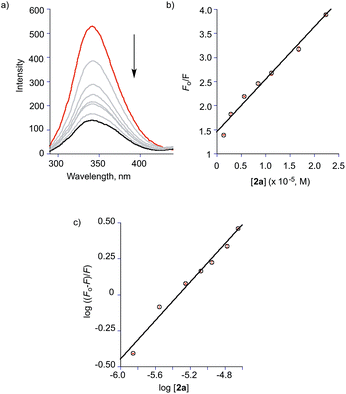
Fig. 9 Emission features of 2a (1.4 μM) upon addition of increasing amounts of BSA in TRIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (96 DMSO (96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4% v:v, pH = 7.4). T = 293 K, λex = 410 nm. 4% v:v, pH = 7.4). T = 293 K, λex = 410 nm. | ||
In this equation, F0 and F are the fluorescence intensities in the absence and presence of the quencher (2a), respectively; kq is the bimolecular quenching constant, τ0 is the lifetime of the fluorophore in the absence of quencher (10−8 s), [Q] is the concentration of the quencher and Ksv is the Stern–Volmer bimolecular quenching constant.54 The Stern–Volmer plot for the titration of BSA with 2a is shown in Fig. 10(b). As kq (1.1 × 1013 M−1 s−1) is much larger than 2 × 1010 M−1 s−1 (the maximum diffusion collision quenching rate constant of various quenchers with the biopolymer), it is likely that the quenching in this case is static in nature.54 The binding constant Ka and number of binding sites n were calculated from the modified Stern–Volmer plot (Fig. 10(c)) for the titrations of BSA with 2a according to eqn (2):
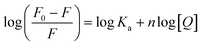 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka as the intercept and n as the slope (Fig. 10(c)). The binding constant Ka and binding sites n for 2a with BSA and HSA are listed in Table 1. Notably, only one equivalent of 2a binds to BSA and HSA versus two equivalents of the structurally similar but smaller 3-hydroxyflavone (3-HflH).54,55 The binding constant for 2a is ∼45-fold lower than that of 3-HflH and is also significantly lower than that of hydroxyl-substituted 3-HflH derivatives that are naturally occurring compounds (e.g., quercetin and morin; Table 1).
Ka as the intercept and n as the slope (Fig. 10(c)). The binding constant Ka and binding sites n for 2a with BSA and HSA are listed in Table 1. Notably, only one equivalent of 2a binds to BSA and HSA versus two equivalents of the structurally similar but smaller 3-hydroxyflavone (3-HflH).54,55 The binding constant for 2a is ∼45-fold lower than that of 3-HflH and is also significantly lower than that of hydroxyl-substituted 3-HflH derivatives that are naturally occurring compounds (e.g., quercetin and morin; Table 1).
| Compound | Binding constant (Ka, M−1) | n | Ref. |
|---|---|---|---|
| 2a-BSA | 3.2 × 103 | 0.66 | This work |
| 2a-BSA (Zn2+) | 1.9 × 102 | 0.48 | This work |
| [(bpy)Zn(2a−)]ClO4 | 1.0 × 102 | 0.39 | This work |
| 2a-BSA (Ca2+) | 3.2 × 103 | 0.71 | This work |
| 2a-BSA (Mg2+) | 2.5 × 103 | 0.69 | This work |
| 2a-BSA (warfarin) | 2.8 × 102 | 0.46 | This work |
| 2a-BSA (ibuprofen) | 2.5 × 103 | 0.67 | This work |
| 2a-HSA | 3.0 × 103 | 0.7 | This work |
| 2a-HSA (warfarin) | 2.5 × 103 | 0.66 | This work |
| 2a-HSA (ibuprofen) | 3.0 × 103 | 0.67 | This work |
| 3-HFlH-BSA | 1.1–1.3 × 105 | 2 | 54 |
| 3-HflH-HSA | 7.2 × 105; 2.5 × 105 | 2 | 55 |
| Quercetin-BSA | 3.65 × 107 | 1.29 | 56 |
| Quercetin-BSA | 1.00 × 105 | 0.84 | 57 |
| Quercetin-BSA | 4.85 × 105 | 1.19 | 58 |
| Quercetin-HSA | 2.30 × 104 | 1.10 | 58 |
| Morin-HSA | 1.13 × 105 | 1.06 | 59 |
| 3a-BSA | 8.5 × 107 | 1.7 | This work |
| 3a-BSA (warfarin) | 6.7 × 103 | 0.9 | This work |
| 3a-BSA (ibuprofen) | 2.5 × 105 | 1.2 | This work |
| BSA-aspirin | 5.0 × 103 | 64 |
Most drug molecules bind to BSA in subdomains IIA and IIIA. Site I (subdomain IIA) is where the blood thinner drug warfarin binds whereas Site II (subdomain IIIA) is the location of ibuprofen binding.63 To gain insight into the binding site of 2a in BSA, displacement studies were performed with warfarin and ibuprofen. Pre-equilibration of a BSA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) warfarin (1
warfarin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixture for 1 h followed by titration with 2a resulted in the loss of tryptophan fluorescence intensity. The binding constant for 2a determined from this data (2.8 × 102 M−1; Fig. S4(a)†) is lower than that observed in the absence of warfarin, suggesting competition for the binding site. A similar experiment involving ibuprofen (Fig. S4(b)†) produced almost no change in the binding constant for 2a, indicating that no binding of this compound occurs in Site II.
1) mixture for 1 h followed by titration with 2a resulted in the loss of tryptophan fluorescence intensity. The binding constant for 2a determined from this data (2.8 × 102 M−1; Fig. S4(a)†) is lower than that observed in the absence of warfarin, suggesting competition for the binding site. A similar experiment involving ibuprofen (Fig. S4(b)†) produced almost no change in the binding constant for 2a, indicating that no binding of this compound occurs in Site II.
To further probe the binding site of 2a with BSA we performed in silico docking studies using AutoDock Vina.49 Two different high affinity binding conformations (Fig. 11) were identified near Site I in subdomain IIA. One of these conformations includes hydrogen-bonding interactions involving the ketone moiety of 2a (Fig. 11(a)). Because these interactions would preclude the formation of the tautomeric excited state form of 2a, which is observed by fluorescence, the other conformation (Fig. 11(b)) is more likely. In this binding mode, the hydroxyl oxygen atom, which would be deprotonated in the tautomeric form, is stabilized by the presence of multiple hydrogen bonding interactions.
 | ||
| Fig. 11 Views of the binding of 2a to BSA near Site I in subdomain IIA. Both conformations have similar energies. | ||
Human serum albumin (HSA) contains only a single tryptophan residue near Site I. The binding properties for 2a to HSA are similar to that observed for BSA, with a single molecule binding with a binding constant of 3.0 × 103 M−1 (Fig. S3(a–c)†). Warfarin and ibuprofen experiments with 2a produced a less pronounced effect relative to those observed for BSA (Fig. S5(a) and (b)†), with a modest affect on binding of 2a only being observed in the presence of warfarin. Overall, these combined results strongly suggest that serum albumin proteins bind 2a at Site I.
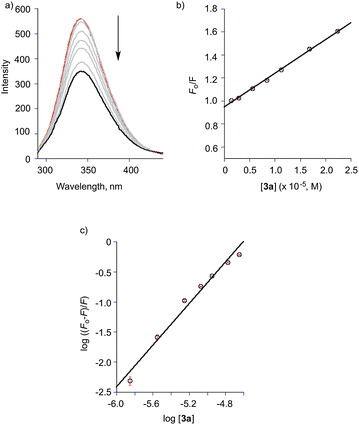
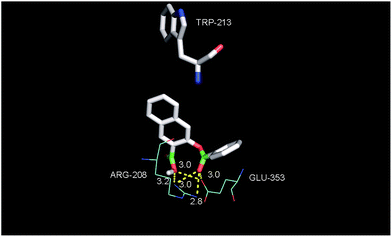
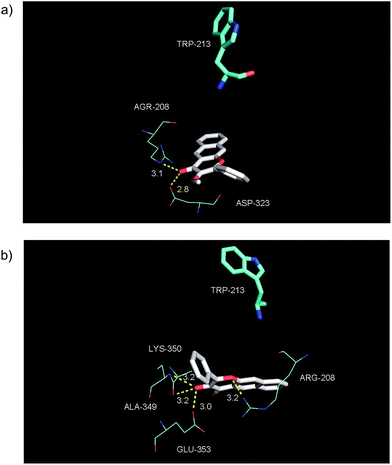
The binding properties of the O-benzoylsalicylate CO release product 3a with BSA were also briefly examined. As shown in Fig. 12(a), this molecule also quenches the intrinsic fluorescence of the protein. A modified Stern–Volmer analysis (Fig. 12(c)) revealed that the binding affinity of 3a to BSA is significantly higher (Ka ∼107 M−1) than that of 2a. Two molecules of 3a bind to BSA. Competitive binding studies performed using warfarin and ibuprofen (Fig. S6†) show significant changes in the binding constant for 3a in the presence of both of these site markers. Docking of 3a with BSA resulted in the identification of two binding sites, with the first being Site I in subdomain IIA and the second being a hydrophobic pocket in subdomain IB. As shown in Fig. 13, binding for O-benzoylsalicylate derivative in Site I, which is likely the higher affinity binding site, involves hydrogen-bonding interactions with two nearby residues (ARG-208, Glu-353). In subdomain IB, which is a secondary binding site for several drugs including AZT,65,66 the interactions appear to be exclusively hydrophobic. Overall, the distinctly different serum albumin binding properties of 2a and 3a place these compounds near the bookends of the range of many known drug molecules that exhibit binding constants of 104–107 M−1 at Site I in HSA.65
Our studies with [(bpy)Zn(2a−)]ClO4 (4) demonstrate that Zn2+ will form coordination complexes with the anion 2a−. To examine the effect of metal ions on the protein binding properties of 2a in aqueous buffer, titrations of solutions of BSA (1.4 μM) containing 150 μM Mn+ (Mn+ = Mg2+, Ca2+, and Zn2+) with 2a were performed. As shown in Table 1, the presence of Zn2+ decreased the binding affinity of 2a for BSA to a value similar to that observed for the zinc complex [(bpy)Zn(2a−)]ClO4 (4) (Table 1).
CO release reactivity
With an understanding of the solution properties of 2a under a variety of aqueous conditions, we next examined the efficiency of the visible light-induced CO release reaction (Scheme 1) under the conditions listed in Table 2. The quantum yields for CO release from 2a/2a− ranges from 0.006 to 0.010, with binding of 2a to BSA producing a ∼10-fold lower quantum yield than that found for the free molecule in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) buffer or CH3CN solutions. Thus the reaction quantum yields found for 2a do not change appreciably as a function of solvent. This is similar to the quantum yields found for CO release from a Zn(II) 3-hydroxyflavone complex wherein a change of solvent from CH3CN to H2O
buffer or CH3CN solutions. Thus the reaction quantum yields found for 2a do not change appreciably as a function of solvent. This is similar to the quantum yields found for CO release from a Zn(II) 3-hydroxyflavone complex wherein a change of solvent from CH3CN to H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (1
DMSO (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) resulted in only a two-fold change in reaction quantum yield.67 For 2a− the presence of Zn2+ in the form of the [(bpy)Zn(2a−)]ClO4 (4) complex produces a slightly higher quantum yield (0.017(1)). We note that the quantum yields for CO release obtained for 2a under all of these conditions are at least five-fold greater than those of the BODIPY derivatives 1a and 1b under aerobic conditions.39
1) resulted in only a two-fold change in reaction quantum yield.67 For 2a− the presence of Zn2+ in the form of the [(bpy)Zn(2a−)]ClO4 (4) complex produces a slightly higher quantum yield (0.017(1)). We note that the quantum yields for CO release obtained for 2a under all of these conditions are at least five-fold greater than those of the BODIPY derivatives 1a and 1b under aerobic conditions.39
| Conditions (pH = 7.4) | Quantum yield |
|---|---|
| CH3CN | 0.007(3) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO 1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TRIS TRIS |
0.006(3) |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO 1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS PBS |
0.010(3) |
4% DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS + CTAB PBS + CTAB |
0.0063(1) |
BSA (40 eq.) in 3.3% DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TRIS TRIS |
0.0006(1) |
[(bpy)Zn(2a−)]ClO4 in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO 1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O H2O |
0.017(1) |
Discussion and conclusions
The development of carbon monoxide releasing molecules (CORMs) for therapeutic applications is an area of significant current interest.2–8,15,16 While the majority of CORMs evaluated to date are based on a metal carbonyl moiety as the CO release unit, challenges regarding incorporation of drug-like structural components, combined with concerns about the possible side reactivity and toxicity of such structures in therapeutic applications, have hampered further development. Metal-free CO-releasing molecules, particularly those that can be controlled in terms of the timing and location of CO release, are attractive as such moieties are likely to be more amenable to standard medicinal chemistry approaches.4 In this regard, CO-releasing organic molecules that can be triggered by visible light are of particular current interest. Two novel metal-free, visible light-induced CO release structural motifs have been recently reported. These are based on BODIPY (1a/1b)39 and extended flavonol (2a–d)42-type structural motifs. Both types of compounds offer attractive features, including low-to-medium toxicity and strong fluorescent emission features that enable tracking in a biological environment. An additional advantage of the BODIPY-based compounds is the structural tunability that enables the use of light of >700 nm to induce CO release. Weaknesses of the BODIPY motif include the requirement of anaerobic conditions to achieve quantitative CO release and the lack of full characterization and toxicity evaluation of the CO release products. The extended flavonol motif offers several notable features that are desirable toward the further development of visible light induced CO release compounds. We previously reported the structural tunability of the flavonol motif in 2a–d that enables modulation of the wavelength of light that can be used to induce CO release as well as tune the quantum yield for this reaction.42 We discovered that one derivative (2d) offers the opportunity to access anaerobic CO release reactivity. Another distinct advantage of the framework in 2a is the ease of synthesis, which involves a one-pot reaction from commercially available starting materials.In this contribution, we outline details of the solution properties of 2a in aqueous buffer environments at physiological pH (7.4) and in the presence of charged or biologically relevant molecules. Our results indicate that 2a can undergo partial ionization to 2a− under certain conditions (e.g., in PBS buffer or in the presence of a cationic surfactant). We have found that the presence of Zn2+ will enhance the quantum yield for CO release from 2a. Overall this knowledge is important with regard to using 2a in biological environments.
As we have described herein, compound 2a is the most reactive metal-free photoCORM motif reported to date under a variety of conditions. The ease of synthesis and structural modification of the extended flavonol framework of 2a, coupled with the reliability of the molecule in terms of visible light induced CO release reactivity under aqueous conditions, makes it especially well-suited for the further development of a family of CO-releasing molecules with modifications to enhance biological properties, functionality, and targeting. Such efforts are currently underway in our laboratory.
Acknowledgements
We thank the US National Science Foundation (CHE-1301092 to LMB; CHE-1429195 for Bruker Avance III HD Ascend-500 spectrometer) for funding.References
- G. Kikuchi, T. Yoshida and M. Noguchi, Biochem. Biophys. Res. Commun., 2005, 338, 558 CrossRef CAS PubMed.
- R. Motterlini and L. E. Otterbein, Nat. Rev. Drug Discovery, 2010, 9, 728 CrossRef CAS PubMed.
- C. Steiger, C. Hermann and L. Meinel, Eur. J. Pharm. Biopharm., 2016 DOI:/10.1016/j.ejpb.2016.11.002.
- X. Ji, K. Damera, Y. Zheng, B. Yu, L. E. Otterbein and B. Wang, J. Pharm. Sci., 2016, 105, 406 CrossRef CAS PubMed.
- C. Szabo, Nat. Rev. Drug Discovery, 2016, 15, 185 CrossRef CAS PubMed.
- K. Nakahira and A. M. K. Choi, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2015, 309, L1387 CrossRef CAS PubMed.
- O. Sulaieva and J. L. Wallace, Curr. Opin. Pharmacol., 2015, 25, 1 CrossRef CAS PubMed.
- C. C. Ramão, W. A. Blattler, J. D. Seixas and G. J. L. Bernardes, Chem. Soc. Rev., 2012, 41, 3571 RSC.
- A. Belhaj, Role of Carbon Monoxide in the Ex Vivo Lung Perfusion Reconditioning, in ClinicalTrials.Gov, University Hospital of Mont-Godinne, National Library of Medicine (US), Bethesda (MD), 2014, present. NLM Identifier: NCT02032082 Search PubMed.
- I. O. Rosas, Study of Inhaled Carbon Monoxide to Treat Idiopathic Pulmonary Fibrosis, in ClinicalTrials.Gov, National Library of Medicine (US), Bethesda (MD), 2010, present. NLM Identifier: NCT01214187 Search PubMed.
- Weill Medical College of Cornell University, Safety Study of Inhaled Carbon Monoxide to Treat Acute Respiratory Distress Syndrome (ARDS), in ClinicalTrials.Gov, National Library of Medicine (US), Bethesda (MD), 2015, present. NLM Identifier: NCT02425579 Search PubMed.
- Medical University of South Carolina, Carbon Monoxide Saturated Medium for Islet Isolation, in ClinicalTrials.Gov, National Library of Medicine (US), Bethesda (MD), 2015, present. NLM Identifier: NCT02567240 Search PubMed.
- R. Machado, Carbon Monoxide Therapy for Severe Pulmonary Arterial Hypertension (CO in PAH), in ClinicalTrials.Gov, University of Illinois at Chicago, National Library of Medicine (US), Bethesda (MD), 2012, present. NLM Identifier: NCT01523548 Search PubMed.
- C. Wang, Daping Hospital and the Research Institute of Surgery of the Third Military Medical University, The Safety and Adverse Reaction Study of Neonatal to Inhaled Carbon Monoxide (CO), in ClinicalTrials.Gov, National Library of Medicine (US). Bethesda (MD), 2013, present. NLM Identifier: NCT01818843 Search PubMed.
- F. Zobi, Future Med. Chem., 2013, 5, 175 CrossRef CAS PubMed.
- U. Schatzschneider, Br. J. Pharmacol., 2015, 172, 1638 CrossRef CAS PubMed.
- B. E. Mann, Top. Organomet. Chem., 2010, 32, 247 CrossRef CAS.
- I. Chakraborty, S. J. Carrington and P. K. Mascharak, Acc. Chem. Res., 2014, 47, 2603 CrossRef CAS PubMed.
- M. A. Gonzales and P. K. Mascharak, J. Inorg. Biochem., 2014, 133, 127 CrossRef CAS PubMed.
- S. J. Carrington, I. Chakraborty and P. K. Mascharak, Dalton Trans., 2015, 44, 13828 RSC.
- R. Mede, M. Klein, R. A. Claus, S. Krieck, S. Quickert, H. Görls, U. Neugebauer, M. Schmitt, G. Gessner, S. H. Heinemann, J. Popp, M. Bauer and M. Westerhausen, Inorg. Chem., 2016, 55, 104 CrossRef CAS PubMed.
- R. Mede, J. Traber, M. Klein, H. Görls, G. Gessner, P. Hoffmann, M. Schmitt, J. Popp, S. H. Heinemann, U. Neugebauer and M. Westerhausen, Dalton Trans., 2017, 46, 1684 RSC.
- M. Tinajero-Trejo, N. Rana, C. Nagel, H. E. Jesse, T. W. Smith, L. K. Wareham, M. Hippler, U. Schatzschneider and R. K. Poole, Antioxid. Redox Signaling, 2016, 24, 765 CrossRef CAS PubMed.
- A. E. Pierri, A. Pallaoro, G. Wu and P. C. Ford, J. Am. Chem. Soc., 2012, 134, 18197 CrossRef CAS PubMed.
- M. A. Wright and J. A. Wright, Dalton Trans., 2016, 45, 6801 RSC.
- E. Stamellou, D. Storz, S. Botov, E. Ntasis, J. Wedel, S. Sollazzo, B. K. Krämer, W. van Son, M. Seelen, G. H. Schmalz, A. Schmidt, M. Hafner and B. A. Yard, Redox Biol., 2014, 2, 739 CrossRef CAS PubMed.
- S. Romanski, E. Stamellou, J. T. Jaraba, D. Storz, B. K. Krämer, M. Hafner, S. Amslinger, H. G. Schmalz and B. A. Yard, Free Radical Biol. Med., 2013, 65, 78 CrossRef CAS PubMed.
- S. Romanski, B. Kraus, M. Guttentag, W. Schlundt, H. Rücker, A. Adler, J. M. Neudörfl, R. Alberto, S. Amslinger and H. G. Schmalz, Dalton Trans., 2012, 41, 13862 RSC.
- S. Romanski, B. Kraus, U. Schatzschneider, J. M. Neudörfl, S. Amslinger and H. G. Schmalz, Angew. Chem., Int. Ed., 2011, 50, 4038 CrossRef CAS.
- H. Meyer, M. Brenner, S. P. Höfert, T.-O. Knedel, P. C. Kunz, A. M. Schmidt, A. Hamacher, M. U. Kassack and C. Janiak, Dalton Trans., 2016, 45, 7605 RSC.
- P. C. Kunz, H. Meyer, J. Barthel, S. Sollazzo, A. M. Schmidt and C. Janiak, Chem. Commun., 2013, 49, 4896 RSC.
- H. Meyer, F. Winkler, P. Kunz, A. M. Schmidt, A. Hamacher, M. U. Kassack and C. Janiak, Inorg. Chem., 2015, 54, 11236 CrossRef CAS PubMed.
- T. Santos-Silva, A. Mukhopadhyay, J. D. Seixas, G. J. L. Bernardes, C. C. Romão and M. J. Romão, J. Am. Chem. Soc., 2011, 133, 1192 CrossRef CAS PubMed.
- M. Chaves-Ferreira, I. S. Albuquerque, D. Matak-Vinkovic, A. C. Coelho, S. M. Carvalho, L. M. Saraiva, C. C. Ramão and G. J. L. Bernardes, Angew. Chem., Int. Ed., 2015, 54, 1172 CrossRef CAS PubMed.
- A. A. Petruk, A. Vergara, D. Marasco, D. Bikiel, F. Doctorovich, D. A. Estrin and A. Merlino, Inorg. Chem., 2014, 53, 10456 CrossRef CAS PubMed.
- J. D. Seixas, A. Mukhopadhyay, T. Santos-Silva, L. E. Otterbein, D. J. Gallo, S. S. Rodrigues, B. H. Guerreiro, A. M. L. Goncalves, N. Penacho, A. R. Marques, A. C. Coelho, P. M. Reis, M. J. Romão and C. C. Romão, Dalton Trans., 2013, 42, 5985 RSC.
- X. Ji, C. Zhou, K. Ji, R. E. Aghoghovbia, Z. Pan, V. Chittavong, B. Ke and B. Wang, Angew. Chem., Int. Ed. Engl., 2016, 55, 15846 CrossRef CAS PubMed.
- D. Z. Wang, E. Viennois, K. Ji, K. Damera, A. Draganov, Y. Q. Zheng, C. Dai, D. Merlin and B. Wang, Chem. Commun., 2014, 50, 15890 RSC.
- E. Palao, T. Slanina, L. Muchová, T. Solomek, L. Vítek and P. Klán, J. Am. Chem. Soc., 2016, 138, 126 CrossRef CAS PubMed.
- P. Peng, C. Wang, Z. Shi, V. K. Johns, L. Ma, J. Oyer, A. Copik, R. Igarashi and Y. Liao, Org. Biomol. Chem., 2013, 11, 6671 CAS.
- L. A. P. Antony, T. Slanina, P. Sebej, T. Solomek and P. Klán, Org. Lett., 2013, 15, 4552 CrossRef CAS PubMed.
- S. N. Anderson, J. M. Richards, H. J. Esquer, A. D. Benninghoff, A. M. Arif and L. M. Berreau, ChemistryOpen, 2015, 4, 590 CrossRef CAS PubMed.
- S. Fetzner, Appl. Environ. Microbiol., 2012, 78, 2505 CrossRef CAS PubMed.
- S. N. Anderson, M. T. Larson and L. M. Berreau, Dalton Trans., 2016, 45, 14570 RSC.
- Y. A. Dávila, M. I. Sancho, M. C. Almandoz and S. E. Blanco, J. Chem. Eng. Data, 2013, 58, 1706 CrossRef.
- K. Grubel, S. Saraf, S. N. Anderson, B. J. Laughlin, R. C. Smith, A. M Arif and L. M. Berreau, Inorg. Chim. Acta, 2012, 407, 91 CrossRef.
- H. J. Kuhn, S. E. Braslavsky and R. Schmidt, Pure Appl. Chem., 2004, 76, 2105 CrossRef CAS.
- C. G. Hatchard and C. A. Parker, Proc. R. Soc. London, Ser. A, 1956, 235, 518 CrossRef CAS.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455 CAS.
- B. Dick and N. P. Ernsting, J. Phys. Chem., 1987, 91, 4261 CrossRef CAS.
- R. Lakowicz, Principles of Fluorescence Spectroscopy, Kluwer Academic/Plenum Publishers, New York, 2nd edn, 1999 Search PubMed.
- S. L. Sorenson, M. Popova, A. M. Arif, and L. M. Berreau, submitted for publication.
- M. Sarkar and P. K. Sengupta, Chem. Phys. Lett., 1991, 179, 68 CrossRef CAS.
- J. Guharay, B. Sengupta and P. K. Sengupta, Proteins, 2001, 43, 75 CrossRef CAS.
- A. Sytnik and I. Litvinyuk, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 12959 CrossRef CAS.
- J. Xiao, M. Suzuki, X. Jiang, X. Chen, K. Yamamoto, F. Ren and M. Xu, J. Agric. Food Chem., 2008, 56, 2350 CrossRef CAS PubMed.
- E.-H. Liu, L.-W. Qi and P. Li, Molecules, 2010, 15, 9092 CrossRef CAS PubMed.
- B. Mishra, A. Barik, K. I. Priyadarsini and H. Mohan, J. Chem. Sci., 2005, 117, 641 CrossRef CAS.
- M.-X. Xie, M. Long, Y. Liu, C. Qin and Y.-D. Wang, Biochim. Biophys. Acta, 2006, 1760, 1184 CrossRef CAS PubMed.
- S. Pal and C. Saha, J. Biomol. Struct. Dyn., 2014, 32, 1132 CAS.
- B. Liu, Y. Pang, R. Bouhenni, E. Duah, S. Paruchuri and L. McDonald, Chem. Commun., 2015, 51, 11060 RSC.
- J. Dai, T. Zou, L. Wang, Y. Zhang and Y. Liu, Luminescence, 2014, 29, 1154 CrossRef CAS PubMed.
- S. Pal, C. Saha, M. Hossain, S. K. Dey and G. S. Kumar, PLoS One, 2012, 7, e43321 CAS.
- A. Sulkowska, B. Bojko, J. Rownicka and W. W. Sulkowski, Biopolymers, 2006, 81, 464 CrossRef CAS PubMed.
- K. Yamasaki, V. T. G. Chuang, T. Maruyama and M. Otagiri, Biochim. Biophys. Acta, 2013, 1830, 5435 CrossRef CAS PubMed.
- F. Zsila, Mol. Pharmaceutics, 2013, 10, 1668 CrossRef CAS PubMed.
- S. N. Anderson, M. Noble, K. Grubel, B. Marshall, A. M. Arif and L. M. Berreau, J. Coord. Chem., 2014, 67, 4061–4075 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1523782. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7ra02653f |
| This journal is © The Royal Society of Chemistry 2017 |