Open Access Article
Open Access ArticleScaling the effect of hydrophobic chain length on gene transfer properties of di-alkyl, di-hydroxy ethylammonium chloride based cationic amphiphiles†
Ankita A. Hiwale‡
a,
Chandrashekhar Voshavar‡b,
Priya Dharmalingamc,
Ashish Dhayania,
Rajesh Mukthavaramd,
Rasajna Nadellac,
Omprakash Sunnapue,
Sivaraman Gandhia,
V. G. M. Naidue,
Arabinda Chaudhuri f,
Srujan Marepally
f,
Srujan Marepally *ac and
Praveen Kumar Vemula
*ac and
Praveen Kumar Vemula *a
*a
aInstitute for Stem Cell Biology and Regenerative Medicine (inStem), GKVK-post, Bellary Road, Bengaluru 560065, India. E-mail: Praveenv@instem.res.in
bBioSatva Technologies, Golnaka, Hyderabad 500013, India
cCentre for Stem Cell Research, Christian Medical College Campus, Bagayam, Vellore 632002, India. E-mail: Srujankm@cmcvellore.ac.in
dTranslational Neuro-oncology Laboratories, Moores Cancer Center, University of California San Diego, La Jolla, CA, USA
eNational Institute for Pharmaceutical Education and Research, Balanagar, Hyderabad 500018, India
fBiomaterials Group, CSIR-Indian Institute of Chemical Technology, Hyderabad 500 007, India
First published on 11th May 2017
Abstract
The success of gene therapy critically depends on the availability of efficient transfection vectors. Cationic lipids are the most widely studied non-viral vectors. The molecular architecture of the cationic lipid determines its transfection efficiency. Variations in alkyl chain lengths of lipids influence self-assembly and liposomal fusion with the cell membrane. These factors determine the transfection ability of the lipid. Thus, to probe the effect of asymmetry in hydrophobic chains on transfection efficiency, we designed and synthesized a series of cationic lipids by systematically varying one of the two alkyl chains linked to the quaternary nitrogen centre from C18 to C10 and keeping the other alkyl C18 chain constant (Lip1818-Lip1810). Transfection studies in multiple cultured mammalian cells (CHO, B16F10 and HeLa) revealed that the lipids with C18:C14 and C18:C12 alkyl chains (Lip1814 & Lip1812) showed 20–30% higher transfection efficacies than their counterparts at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 4
1 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lipid to pDNA charge ratios. Cryo-transmission electron images showed unilamellar vesicle structures for the liposomes of lipids. Mechanistic studies involving Small Angle X-ray Scattering (SAXS) revealed that asymmetry in the hydrophobic region has a significant impact on liposomal fusion with the plasma membrane model. Collectively, these findings demonstrate that chain length asymmetry in the hydrophobic region of cationic lipids has an important role in their liposome–DNA interactions at optimal 2
1 lipid to pDNA charge ratios. Cryo-transmission electron images showed unilamellar vesicle structures for the liposomes of lipids. Mechanistic studies involving Small Angle X-ray Scattering (SAXS) revealed that asymmetry in the hydrophobic region has a significant impact on liposomal fusion with the plasma membrane model. Collectively, these findings demonstrate that chain length asymmetry in the hydrophobic region of cationic lipids has an important role in their liposome–DNA interactions at optimal 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 4
1 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lipid to pDNA charge ratios, which in turn modulates their gene transfer properties.
1 lipid to pDNA charge ratios, which in turn modulates their gene transfer properties.
Introduction
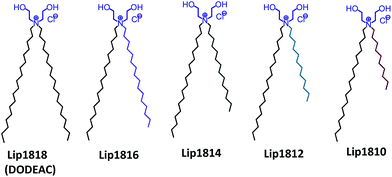
Intracellular delivery of nucleic acids is one of the promising strategies for the development of therapeutic modalities for treating genetic diseases.1–3 Use of cationic lipids has received significant interest to deliver nucleic acids to the specific sites in a targeted manner.4,5 The success of nucleic acid-based therapies critically depends on the efficiency of the carrier.6 Towards developing efficient liposomal systems, structure–activity studies varying the hydrophilic head group, linker functionality and hydrophobic tail of cationic lipids have been performed.7,8 However, these studies indicated a general trend of increased transfection activity as the saturated aliphatic chains increased from C14 to C18.9 Prior findings demonstrated that alkyl chain length had a major role in determining the rigidity of the liposomes.10,11 Longer alkyl chains such as C18 imparts rigidity due to its higher hydrophobicity and ordered packing, while shorter chains such as C10–C14 increases the membrane fluidity.9 Hence, finding the optimal chain length to achieve higher transfection efficiency is a challenging task.12 However, the possible best results can be achieved with a balance between hydrophilic moiety and hydrophobic chain lengths along with a spacer, if any.13 Prior findings, including our own, demonstrated that cationic lipids with the shorter aliphatic chains (C12 & C14) found to be more efficient under in vitro conditions while longer aliphatic chains (C16 & C18) are active for in vivo nucleic acid delivery.14 However, it is difficult to generalize the phenomenon as other factors such as head group and linker play key roles in determining the transfection efficacies both in vitro and in vivo. Understanding the effect of variation in the hydrophobic region of cationic amphiphiles is critical for the development of efficient transfection reagents.15–21 First developments of cationic lipids were reported with identical hydrophobic alkyl chains (symmetric chains).13,22 Zhi et al., demonstrated that transfection could be enhanced by doping two cationic lipids with the same head group and symmetric chains with varying lengths.17,23 Such improved transfection could be due to enhanced inter-membrane mixing of different hydrophobic chains during endocytosis or early endosomal escape of nucleic acids.16 Prior studies, including our own, convincingly demonstrated that the lipids with asymmetry in the hydrophobic region had higher transfection properties than their symmetric counterparts.16,19,20,24 Nantz group demonstrated that the hydrophobic chain asymmetry varying from myristoyl to lauroyl (2 carbons variation) in the liposomal system significantly enhanced the in vivo transfection efficiency of non-glyceryl based lipids.19 However, to the best of our knowledge, in-depth structure–activity investigations were not yet reported addressing the effect in non-glyceryl based lipids or lipids without linkers so far. More importantly, the role of different chain lengths in liposome and DNA interactions with varying charge ratios are not clearly understood. Given the strong modulation effects of asymmetry in the chain lengths on transfection efficacies, we planned to explore the role of variation in hydrophobic chains with a systematic structure-efficiency investigation. To this end, we have used one of our previously reported efficient transfection reagent, DODEAC (N,N-di-n-octadecyl-N,N-dihydroxyethyl ammonium chloride) as a symmetric core/model lipid.25 This cationic lipid contains two C18 chains attached to a quaternary nitrogen-containing two hydroxyl ethylene head groups.To scale the effect of asymmetry within the hydrophobic region of our DODEAC cationic amphiphile, we have synthesized a series of cationic amphiphiles with C18 as a common chain in all the cationic amphiphiles, while varying the other chain from C16–C10 (Fig. 1). We have used DODEAC (with two C18 chains) as control symmetric lipid. Transfection studies in cultured mammalian cell lines showed 20–30% higher transfection efficiency for Lip1814 and Lip1812 in the series. Herein, we present systematic structure–activity investigation on the role of asymmetric hydrophobic chains within a di-alkyl, di-hydroxy ethyl ammonium chloride based cationic amphiphile in influencing transfection activity. Our study includes design, synthesis of lipid series (Lip1818-Lip1810, Fig. 1), physico-chemical characterization and transfection studies using liposomes of synthesized lipids in cultured mammalian cell lines.
Results and discussion
A number of structure–activity investigations have been carried out on the core template/moieties of a cationic amphiphile with varying head groups, charges, spacer effects and hydrophobic cores for enhancing transfection efficiency.22 However, chain length alterations within the hydrophobic core of cationic amphiphiles were not well understood. Such modification studies on the hydrophobic core involved the use of either different alkyl chain lengths or steroidal backbones such as cholesterol and bile acids. To this end, we wanted to evaluate the effect of length of asymmetric chains within the hydrophobic core of a successful transfection reagent, DODEAC.25 We hypothesized that balanced asymmetric chain lengths within di-alkyl, di-hydroxy ethyl ammonium chloride based cationic amphiphiles would cause membrane perturbation resulting in enhanced endocytosis and ultimately leading to a higher transfection activity.Synthesis of lipids (Lip1818-Lip1810)
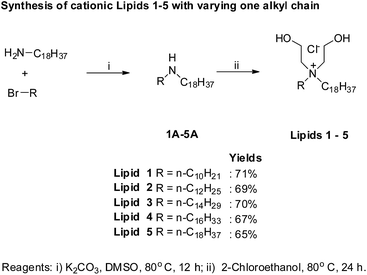
To begin with, we have synthesized a series of lipids (Fig. 1) with variation in one of the two alkyl chains attached to a quaternary nitrogen centre of a cationic lipid, DODEAC. Each lipid possessed one C18 alkyl chain while varying the length of second alkyl chain from C18 to C10 in the hydrophobic region of the lipid. All the lipids were synthesized following 2-step reactions. Initially, individual asymmetric chain containing secondary amines were prepared by N-alkylation of stearyl amine using individual alkyl bromides (C16–C10) along with symmetric C18 bromide. All the secondary amines were subjected for quaternization using 2-chloroethanol to yield final compounds (Lip1818-Lip1810, Fig. 2). All the lipids were purified by re-crystallization and characterized using NMR (1H & 13C) and ESI-MS. For both secondary amine intermediates (1A–5A, Fig. 2) and final lipids (Lip1818-Lip1810), there was no significant variation in the yields with a decrease in the chain length.Physico-chemical characterization
One of the major issues in the transfections is the stability of lipoplexes and their membrane interaction, which plays a key role in cellular uptake and strongly modulate the transfection efficacies. Such modulating factors are likely to depend on the size and global surface charges of lipoplexes. Self-assembly of cationic lipids (Lip1818-Lip1810, Fig. 1) produced unilamellar vesicles that were characterized using dynamic light scattering (DLS) and cryo-TEM. Liposomes of these lipids showed hydrodynamic diameter in the range of 125 to 150 nm (Table S1, ESI†), while zeta potentials were within +45 to +49 except Lip1810 which showed +35.7 (Table S1, ESI†). Size & surface potential measurements of liposomes did not show any difference among lipids, Lip1818-Lip1810 possibly due to single positive charge in all five lipids. Furthermore, liposomes made from all five lipids were complexed with plasmid DNA (pDNA), and size & zeta potentials of the resulting lipoplexes were measured (Tables S2 and S3, ESI†). Data suggests that lipoplexes (lipid–DNA complexes) have higher size (400–1900 nm) compared to liposomes (125–150 nm). Interestingly, lipid/DNA charge ratio seems to have a significant effect on size and potentials of lipoplexes. Decreasing lipid/DNA charge ratio from 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 increased the size of lipoplexes (Table S2, ESI†) while zeta potential has decreased (Table S3, ESI†). Surface potentials of lipoplexes at 8
1 increased the size of lipoplexes (Table S2, ESI†) while zeta potential has decreased (Table S3, ESI†). Surface potentials of lipoplexes at 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–4
1–4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lipid
1 lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratios are positive due to higher lipid ratio. While, at lower charge ratios 2
DNA charge ratios are positive due to higher lipid ratio. While, at lower charge ratios 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 varied with a low positive charge to mostly negative charge for all the lipids (Table S3, ESI†). Overall, data from global surface charges and potentials indicate that there is no clear correlation with transfection efficiencies of lipids, Lip1818-Lip1810. It also rules out the possibility of their major role in influencing the transfection efficacies. Next, we measured the DNA binding ability for lipids Lip1818-Lip1810 across lipid
1 varied with a low positive charge to mostly negative charge for all the lipids (Table S3, ESI†). Overall, data from global surface charges and potentials indicate that there is no clear correlation with transfection efficiencies of lipids, Lip1818-Lip1810. It also rules out the possibility of their major role in influencing the transfection efficacies. Next, we measured the DNA binding ability for lipids Lip1818-Lip1810 across lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratios 8
DNA charge ratios 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to understand whether any difference in the lipid DNA interactions using conventional agarose gel retardation assay. Liposomes of all the lipids were complexed with pDNA and subjected to gel retardation assay in 1% agarose gel with ethidium bromide as intercalating agent. The lipid
1 to understand whether any difference in the lipid DNA interactions using conventional agarose gel retardation assay. Liposomes of all the lipids were complexed with pDNA and subjected to gel retardation assay in 1% agarose gel with ethidium bromide as intercalating agent. The lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA ratios are indicated at the top of each lane.
DNA ratios are indicated at the top of each lane.
Results from gel-retardation assay showed that DNA was completely complexed with the liposomes of Lip1818-Lip1810 from lipid/DNA charge ratios, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 2
1 to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 while 1
1 while 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 had some unbound DNA (Fig. 3). The conventional gel retardation assay indicates strong lipid
1 had some unbound DNA (Fig. 3). The conventional gel retardation assay indicates strong lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA binding interactions for all the lipids, reveals that the hydrophobic chain length variation has no role in determining the lipid–DNA interactions. Following DLS measurements and DNA binding studies, we have employed the cryo-TEM technique to observe the structural characteristics of liposomes of Lip1818-Lip1810. Cryo-TEM images showed that liposomes of individual lipids possess similar structural and size patterns (∼100–150 nm) revealing their unilamellar nature (Fig. 4). Physico-chemical data for liposomes and lipoplexes of Lip1818-Lip1810 obtained from DNA binding, DLS studies and cryo-TEM collectively demonstrated that all the lipids formed unilamellar liposomes and showed efficient complexation with pDNA.
DNA binding interactions for all the lipids, reveals that the hydrophobic chain length variation has no role in determining the lipid–DNA interactions. Following DLS measurements and DNA binding studies, we have employed the cryo-TEM technique to observe the structural characteristics of liposomes of Lip1818-Lip1810. Cryo-TEM images showed that liposomes of individual lipids possess similar structural and size patterns (∼100–150 nm) revealing their unilamellar nature (Fig. 4). Physico-chemical data for liposomes and lipoplexes of Lip1818-Lip1810 obtained from DNA binding, DLS studies and cryo-TEM collectively demonstrated that all the lipids formed unilamellar liposomes and showed efficient complexation with pDNA.
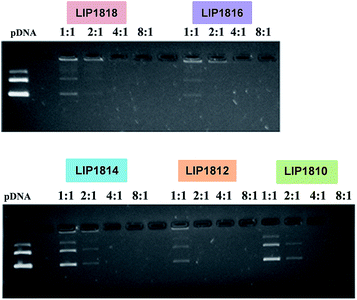 | ||
| Fig. 3 Electrophoretic gel patterns for lipid-associated DNA in gel retardation assay for lipids (Lip1818-Lip1810). | ||
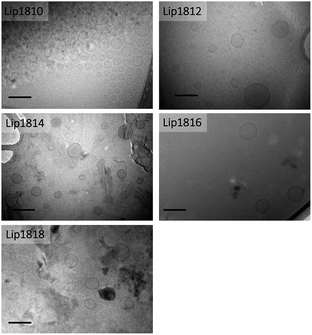 | ||
| Fig. 4 Cryo-transmission electron microscopic images of liposomes of lipids (Lip1818-Lip1810) (scale 0.1 μm). | ||
Cytotoxicity assay
MTT-based colorimetric cell viability assay was performed in representative HeLa cells with lipoplexes of lipids (Lip1818-Lip1810) and plasmid DNA for 24 h. Results showed high percent cell viabilities (>75%) for lipids Lip1818-Lip1810 within all the lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratios (Fig. S1, ESI†). HeLa cells treated with lipoplexes of lipids Lip1818-Lip1810 at 1
DNA charge ratios (Fig. S1, ESI†). HeLa cells treated with lipoplexes of lipids Lip1818-Lip1810 at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratio revealed remarkably high cell viabilities (>95% and >90% for 1
1 charge ratio revealed remarkably high cell viabilities (>95% and >90% for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 & 2
1 & 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratios, respectively). Percent cell viabilities at 4
1 charge ratios, respectively). Percent cell viabilities at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 8
1 and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratios of lipids, Lip1818-Lip1810 were found to be low within the range of 90–75% as depicted in Fig. S1, ESI.† Since all the lipids showed comparable cell viability profiles across varying lipid
1 charge ratios of lipids, Lip1818-Lip1810 were found to be low within the range of 90–75% as depicted in Fig. S1, ESI.† Since all the lipids showed comparable cell viability profiles across varying lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratios, the enhanced and/or compromised transfection efficacies of individual lipids Lip1818-Lip1810 cannot be attributed to their cytotoxicity.
DNA charge ratios, the enhanced and/or compromised transfection efficacies of individual lipids Lip1818-Lip1810 cannot be attributed to their cytotoxicity.
Transfection studies
In vitro transfection efficacies were evaluated to compare the enhancement in their activities due to change in the hydrophobic chain lengths for the series of lipids, Lip1818-Lip1810 in cultured animal cell lines, CHO and B16F10 using pCMV-SPORT-β-gal plasmid DNA encoding a reporter gene/enzyme ‘β-galactosidase’. Transfection studies in both cell lines i.e. CHO & B16F10 showed an interesting trend of transfection profiles among lipids Lip1818-Lip1810 with an inverted ‘V’ curve (Fig. 5).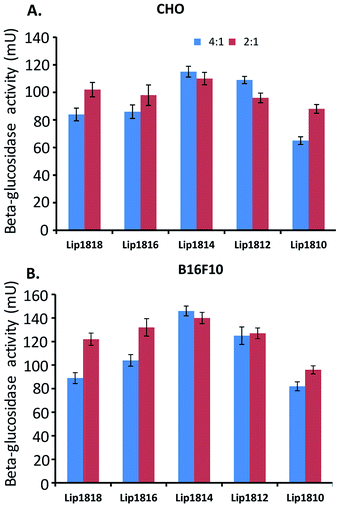 | ||
Fig. 5 In vitro transfection studies of lipids (Lip1818-Lip1810). Transfection efficiencies of lipids were evaluated in CHO and B16F10 cell lines at lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratio of 2 DNA charge ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 4 1 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Absorption at 405 nm was converted to β-galactosidase units using a calibration curve constructed with pure commercial β-galactosidase enzyme. Units of β-galactosidase activity were plotted against the lipids. The transfection values shown are the average of triplicate experiments performed with 4–6 data points.§ 1. Absorption at 405 nm was converted to β-galactosidase units using a calibration curve constructed with pure commercial β-galactosidase enzyme. Units of β-galactosidase activity were plotted against the lipids. The transfection values shown are the average of triplicate experiments performed with 4–6 data points.§ | ||
We found that transfection efficiencies increased as the chain length decreased from C18 to C14 i.e. from Lip1818 to Lip1814 followed by decrease in the transfection with further reduction in the chain lengths, C12 to C10 i.e. Lip1812 to Lip1810 in all the lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratios except 8
DNA charge ratios except 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 5 and S2, ESI†). At 8
1 (Fig. 5 and S2, ESI†). At 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratio, we observed a slight increase in the transfection from Lip1818 to Lip1816 followed by decrease in the transfection (Fig. S2, ESI†). In CHO cells, highest transfection was observed at 4
1 charge ratio, we observed a slight increase in the transfection from Lip1818 to Lip1816 followed by decrease in the transfection (Fig. S2, ESI†). In CHO cells, highest transfection was observed at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lipid
1 lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratio for all the lipids (Fig. 5A). In this, Lip1818 & Lip1816 showed comparatively similar transfection efficacy (with ∼85 mU of β-galactosidase activity) while Lip1814 showed increased activity of ∼120 mU. Lip1812 & Lip1810 showed ∼110 and ∼65 mU of activity respectively. Similar profiles were observed in B16F10 cells at 4
DNA charge ratio for all the lipids (Fig. 5A). In this, Lip1818 & Lip1816 showed comparatively similar transfection efficacy (with ∼85 mU of β-galactosidase activity) while Lip1814 showed increased activity of ∼120 mU. Lip1812 & Lip1810 showed ∼110 and ∼65 mU of activity respectively. Similar profiles were observed in B16F10 cells at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio which showed the highest transfection among all the lipid
1 ratio which showed the highest transfection among all the lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratios. Lip1818 showed ∼90 mU and Lip1816 exhibited slightly increased activity with ∼105 mU. However, Lip1814 showed high transfection with ∼145 mU while Lip1812 & Lip1810 showed ∼125 and ∼80 mU of activity respectively (Fig. 5B). Results from transfection studies revealed that the asymmetry has a pattern at 4
DNA charge ratios. Lip1818 showed ∼90 mU and Lip1816 exhibited slightly increased activity with ∼105 mU. However, Lip1814 showed high transfection with ∼145 mU while Lip1812 & Lip1810 showed ∼125 and ∼80 mU of activity respectively (Fig. 5B). Results from transfection studies revealed that the asymmetry has a pattern at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 & 2
1 & 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lipid to DNA charge ratio in both CHO & B16F10 cells. In summary, among 5 lipids, Lip1814 and Lip1812 found to be effective in delivering genes compared to Lip1818, Lip1816 and Lip1810 in multiple cultured cell lines. This demonstrated that optimal variation of alkyl chain lengths in di-alkyl, di-hydroxy ethylammonium chloride based cationic amphiphiles are 1814 and 1812. These results suggest that the optimal chain length variation in the hydrophobic core of the lipids is 18
1 lipid to DNA charge ratio in both CHO & B16F10 cells. In summary, among 5 lipids, Lip1814 and Lip1812 found to be effective in delivering genes compared to Lip1818, Lip1816 and Lip1810 in multiple cultured cell lines. This demonstrated that optimal variation of alkyl chain lengths in di-alkyl, di-hydroxy ethylammonium chloride based cationic amphiphiles are 1814 and 1812. These results suggest that the optimal chain length variation in the hydrophobic core of the lipids is 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14 and 18
14 and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12, a further increase in the alkyl chain length variations leads to a decrease in the transfection activity that we found in Lip1810 (Fig. 5).
12, a further increase in the alkyl chain length variations leads to a decrease in the transfection activity that we found in Lip1810 (Fig. 5).
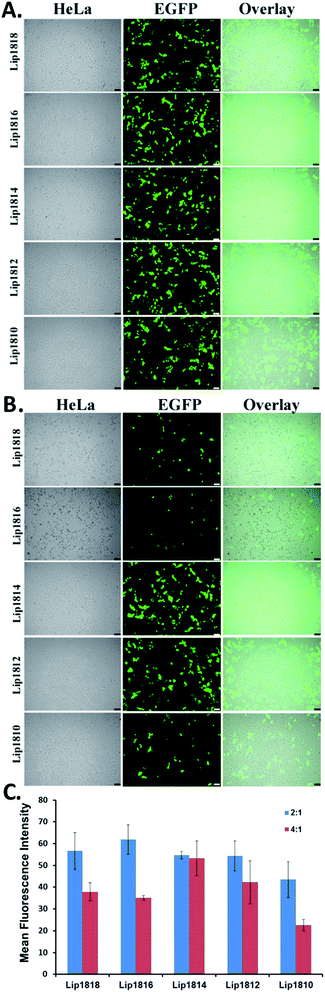
Cellular uptake and expression studies
Following the transfection experiments, we have performed cellular uptake and expression studies by microscopy imaging at lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratio of 4
DNA charge ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 & 2
1 & 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in representative HeLa cells. To this end, we have transfected HeLa cells with lipoplexes of lipids Lip1818-Lip1810 and pα5GFP plasmid DNA encoding Green Fluorescent Protein (GFP) (Fig. 6). Results from imaging studies are in-line with the transfection data obtained in CHO & B16F10 cells (compare Fig. 5 with Fig. 6). HeLa cells showed high GFP expression for Lip1814 and Lip1812 followed by Lip1816 & Lip1810. Lip1818 showed the least fluorescence among all the lipids (Fig. 6A and B). Imaging data obtained is consistent with the transfection data.
1 in representative HeLa cells. To this end, we have transfected HeLa cells with lipoplexes of lipids Lip1818-Lip1810 and pα5GFP plasmid DNA encoding Green Fluorescent Protein (GFP) (Fig. 6). Results from imaging studies are in-line with the transfection data obtained in CHO & B16F10 cells (compare Fig. 5 with Fig. 6). HeLa cells showed high GFP expression for Lip1814 and Lip1812 followed by Lip1816 & Lip1810. Lip1818 showed the least fluorescence among all the lipids (Fig. 6A and B). Imaging data obtained is consistent with the transfection data.
SAXS studies
Interaction of liposomes with cellular membrane plays a key role in the cellular uptake of lipoplexes (lipid–DNA complexes). Liposomes are known to perturb the cell membranes inducing spontaneous curvature to facilitate the endocytosis.26,27 The closer arrangement of the individual lipids imparts rigidity to the liposome that in turn affects membrane perturbation properties. To gain insight on the role of the hydrophobic core of each lipid (Lip1818-Lip1810) with the cell membrane, we employed Small Angle X-ray Scattering (SAXS) technique.28 In this experiment, a model membrane was prepared with DOPC, DOPE, DOPS and cholesterol at 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, w/w ratio. Liposomes of each lipid were combined with the model membrane at 1
15, w/w ratio. Liposomes of each lipid were combined with the model membrane at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio at 25 °C was used to record the scattering patterns. SAXS diagrams show a lamellar arrangement for lipids, Lip1818-Lip1810 alone. Scattering curves obtained from SAXS experiment shows perturbation of model membrane with liposomes of lipids Lip1818-Lip1810 (Fig. S3, ESI†). Although it is not clear, we observed sequential variation in the lamellar phase (phase transition) for lipids, Lip1818-Lip1810 with membrane model (Fig. S3, ESI†). However, due to irregular spacing/peaks in the SAXS patterns, it is hard to identify and assign the transitioned phase. The degree of variation in phase transitions for Lip1816, Lip1814 & Lip1812 is more prominent in the SAXS diagrams than for Lip1818 & Lip1810. In addition, liposomes of lipids, Lip1816, Lip1814 & Lip1812 exhibit strong electrostatic interactions over liposomes of lipids, Lip1818 & Lip1810 as revealed by their larger variation in the observed intensities (Y-axis in Fig. S3, ESI†). However, further scattering measurements on the interaction of lipoplexes (lipid–DNA complex) of lipids Lip1818-Lip1810 with model biomembrane might shed more light on their membrane perturbation and cellular internalization.
1 molar ratio at 25 °C was used to record the scattering patterns. SAXS diagrams show a lamellar arrangement for lipids, Lip1818-Lip1810 alone. Scattering curves obtained from SAXS experiment shows perturbation of model membrane with liposomes of lipids Lip1818-Lip1810 (Fig. S3, ESI†). Although it is not clear, we observed sequential variation in the lamellar phase (phase transition) for lipids, Lip1818-Lip1810 with membrane model (Fig. S3, ESI†). However, due to irregular spacing/peaks in the SAXS patterns, it is hard to identify and assign the transitioned phase. The degree of variation in phase transitions for Lip1816, Lip1814 & Lip1812 is more prominent in the SAXS diagrams than for Lip1818 & Lip1810. In addition, liposomes of lipids, Lip1816, Lip1814 & Lip1812 exhibit strong electrostatic interactions over liposomes of lipids, Lip1818 & Lip1810 as revealed by their larger variation in the observed intensities (Y-axis in Fig. S3, ESI†). However, further scattering measurements on the interaction of lipoplexes (lipid–DNA complex) of lipids Lip1818-Lip1810 with model biomembrane might shed more light on their membrane perturbation and cellular internalization.
Conclusion
The role of asymmetric chain lengths within the hydrophobic core of cationic lipids in influencing transfection efficacies was evaluated in the present investigation. We designed and synthesized a series of cationic lipids by varying one of the two alkyl chains linked to quaternary nitrogen centre from C10 to C16 and keeping the other alkyl chain C18 as constant. All the lipids, Lip1818-Lip1810 showed significant transfection efficiencies in delivering reporter genes into multiple cultured mammalian cells including CHO, B16F10 and HeLa. Interestingly, Lip1814 and Lip1812 (with 18, 14 & 18, 12 alkyl chains respectively) showed higher transfections than the other three lipids at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lipid to DNA charge ratio. The transfection profiles revealed that the interaction among lipids with DNA varied with a variation in the asymmetry. Dynamic laser light scattering studies revealed that no significant size difference among the liposomes of lipids, Lip1818-Lip1810. However, liposomes of Lip1814 and Lip1812 exhibited strong DNA condensation at 4
1 lipid to DNA charge ratio. The transfection profiles revealed that the interaction among lipids with DNA varied with a variation in the asymmetry. Dynamic laser light scattering studies revealed that no significant size difference among the liposomes of lipids, Lip1818-Lip1810. However, liposomes of Lip1814 and Lip1812 exhibited strong DNA condensation at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lipid to DNA charge ratio. These findings were further confirmed by SAXS experiments. Thus, cumulative results suggest that the varying chain length asymmetry within the hydrophobic core of cationic amphiphile has an important role in DNA interaction of its liposome, in turn, affects the transfection properties. Collectively, the present findings further enriched our understanding of the lipid asymmetry and its superior influence on DNA binding properties, consequently determining/enhancing the gene transfer efficacies.
1 lipid to DNA charge ratio. These findings were further confirmed by SAXS experiments. Thus, cumulative results suggest that the varying chain length asymmetry within the hydrophobic core of cationic amphiphile has an important role in DNA interaction of its liposome, in turn, affects the transfection properties. Collectively, the present findings further enriched our understanding of the lipid asymmetry and its superior influence on DNA binding properties, consequently determining/enhancing the gene transfer efficacies.
Materials and methods
General procedures and reagents
1H & 13C NMR spectra were recorded on an AV 300 MHz NMR spectrometer. Mass spectral data were acquired by using a commercial LCQ ion trap mass spectrometer (ThermoFinnigan, SanJose, CA, USA) equipped with an ESI source or micromass Quatro LC triple quadrupole mass spectrometer for ESI-mass analysis. Column chromatography was performed with silica gel (Acme Synthetic Chemicals, India, 60–120 mesh). All the reagents were purchased from Sigma-Aldrich, St. Louis, USA unless otherwise stated. NP-40, antibiotics, and agarose were procured from Hi-media, India. HeLa, B16F10 and CHO cells were procured from the National Centre for Cell Sciences (NCCS), Pune, India. Cells were grown at 37 °C in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS in a humidified atmosphere containing 5% CO2/95% air.Synthesis
Detailed procedures for synthesizing the di-alkyl, di-hydroxy ethyl ammonium chloride based cationic amphiphiles are given below. Structures of all lipids and their synthetic intermediates were confirmed by 1H, 13C NMR and ESI-MS. Synthesis of cationic lipids was shown Fig. 2.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 methanol
95 methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chloroform, v/v). The yield of 1A: 87%; yield of 2A: 85%; yield of 3A: 85%; yield of 4A: 79%; yield of 5A: 80%.
chloroform, v/v). The yield of 1A: 87%; yield of 2A: 85%; yield of 3A: 85%; yield of 4A: 79%; yield of 5A: 80%.
(i) N-Octadecyloctadecan-1-amine (intermediate 1A, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.83–0.89 (t, 6H), 1.19–1.32 (m, 60H); 1.39–1.42 (m, 4H), 1.71–1.76 (bs, 1H), 2.50–2.58 (m, 4H).
ESI-MS: C36H75N, calculated 521.59; found 522 [M+].
(ii) N-Hexadecyloctadecan-1-amine (intermediate 2A, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.82–0.89 (t, 6H), 1.21–1.30 (m, 56H); 1.39–1.43 (m, 4H), 1.73–1.80 (bs, 1H), 2.48–2.55 (m, 4H).
ESI-MS: C34H71N, calculated 493.56; found 494 [M+].
(iii) N-Tetradecyloctadecan-1-amine (intermediate 3A, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.85–0.91 (t, 6H), 1.21–1.30 (m, 52H); 1.41–1.46 (m, 4H), 1.73–1.82 (bs, 1H), 2.53–2.60 (m, 4H).
ESI-MS: C32H67N, calculated 465.53; found 466 [M+].
(iv) N-Dodecyloctadecan-1-amine (intermediate 4A, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.81–0.89 (t, 6H), 1.15–1.23 (m, 48H); 1.38–1.44 (m, 4H), 1.70–1.78 (bs, 1H), 2.51–2.57 (m, 4H).
ESI-MS: C30H63N, calculated 437.50; found 438 [M+].
(v) N-Decyloctadecan-1-amine (intermediate 5A, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.85–0.91 (t, 6H), 1.21–1.30 (m, 44H); 1.41–1.46 (m, 4H), 1.73–1.82 (bs, 1H), 2.53–2.60 (m, 4H).
ESI-MS: C28H59N, calculated 409.46; found 410 [M+].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 methanol
95 methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chloroform, v/v). The yield of Lip1818: 71%; yield of Lip1816: 69%; yield of Lip1814: 70%; yield of Lip1812: 67%; yield of Lip1810: 65%.
chloroform, v/v). The yield of Lip1818: 71%; yield of Lip1816: 69%; yield of Lip1814: 70%; yield of Lip1812: 67%; yield of Lip1810: 65%.
(i) 2,2′-(Dioctadecyl-L4-azanediyl)bis(ethan-1-ol), chloride salt (Lip1818, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.9 (t, 6H), 1.2 (m, 64H), 1.7 (m, 4H), 3.5 (m, 4H), 3.8 (m, 4H), 4.2 (m, 4H).
ESI-MS: C40H84NO2, calculated 610.65; found 610 [M+].
(ii) 2,2′-(Hexadecyl(octadecyl)-λ4-azanediyl)bis(ethan-1-ol), chloride salt (Lip1816, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.83–0.92 (t, 6H), 1.12–1.40 (m, 56H); 1.61–1.72 (m, 4H), 3.38–3.48 (m, 4H), 3.63–3.73 (m, 4H), 4.04–4.15 (m, 4H), 5.47–5.58 (m, 2H).
ESI-MS: C38H80NO2, calculated 582.62; found 583 [M+].
(iii) 2,2′-(Tetradecyl(octadecyl)-λ4-azanediyl)bis(ethan-1-ol), chloride salt (Lip1814, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.85–0.93 (t, J = 7.2 Hz, 6H), 1.17–1.41 (m, 56H); 3.00–3.20 (m, 4H), 3.38–3.49 (m, 2H), 3.65–3.73 (m, 2H), 3.96–4.15 (m, 4H), 5.38–5.48 (bs, 2H).
ESI-MS: C36H76NO2, calculated 554.59; found 555 [M+].
(iv) 2,2′-(Dodecyl(octadecyl)-λ4-azanediyl)bis(ethan-1-ol), chloride salt (Lip1812, Fig. 2). 1H NMR (CDCl3, 300 MHz): δ 0.82–0.93 (t, 6H), 1.16–1.41 (m, 52H); 3.35–3.49 (m, 4H), 3.63–3.73 (m, 4H), 4.03–4.15 (m, 4H), 5.35–5.77 (m, 2H).
ESI-MS: C34H72NO2, calculated 526.56; found 527 [M+].
(v) 2,2′-(Decyl(octadecyl)-λ4-azanediyl)bis(ethan-1-ol), chloride salt (Lip1810, Fig. 2). 1H NMR (CDCl3, 500 MHz): δ 0.85–0.91 (t, J = 7.0 Hz, 6H), 1.19–1.32 (m, 36H); 1.32–1.40 (bs, 8H), 1.61–1.72 (bs, 4H), 3.39–3.47 (m, 4H), 3.64–3.71 (m, 4H), 4.05–4.13 (m, 4H), 5.47–5.54 (m, 2H).
ESI-MS: C32H68NO2, calculated 498.53; found 499 [M+].
Preparation of liposomes and plasmid DNA
1 mM liposomes were prepared with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mole ratios of each lipid and cholesterol. Briefly, the cationic lipids and cholesterol in the appropriate mole ratios were dissolved in chloroform (500 μl) in a glass vial. The solvent was removed with a thin flow of moisture-free nitrogen gas and the lipid film was kept for drying under high vacuum for 6 h. 1 ml of sterile deionized water was added to the vacuum dried lipid films and the mixtures were allowed to swell overnight. The vials were then vortexed for 2–3 minutes at room temperature to produce multilamellar vesicles (MLVs). MLVs were then sonicated initially in a water bath followed by an ice bath until clarity using a Branson 450 Sonifier at 100% duty cycle and 25 W output power to produce small unilamellar vesicles (SUVs). The p-CMV-SPORT-β-gal plasmid was amplified in DH5α-strain of Escherichia coli, isolated by alkaline lysis procedure and finally purified by PEG-8000 precipitation as described previously.29 The purity of plasmid was checked by A260/A280 ratio (around 1.9) and 1% agarose gel electrophoresis.
1 mole ratios of each lipid and cholesterol. Briefly, the cationic lipids and cholesterol in the appropriate mole ratios were dissolved in chloroform (500 μl) in a glass vial. The solvent was removed with a thin flow of moisture-free nitrogen gas and the lipid film was kept for drying under high vacuum for 6 h. 1 ml of sterile deionized water was added to the vacuum dried lipid films and the mixtures were allowed to swell overnight. The vials were then vortexed for 2–3 minutes at room temperature to produce multilamellar vesicles (MLVs). MLVs were then sonicated initially in a water bath followed by an ice bath until clarity using a Branson 450 Sonifier at 100% duty cycle and 25 W output power to produce small unilamellar vesicles (SUVs). The p-CMV-SPORT-β-gal plasmid was amplified in DH5α-strain of Escherichia coli, isolated by alkaline lysis procedure and finally purified by PEG-8000 precipitation as described previously.29 The purity of plasmid was checked by A260/A280 ratio (around 1.9) and 1% agarose gel electrophoresis.
DNA binding assay
The DNA binding ability of the lipids were assessed by their gel retardation assay on a 1% agarose gel (pre-stained with ethidium bromide) across the varying lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratios of 8
DNA charge ratios of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. pCMV-β-gal (0.3 μg) was complexed with the varying amount of cationic lipids in a total volume of 30 μl in HEPES buffer (pH 7.4) and incubated at room temperature for 20–25 minutes. 4 μl of 6× loading buffer (0.25% bromophenol blue in 40% (w/v) sucrose with sterile H2O) was added to it and from the resulting solution 30 μl was loaded on each well. The samples were electrophoresed at 80 V for 45 minutes and the DNA bands were visualized in the gel documentation unit.
1. pCMV-β-gal (0.3 μg) was complexed with the varying amount of cationic lipids in a total volume of 30 μl in HEPES buffer (pH 7.4) and incubated at room temperature for 20–25 minutes. 4 μl of 6× loading buffer (0.25% bromophenol blue in 40% (w/v) sucrose with sterile H2O) was added to it and from the resulting solution 30 μl was loaded on each well. The samples were electrophoresed at 80 V for 45 minutes and the DNA bands were visualized in the gel documentation unit.
Zeta potential (ξ) and size measurements
The sizes and the zeta potentials (surface charges) of neat liposomes and lipoplexes with varying charge ratios (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were measured by photon correlation spectroscopy and electrophoretic mobility on a Zeta sizer 3000HSA (Malvern UK). The sizes and potentials of liposomes were measured in deionised water with a sample refractive index of 1.59 and a viscosity of 0.89. Liposomes of lipids, Lip1818-Lip1810 were complexed with DNA in plain DMEM for size and potential measurements of lipoplexes. The system was validated by using the 200 nm + 5 nm polystyrene polymer (Duke Scientific Corps. Palo Alto, CA). The diameters of liposomes and lipoplexes were calculated by using the automatic mode. The zeta potential was measured using the following parameters: viscosity, 0.89 cP; dielectric constant, 79; temperature, 25 °C; F(Ka), 1.50 (Smoluchowski); maximum voltage of the current, V. Using DTS0050 standard from Malvern, UK validated the system. All the size measurements were done 10 times in triplicate with the zero field correction and values represented as the average of triplicate measurements. The potentials were measured 10 times and represented as their average values as calculated by using the Smoluchowski approximation.
1) were measured by photon correlation spectroscopy and electrophoretic mobility on a Zeta sizer 3000HSA (Malvern UK). The sizes and potentials of liposomes were measured in deionised water with a sample refractive index of 1.59 and a viscosity of 0.89. Liposomes of lipids, Lip1818-Lip1810 were complexed with DNA in plain DMEM for size and potential measurements of lipoplexes. The system was validated by using the 200 nm + 5 nm polystyrene polymer (Duke Scientific Corps. Palo Alto, CA). The diameters of liposomes and lipoplexes were calculated by using the automatic mode. The zeta potential was measured using the following parameters: viscosity, 0.89 cP; dielectric constant, 79; temperature, 25 °C; F(Ka), 1.50 (Smoluchowski); maximum voltage of the current, V. Using DTS0050 standard from Malvern, UK validated the system. All the size measurements were done 10 times in triplicate with the zero field correction and values represented as the average of triplicate measurements. The potentials were measured 10 times and represented as their average values as calculated by using the Smoluchowski approximation.
Cryo-transmission electron microscopy of liposomes and lipoplexes
The sample (5 μl) was snap frozen using Tecnai Vitrobot (FEI Company, Hillsboro, OR, USA) in liquid ethane using holey carbon grids. Sample preparation was done at a constant temperature of 22 °C. To relative humidity was maintained in the chamber to prevent solvent evaporation and changes in solvent concentration. The parameters used were as follows: blot time (s) – 1.0, blot force – 0, wait time (s) – 1.0, blot total – 1, drain time (s) – 0.5. Until imaging, the vitrified specimens were stored under liquid nitrogen. Imaging of the cryo-samples was done using Tecnai G2 Spirit Bio-TWIN Transmission Electron Microscope (FEI Company, Hillsboro, OR, USA) at 100 kV with 4k × 2k GatanOrius side mount CCD camera (Gatan, Pleasanton, CA).Small angle X-ray scattering (SAXS) measurements
The Small Angle X-ray Scattering (SAXS) system was used to decipher the physical and structural interactions of liposomes of lipids, Lip1818-Lip1810 with the model membrane in solution. SAXS experiments were performed at Centre for Cellular and Molecular Biology, Hyderabad, India. The SAXS facility at CCMB is S3-MICRO Point-Focus system, Hecus X-ray systems, GmbH with 50 watt source power and Cu-Kα (λ = 1.54 Å) X-rays. The beam size at the sample is 50 × 200 μm2 with photon flux upto 2 × 108 photons per s. Pilatus 100k detector with a pixel size of 172 × 172 μm2 with a resolution range of 2000 Å to 30 Å was used. The sample-detector distance was maintained at 300 mm and Q-range of 0.003 to 0.6 Å−1 that enables to cover a distance range of 2000 to 11 Å. Samples were filled into glass capillaries and flame-sealed. Scans were performed at 22 °C with an exposure time of 1 h. Data was collected by using a Pilatus 100k detector. Diffraction intensity vs. Q plots were obtained by radial integration of the 2D patterns by using the interactive data evaluating program FIT2D followed by ATSAS program suite.Transfection studies
Cells were seeded at a density of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells (CHO, B16F10) per well in a 96-well plate 12–18 h before the transfection. 0.3 μg of plasmid DNA was complexed with varying amounts of lipids (0.9–7.2 nmol) in complete DMEM/MEM medium (total volume made up to 100 μl) for 30 minutes. The lipid
000 cells (CHO, B16F10) per well in a 96-well plate 12–18 h before the transfection. 0.3 μg of plasmid DNA was complexed with varying amounts of lipids (0.9–7.2 nmol) in complete DMEM/MEM medium (total volume made up to 100 μl) for 30 minutes. The lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA (±) charge ratios were from 8
DNA (±) charge ratios were from 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 over these ranges of the lipids. The complexes were then added to the cells and incubated. The reporter gene activity was estimated between 36–48 h. The cells were washed with PBS (2 × 100 μl) and lysed with 50 μl lysis buffer [0.25 M Tris–HCl pH 8.0, 0.5% NP40]. Care was taken to ensure complete lysis. The β-galactosidase activity per well was estimated by adding 50 μl of 2×-substrate solution [1.33 mg ml−1 of ONPG, 0.2 M sodium phosphate (pH 7.3) and 2 mM magnesium chloride] to the lysate in a 96-well plate. Absorption at 405 nm was converted to β-galactosidase units using a calibration curve constructed with the pure commercial β-galactosidase enzyme. The transfection experiments are the average of triplicate experiments with at least 3–4 values in each experiment.
1 over these ranges of the lipids. The complexes were then added to the cells and incubated. The reporter gene activity was estimated between 36–48 h. The cells were washed with PBS (2 × 100 μl) and lysed with 50 μl lysis buffer [0.25 M Tris–HCl pH 8.0, 0.5% NP40]. Care was taken to ensure complete lysis. The β-galactosidase activity per well was estimated by adding 50 μl of 2×-substrate solution [1.33 mg ml−1 of ONPG, 0.2 M sodium phosphate (pH 7.3) and 2 mM magnesium chloride] to the lysate in a 96-well plate. Absorption at 405 nm was converted to β-galactosidase units using a calibration curve constructed with the pure commercial β-galactosidase enzyme. The transfection experiments are the average of triplicate experiments with at least 3–4 values in each experiment.
Cytotoxicity assay
The cytotoxicity of lipids Lip1818-Lip1810 was evaluated in representative HeLa cells across the lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratios of 8
DNA charge ratios of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) based reduction assay as described earlier.29 The cytotoxicity assay was performed in 96-well plates by maintaining the same ratio of a number of cells to an amount of cationic lipid, as used in the previously described transfection experiments. Briefly, 4 h after the addition of lipoplexes, media was replaced with complete media and incubated for 24 h. After 24 h, MTT (5 mg ml−1 in PBS) was added to cells and incubated for 4 h at 37 °C. Results were expressed as percent viability = [A540(treated cells) − background/A540(untreated cells) − background] × 100.
1 using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) based reduction assay as described earlier.29 The cytotoxicity assay was performed in 96-well plates by maintaining the same ratio of a number of cells to an amount of cationic lipid, as used in the previously described transfection experiments. Briefly, 4 h after the addition of lipoplexes, media was replaced with complete media and incubated for 24 h. After 24 h, MTT (5 mg ml−1 in PBS) was added to cells and incubated for 4 h at 37 °C. Results were expressed as percent viability = [A540(treated cells) − background/A540(untreated cells) − background] × 100.
Cellular uptake studies by epifluorescence microscopy
For fluorescence microscopy experiments, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells (HeLa) were seeded in each well of a 96-well plate (Corning Inc., Corning, NY) 12 h in 500 μl of growth medium such that the well became 30–50% confluent at the time of transfection. Liposomes of lipids Lip1818-Lip1810 were complexed with pCMV-α5GFP (0.3 μg per well) at 4
000 cells (HeLa) were seeded in each well of a 96-well plate (Corning Inc., Corning, NY) 12 h in 500 μl of growth medium such that the well became 30–50% confluent at the time of transfection. Liposomes of lipids Lip1818-Lip1810 were complexed with pCMV-α5GFP (0.3 μg per well) at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 lipid
1 lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA charge ratio in a total volume of 100 μl DMEM for 15–20 min. The complexes were then added to the cells. After 4 h incubation, media was replaced with complete media and incubated for 36–48 h. After incubation, cells were washed with PBS (2 × 100 μl) and fixed with 3.8% paraformaldehyde in PBS at room temperature for 10 min. The green fluorescent cells were detected under an inverted fluorescence microscope (Nikon, Japan).
DNA charge ratio in a total volume of 100 μl DMEM for 15–20 min. The complexes were then added to the cells. After 4 h incubation, media was replaced with complete media and incubated for 36–48 h. After incubation, cells were washed with PBS (2 × 100 μl) and fixed with 3.8% paraformaldehyde in PBS at room temperature for 10 min. The green fluorescent cells were detected under an inverted fluorescence microscope (Nikon, Japan).
Conflict of interest
Authors declare no conflict of interest.Abbreviations
| DOPC | 1,2-Dioleoyl-sn-glycero-3-phosphocholine |
| DOPE | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine |
| DOPS | 1,2-Dioleoyl-sn-glycero-3-phospho-L-serine |
Acknowledgements
PKV thanks Department of Biotechnology, India for Ramalingaswami Re-Entry fellowship, and DST for funding support (INT/SWISS/SNSFP-51/2015). AD thanks the Indian University Grant Commission for Junior Research Fellowship. MS thanks SERB-Fast track grant for the fellowship (SB/FT/CS-198/2013), Department of Science and Technology (DST). We thank Cellular Imaging Facility at inStem/NCBS, Bangalore and CSCR/CMC, Vellore. We thank Dr Sankaranarayanan, Ms Rukmini Raju & Mr Mallesh K, CCMB, Hyderabad, India for SAXS studies.Notes and references
- B. Draghici and M. A. Ilies, J. Med. Chem., 2015, 58, 4091–4130 CrossRef CAS PubMed.
- M. Jafari, M. Soltani, S. Naahidi, D. N. Karunaratne and P. Chen, Curr. Med. Chem., 2012, 19, 197–208 CrossRef CAS PubMed.
- M. Elsabahy, A. Nazarali and M. Foldvari, Curr. Drug Delivery, 2011, 8, 235–244 CrossRef CAS.
- K. K. Ewert, A. Zidovska, A. Ahmad, N. F. Bouxsein, H. M. Evans, C. S. McAllister, C. E. Samuel and C. R. Safinya, Top. Curr. Chem., 2010, 296, 191–226 CrossRef CAS PubMed.
- C. R. Safinya, K. K. Ewert, R. N. Majzoub and C. Leal, New J. Chem., 2014, 38, 5164–5172 RSC.
- P. P. Karmali and A. Chaudhuri, Med. Res. Rev., 2007, 27, 696–722 CrossRef CAS PubMed.
- W. Li and F. C. Szoka Jr, Pharm. Res., 2007, 24, 438–449 CrossRef CAS PubMed.
- S. Bhattacharya and A. Bajaj, Chemical Commun., 2009, 4632–4656, 10.1039/b900666b.
- V. V. Kumar, R. S. Singh and A. Chaudhuri, Curr. Med. Chem., 2003, 10, 1297–1306 CrossRef CAS PubMed.
- N. Monteiro, A. Martins, R. L. Reis and N. M. Neves, J. R. Soc., Interface, 2014, 11, 20140459 CrossRef.
- S. Duangjit, B. Pamornpathomkul, P. Opanasopit, T. Rojanarata, Y. Obata, K. Takayama and T. Ngawhirunpat, Int. J. Nanomed., 2014, 9, 2005–2017 CrossRef PubMed.
- Y. Zhao and L. Huang, Adv. Genet., 2014, 88, 13–36 Search PubMed.
- R. Mukthavaram, S. Marepally, M. Y. Venkata, G. N. Vegi, R. Sistla and A. Chaudhuri, Biomaterials, 2009, 30, 2369–2384 CrossRef CAS PubMed.
- R. P. Balasubramaniam, M. J. Bennett, A. M. Aberle, J. G. Malone, M. H. Nantz and R. W. Malone, Gene Ther., 1996, 3, 163–172 CAS.
- B. G. Tenchov, L. Wang, R. Koynova and R. C. MacDonald, Biochim. Biophys. Acta, 2008, 1778, 2405–2412 CrossRef CAS PubMed.
- L. Wang and R. C. MacDonald, Gene Ther., 2004, 11, 1358–1362 CrossRef CAS PubMed.
- D. Zhi, S. Zhang, B. Wang, Y. Zhao, B. Yang and S. Yu, Bioconjugate Chem., 2010, 21, 563–577 CrossRef CAS PubMed.
- H. S. Kim, J. Moon, K. S. Kim, M. M. Choi, J. E. Lee, Y. Heo, D. H. Cho, D. O. Jang and Y. S. Park, Bioconjugate Chem., 2004, 15, 1095–1101 CrossRef CAS PubMed.
- M. H. Nantz, C. W. Dicus, B. Hilliard, S. Yellayi, S. Zou and J. G. Hecker, Mol. Pharmaceutics, 2010, 7, 786–794 CrossRef CAS PubMed.
- R. Koynova, B. Tenchov, L. Wang and R. C. Macdonald, Mol. Pharm., 2009, 6, 951–958 CrossRef CAS PubMed.
- R. Koynova, L. Wang and R. C. MacDonald, J. Phys. Chem. B, 2007, 111, 7786–7795 CrossRef CAS PubMed.
- R. Srinivas, S. Samanta and A. Chaudhuri, Chem. Soc. Rev., 2009, 38, 3326–3338 RSC.
- R. R. Meka, S. Godeshala, S. Marepally, K. Thorat, H. K. R. Rachamalla, A. Dhayani, A. Hiwale, R. Banerjee, A. Chaudhuri and P. K. Vemula, RSC Adv., 2016, 6, 77841–77848 RSC.
- V. Chandrashekhar, M. Srujan, R. Prabhakar, R. C. Reddy, B. Sreedhar, K. K. Rentam, S. Kanjilal and A. Chaudhuri, Bioconjugate Chem., 2011, 22, 497–509 CrossRef CAS PubMed.
- R. S. Singh, K. Mukherjee, R. Banerjee, A. Chaudhuri, S. K. Hait, S. P. Moulik, Y. Ramadas, A. Vijayalakshmi and N. M. Rao, Chemistry, 2002, 8, 900–909 CrossRef CAS PubMed.
- J. Zimmerberg and M. M. Kozlov, Nat. Rev. Mol. Cell Biol., 2006, 7, 9–19 CrossRef CAS PubMed.
- K. Stiasny and F. X. Heinz, J. Virol., 2004, 78, 8536–8542 CrossRef CAS PubMed.
- Y. Onuki, Y. Obata, K. Kawano, H. Sano, R. Matsumoto, Y. Hayashi and K. Takayama, Mol. Pharmaceutics, 2016, 13, 369–378 CrossRef CAS PubMed.
- M. Srujan, V. Chandrashekhar, R. C. Reddy, R. Prabhakar, B. Sreedhar and A. Chaudhuri, Biomaterials, 2011, 32, 5231–5240 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: 1H & 13C NMR and ESI-MS spectral characterizations for all the cationic amphiphiles, RP-HPLC chromatograms for target lipids, size and surface potential of liposome & lipoplexes, cytotoxicity & transfection data. See DOI: 10.1039/c7ra02271a |
| ‡ Equally contributing authors. |
| § Statistical analysis: Data in individual graphs is represented as the mean ± SD of obtained values from each experiment. Data from each group were compared with other groups within the graph using Student t test. p < 0.05 was considered as significant. |
| This journal is © The Royal Society of Chemistry 2017 |