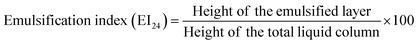Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Monitoring surfactant mediated defence of gastrointestinal Proteus mirabilis DMTMMK1 against pathogenic consortia of Vibrio cholerae†
Suganya Kannana,
Krithika Ashokkumara,
Govindan Krishnamoorthya,
Asha Dhasayanb and
Murugan Marudhamuthu *a
*a
aDepartment of Microbial Technology, School of Biological Sciences, Madurai Kamaraj University, Madurai-625021, Tamil Nadu, India. E-mail: murubio2001@yahoo.com
bAquatic Animal Health and Environment Division, Central Institute of Brackish Water Aquaculture (ICAR-CIBA), Rajaannamalai Puram, Chennai 600 028, Tamil Nadu, India
First published on 12th April 2017
Abstract
In recent times gastrointestinal infections caused by bacterial pathogens have become potentially life-threatening. Particularly, biofilm assembly participates in stimulating the ailments. Bacterial biofilms hold a pivotal role in disease dispersion, pathogenesis and consortia formation. This study reports the antimicrobial and anti-biofilm efficacy of the native gut bacterium Proteus mirabilis DMTMMK1 effected by the production of biosurfactant. Biosurfactant-producing isolates were screened by preliminary assays, and a surfactant with high emulsification capacity produced by Proteus mirabilis DMTMMK1 was purified and subjected to FTIR and NMR spectral analysis which gave evidence of the active characteristics of the surfactant. In the presence of such bioactive compounds an exponential reduction in secreted protease, haemolysis, quorum sensing, and phenotypic inhibition of swarming motility in pathogenic Vibrio cholerae was observed. Further, SEM and HCS (high-content screening) analysis revealed the hampered and disrupted nature of biofilm and cytometry results emphasized a sizeable dead cell population in Vibrio cholerae that had been treated with biosurfactant. Cytotoxicity assay on Vero cell lines evaluated the LD50 value (600 μg mL−1), which showed significant reduction in cell viability and proliferation upon exposure up to 48 h. Furthermore, challenge survival studies provided evidence of biosurfactant effectively inhibiting intestinal colonization of Vibrio cholerae in the nematode Caenorhabditis elegans.
Introduction
Human intestine is an extremely nutritive environment which harbours a tremendous number of microbes, the presence of which has significant impact on the health, nourishment and well-being of the host.1 These bacteria greatly expand the host genetic repertoire, performing many functions that are not encoded by host genome.2 Thus, the gut microbiota, often considered as an additional organ, thereby controls both metabolic and physiological activity of the host. The human gut is predominantly governed by bacterial phyla such as Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria, which survive throughout the host's lifetime. Generally, the presence of these microbes in the gut promotes vascularization, villus thickening, mucosal surface widening, mucus production, cellular proliferation, and maintenance of epithelial junctions. More than 99% of the gut microbial population are beneficial, involved in digestion, supplement special nutrients like vitamins and enzymes and stabilize the intestinal environment, and their inheritance triggers the immune system, facilitating a strong defence mechanism against pathogens.3–7The production of metabolites, especially biosurfactants, by normal microbiota is often recognized as a defence weapon against pathogens colonizing in the gastrointestinal, oral and urogenital tract. Biosurfactants are an assorted group of amphiphilic molecules produced largely by microorganisms on their cell surface or released extracellularly. In the modern biotechnological market these microbial surfactants are major substitutes for synthetic surfactants.8,9 The most commonly isolated biosurfactants are glycolipids, rhamnolipids, phospholipids, lipopeptides, and polysaccharide–protein complexes.10,11 This compositional diversity has led to various applications in food and agriculture, cosmetics, enhanced oil recovery and bioremediation. Besides their regular roles, their therapeutic properties, such as antibacterial, antifungal and antiviral activities, have also gained importance in recent years.12 The biosurfactants studied for antimicrobial activity include iturin from Bacillus subtilis,13 mannosylerythritol from Candida antarctica,14 rhamnolipids from Pseudomonas aeruginosa15 and surfactant from probiotic Lactobacillus lactis16 and Streptococcus thermophilus.17 These biosurfactants outcompete the pathogen by modifying hydrophobicity and act as anti-adhesives, thereby enhancing the detachment process.18
Till now most of the biosurfactants with antimicrobial activity have been obtained from organisms isolated from terrestrial, marine and petroleum spill environmental sites. Human normal flora has a better impact on human health; however, only very few reports have highlighted its production of biosurfactants. The biosurfactants produced by Lactobacillus acidophilus inhibit the adherence of Listeria monocytogenes to stainless steel surfaces, and biosurfactant from Lactobacillus fermentum inhibits the adherence of Enterococcus faecalis.19,20 Their role as antimicrobial and anti-biofilm agents is still an untapped area. Considering their therapeutic value, the present study focused on the antimicrobial and anti-adhesive properties of bacterial surfactants against gut pathogens. Thus, the study comprised isolation and purification of glycolipid biosurfactant produced by intestinal origin Proteus mirabilis strain DMTMMK1 and evaluated its antimicrobial and anti-biofilm potential against Vibrio cholerae along with a few other potential pathogens (see Fig. 1 for schematic representation).
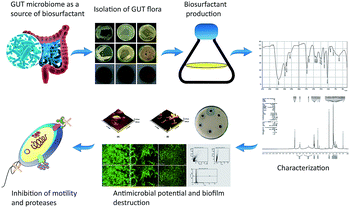 | ||
| Fig. 1 Schematic representation of the biosurfactant production and evaluation of anti-pathogenic potential. GIT, gastrointestinal tract. | ||
Methods
Isolation of gastrointestinal tract microorganisms
Rectal samples were collected from 25 healthy individuals who had not taken any antibiotics during one month before sample collection. Each individual was provided with a self-collectable sterile rectal swab and written instructions for sample collection. Swabs were inserted 3 to 5 cm into the rectum and rotated for 5–10 s and then introduced into the transport medium.21 Samples were refrigerated at 4 °C and used within 1 to 12 h of procurement. One mL of sample was serially diluted in 0.85% saline till a 10−5 dilution factor was reached, plated on de Man Rogosa and Sharpe (MRS) medium and incubated at 37 °C for 24 to 72 h. The tolerance of the bacterial strains to acid (pH 3.0) and bile salt (0.3% w/v) was investigated using MRS broth.22Screening assays for biosurfactant production
Qualitative drop collapse test was initially used to screen the production of biosurfactant.23 Extracellular anionic biosurfactant production was screened by blue agar plates containing cetyltrimethyl ammonium bromide (CTAB) (0.2 mg mL−1) and methylene blue (5 μg mL−1).24 Lipase activity was assessed using tributyrin agar plates. Emulsification activities of the isolates were performed as described by Abouseoud et al.25 Coconut oil, corn oil, sesame oil, olive oil, and cooked oil were also evaluated for emulsification. All the assays were done in triplicate with distilled water as the negative control. After 24 h of incubation, the height of the emulsified layer was measured and the emulsification index (EI24), a measure of emulsion stability, was assessed as follows:Characterization of efficient biosurfactant producer
Among the isolates, DMTMMK1 exhibited high emulsification index and was identified by standard biochemical analysis.26 The susceptibility of the isolate DMTMMK1 to various antibiotics such as ampicillin, chloramphenicol, penicillin G, streptomycin, tetracycline, bacitracin, polymyxin, gentamicin, neomycin, amikacin, ceftriaxone, ofloxacin and ticarcillin was determined using Hexa-G discs (Himedia, India) on Mueller–Hinton Agar (MHA). The genomic DNA was extracted by the CTAB-NaCl method.27 For 16S rRNA gene amplification, PCR analysis was performed by using the 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′), 1492R (5′-CGG TTA CCT TGT TAC GAC TT-3′) primer combination. The 16S rRNA gene sequence obtained from the isolate DMTMMK1 was compared using NCBI BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and multiple sequence alignment was carried out by ClustalW 1.83 (https://www.ebi.ac.uk/cgi-bin/clustalw/). Phylogenetic trees were constructed in MEGA 6.0 (https://www.megasoftware.net) using neighbour joining (NJ), and minimum evolution (ME).Tolerance of the strain DMTMMK1 to simulated human gastrointestinal (GI) tract
Tolerance of the isolate DMTMMK1 to bile salts at low pH was evaluated as described previously. Briefly, overnight culture (18 h) was centrifuged at 4 °C for 15 min (6000 rpm) and washed twice with phosphate-buffered saline (PBS). Simulated gastric juice was prepared by dissolving bile salts (0.3% w/v) in PBS (pH 2.0). Washed DMTMMK1 (0.2 mL) cell suspension was inoculated into 1 mL of simulated gastric juice with 0.3 mL of 0.5% NaCl, and the suspension was incubated at 37 °C. Total viable cell counts were calculated at 0 to 3 h for testing the transit tolerance of the isolate in gastric juice.Production and isolation of biosurfactant
The inoculum was prepared using MRS medium and incubated at 37 °C for 12 h. Three per cent inoculum was inoculated into 100 mL glucose mineral salt production medium in a 500 mL erlenmeyer flask and incubated at 150 rpm at 37 °C for 48 h.28 After incubation, culture was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 15 min to get the cell-free supernatant. The supernatant was then acidified to pH 2.0 with 6 N HCl, and incubated overnight at 4 °C. The precipitate was collected by centrifugation at 15
000 rpm at 4 °C for 15 min to get the cell-free supernatant. The supernatant was then acidified to pH 2.0 with 6 N HCl, and incubated overnight at 4 °C. The precipitate was collected by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min and then redissolved in Milli-Q water. Ethyl acetate and methanol at 4
000 rpm for 20 min and then redissolved in Milli-Q water. Ethyl acetate and methanol at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio was added to the dissolved precipitate, shaken vigorously for several minutes and allowed to settle until phase separation occurred. The organic phase was collected. The above procedure was repeated 2 or 3 times. The excess solvent was removed and concentrated using a rotary evaporator. The resulting product was lyophilized and stored at −20 °C for further studies.
1 ratio was added to the dissolved precipitate, shaken vigorously for several minutes and allowed to settle until phase separation occurred. The organic phase was collected. The above procedure was repeated 2 or 3 times. The excess solvent was removed and concentrated using a rotary evaporator. The resulting product was lyophilized and stored at −20 °C for further studies.
Characterization of biosurfactant
The extracted biosurfactant was analysed by using analytical thin layer chromatography (TLC) on pre-coated silica gel plates (Merck, India). Biosurfactant (0.1 g) was solubilized in 1 mL of methanol and 5 μL was separated on a silica plate with a solvent system consisting of chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water in the ratio of 65
water in the ratio of 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (V/V). Finally the plates were visualized under a shortwave ultraviolet-emitting mercury lamp and the spots were marked for the calculation of Rf value. The biosurfactant extract was characterized by Fourier transform infrared spectroscopy (FTIR) (Shimadzu, Columbia, USA). The IR spectra were recorded with 4 cm−1 resolution, yielding IR traces over the range of 250–4500 cm−1. Further, the biosurfactant was dissolved in deuterated chloroform and its 1H nuclear magnetic resonance (NMR) spectrum was recorded using a Bruker DRX400 spectrophotometer at 27 °C with 5.9 μs pulse duration and 2.6 s acquisition time.
4 (V/V). Finally the plates were visualized under a shortwave ultraviolet-emitting mercury lamp and the spots were marked for the calculation of Rf value. The biosurfactant extract was characterized by Fourier transform infrared spectroscopy (FTIR) (Shimadzu, Columbia, USA). The IR spectra were recorded with 4 cm−1 resolution, yielding IR traces over the range of 250–4500 cm−1. Further, the biosurfactant was dissolved in deuterated chloroform and its 1H nuclear magnetic resonance (NMR) spectrum was recorded using a Bruker DRX400 spectrophotometer at 27 °C with 5.9 μs pulse duration and 2.6 s acquisition time.
Surface tension determination
Surface tension of the biosurfactant was measured using a stalagmometer. The values represent the average of three independent measurements done at room temperature (25 °C). The calculations were made as per the following formula| γ2 = γ1n1ρ1/n2ρ2 |
Antimicrobial assay with biosurfactant
The antimicrobial activity of biosurfactant was determined by agar well diffusion assay. Biosurfactant solutions (1 mg mL−1) were prepared in PBS and filtered through 0.22 μm membrane filters (Whatman, Maidstone, Kent, UK) and stored in glass vials. The susceptibility test was carried out using strains Bacillus cereus MTCC 1272, Escherichia coli MTCC 1698, Pseudomonas aeruginosa MTCC 6458, Salmonella enterica MTCC 1164, Shigella flexneri MTCC 1457, Vibrio cholerae MTCC 3904, Haemophilus influenzae MTCC 3826 and Klebsiella pneumoniae NCCS 2748 on Mueller–Hinton agar (MHA) plates. Bacterial strains were cultured overnight in nutrient broth at 37 °C and the optical density (OD) was adjusted to 0.1. Initial inoculum was inoculated onto MHA plates by evenly flooding the surface of the plates. The wells were made aseptically by sterile well borer; then 20 μL of different biosurfactant stock solutions (concentrations from 5 μg mL−1 to 200 μg mL−1) was added to each well and incubated at 37 °C for 24 h. Zone of inhibition was measured using an antibiotic zone scale (HiMedia). Antibiotics such as streptomycin (Ambistryn S), penicillin (amoxicillin – Amoxil), doxycycline and azithromycin were used as positive control and PBS served as negative control. All tests were done in triplicate and the inhibition zone diameter values were represented as mean value ± SD. Student's t-test was performed to analyse the statistical differences in the zone of inhibition produced by the biosurfactant.Inhibition of Vibrio cholerae biofilm
Biofilm disruption potential of biosurfactant against Vibrio cholerae was quantified in 96-well microtitre plates by measuring the absorbance at 600 nm.29,30 The biofilm inhibition percentages at different biosurfactant concentrations were calculated by the following formula| % biofilm inhibition (c) = [1 − (AC/A0)] × 100 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 15 min followed by washing twice with sterile Milli-Q water. Bacterial cells at logarithmic growth phase, grown in BHI broth, were centrifuged at 5000 rpm for 10 min at 4 °C and 5 μL of 10−3 diluted bacterial cells was spread on the cleaned glass cover slips and incubated at 37 °C. After incubation the cover slips were carefully removed using fine-tip tweezers, washed with Milli-Q water to detach free unbound cells and dried at room temperature for 3 h before imaging. AFM imaging was carried out for both untreated samples and bacterial biofilm treated with biosurfactant. Cells were imaged in air with a tapping mode atomic force microscope (A-100-AFM, APE Research, Italy). A triangular Si cantilever tip with a spring constant of 0.35 N m−1 and a resonance frequency of 18 kHz was used with a scan speed of 0.7–1.5.
1) for 15 min followed by washing twice with sterile Milli-Q water. Bacterial cells at logarithmic growth phase, grown in BHI broth, were centrifuged at 5000 rpm for 10 min at 4 °C and 5 μL of 10−3 diluted bacterial cells was spread on the cleaned glass cover slips and incubated at 37 °C. After incubation the cover slips were carefully removed using fine-tip tweezers, washed with Milli-Q water to detach free unbound cells and dried at room temperature for 3 h before imaging. AFM imaging was carried out for both untreated samples and bacterial biofilm treated with biosurfactant. Cells were imaged in air with a tapping mode atomic force microscope (A-100-AFM, APE Research, Italy). A triangular Si cantilever tip with a spring constant of 0.35 N m−1 and a resonance frequency of 18 kHz was used with a scan speed of 0.7–1.5.
Inhibition of Vibrio cholerae protease secretion and motility by biosurfactant
The secretion of proteases by Vibrio cholerae in the presence of biosurfactant was evaluated using azocasein assay.31 To quantify the proteolytic activity of biofilm formed by Vibrio cholerae in the presence of biosurfactant (10 μL of 5 μg mL−1), an equal quantity of pathogenic biofilm was scraped and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C. The supernatant was taken as crude protease and assayed for protease activity. One millilitre of supernatant was mixed with 480 μL of 1% (w/v) azocasein and 120 μL of 50 mM Tris buffer (pH 7.4). Pathogenic biofilm without biosurfactant treatment served as a control. The reaction mixture was incubated at 37 °C for 30 min. After incubation, 600 μL of 10% trichloroacetic acid (TCA) was added to stop the reaction and incubated at room temperature for 15 min. The mixture was centrifuged at 12
000g for 30 min at 4 °C. The supernatant was taken as crude protease and assayed for protease activity. One millilitre of supernatant was mixed with 480 μL of 1% (w/v) azocasein and 120 μL of 50 mM Tris buffer (pH 7.4). Pathogenic biofilm without biosurfactant treatment served as a control. The reaction mixture was incubated at 37 °C for 30 min. After incubation, 600 μL of 10% trichloroacetic acid (TCA) was added to stop the reaction and incubated at room temperature for 15 min. The mixture was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. Six hundred microlitres of supernatant was dispensed into a fresh microfuge tube, to which 1.8 mL NaOH was added. The absorbance of the clear supernatant was determined at 440 nm against a reaction blank. Inhibition of secreted proteases upon treatment with biosurfactant was calculated by the following formula:
000 rpm for 10 min. Six hundred microlitres of supernatant was dispensed into a fresh microfuge tube, to which 1.8 mL NaOH was added. The absorbance of the clear supernatant was determined at 440 nm against a reaction blank. Inhibition of secreted proteases upon treatment with biosurfactant was calculated by the following formula:| Inhibition (%) = [(AC − AT)/AC] × 100 |
The swarming motility of control and biosurfactant-treated Vibrio cholerae was assessed using soft agar tubes and plates containing 0.5% peptone, 1% glycerol and 0.75% agar. Briefly, culture was inoculated by means of a straight-line stab with a needle into the centre of soft agar, 3 μL of biosurfactant (5 μg mL−1) was added and the tubes were incubated at 28 °C for 24 h. After incubation, turbidity and swarming pattern due to bacterial growth could be observed away from the line of the stab.
Inhibition of haemolysin production
A haemolytic assay was performed using cell-free culture filtrate (CFCF) of Vibrio cholerae grown for 18 h in the presence and absence of biosurfactant (5 μg mL−1). For haemolysis, sheep blood was collected according to the guidelines of Research Animal Resources and the blood collection was approved by the institutional ethical committee of Madurai Kamaraj University [Internal Research and Review Board (IRB), Ethical Clearance (EC), Biosafety and Animal Welfare Committee]. Two per cent sheep blood was suspended in PBS (pH 7.4). Fifty microlitres of cell-free supernatant was added with 450 μL of blood suspension and incubated for 1 h at 37 °C. After incubation the entire suspension was centrifuged at 2000 rpm for 10 min. The supernatant was removed and OD was measured at 530 nm. Percentage inhibition was calculated as described above.Inhibition of quorum sensing (QS)
Fifty microlitres of cell-free supernatant of Vibrio cholerae was added into each well of a 96-well plate with and without 5 μL of biosurfactant (5 μg mL−1), and 60 μL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of 2 M hydroxylamine and 3.5 M NaOH was added into the wells. Subsequently, 100 μL of a 1
1 mixture of 2 M hydroxylamine and 3.5 M NaOH was added into the wells. Subsequently, 100 μL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of ferric chloride (10% in 4 M HCl) and 95% ethanol was added and the plate incubated until the development of dark brown colour, when the plate was read at 520 nm.
1 mixture of ferric chloride (10% in 4 M HCl) and 95% ethanol was added and the plate incubated until the development of dark brown colour, when the plate was read at 520 nm.
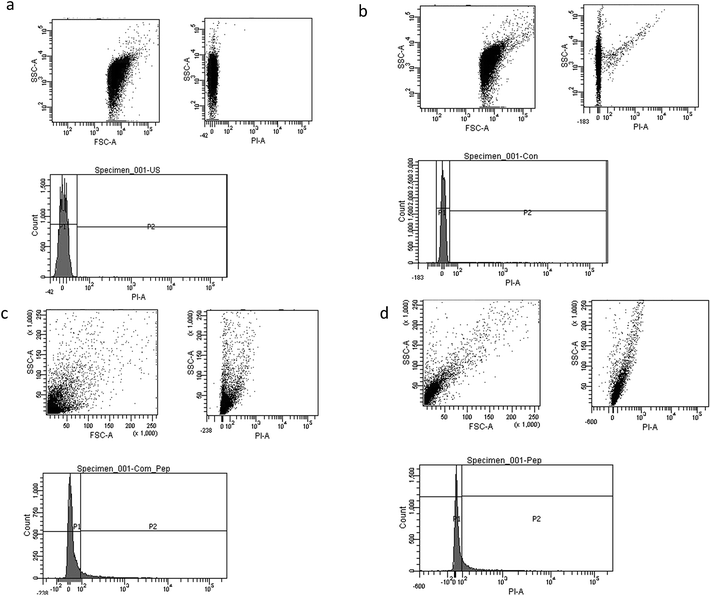
Investigation of Vibrio cholerae viability by flow cytometry
Vibrio cholerae cells were grown in LB broth for 20–24 h and harvested by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min. The cells were washed twice and resuspended in filtered (0.22 μm membrane filter; Millipore, Bedford, USA) phosphate-buffered saline (pH 7.4). One millilitre of the aliquots of bacterial suspension was incubated with biosurfactant (15 μg mL−1) for various exposure times from 0 min to 60 min in order to permeabilize the cell membrane to cause cell death. Staining with propidium iodide (PI; final concentration of 5 μg mL−1) was performed by incubating the aliquots with 10 μL of PI for 10 min at room temperature in the dark. Unstained cells were used as a background control and viability was determined using a fluorescence-activated cell sorter (FACSAria; BD Biosciences, NJ, USA). Cells were initially gated using forward scatter. The flow rate was set at 400–600 events per second up to a total of 10
000g for 15 min. The cells were washed twice and resuspended in filtered (0.22 μm membrane filter; Millipore, Bedford, USA) phosphate-buffered saline (pH 7.4). One millilitre of the aliquots of bacterial suspension was incubated with biosurfactant (15 μg mL−1) for various exposure times from 0 min to 60 min in order to permeabilize the cell membrane to cause cell death. Staining with propidium iodide (PI; final concentration of 5 μg mL−1) was performed by incubating the aliquots with 10 μL of PI for 10 min at room temperature in the dark. Unstained cells were used as a background control and viability was determined using a fluorescence-activated cell sorter (FACSAria; BD Biosciences, NJ, USA). Cells were initially gated using forward scatter. The flow rate was set at 400–600 events per second up to a total of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per sample and analysed for red (610 ± 20 nm) fluorescence using a 488 nm argon laser. All histograms were obtained using logarithmic amplification.
000 events per sample and analysed for red (610 ± 20 nm) fluorescence using a 488 nm argon laser. All histograms were obtained using logarithmic amplification.
Cytotoxicity assays for biosurfactant in eukaryotic cells
The cytotoxicity of the biosurfactant was evaluated by 3-(4,5-dimethlthiozol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, Vero kidney epithelial cell (National Centre for Cell Science, Pune, India) monolayers were maintained in Dulbecco's modified Eagle's medium with phenol red supplemented with 10% foetal bovine serum. One hundred microlitres of the biosurfactant with stock concentration from 5 μg to 700 μg was added to each well in triplicate, and the cells were maintained at 37 °C in a humidified 5% CO2 incubator for 48 h. After incubation, MTT at a final concentration of 5 mg mL−1 was added into the wells and incubated for 4 h, and formazan crystals were observed. The medium was pipetted out without disturbing the formazan crystals. DMSO (100 μL) was added to all the wells and the absorbance was measured in a microplate reader at 620 nm and 50% inhibition of cell viability (IC50) was determined with a standard graph.Challenge survival assays with Vibrio cholerae-infected Caenorhabditis elegans (C. elegans)
Wild-type C. elegans (N2, Bristol) were obtained from C. elegans Genetics Center (CGC, University of Minnesota, USA) and grown on nematode growth medium (NGM) with bacterial culture E. coli OP50 as a feed, at 20 °C for 2 days to obtain gravid adults. For challenge assay an overnight broth culture (10 μL) of Vibrio cholerae was spread onto NGM agar plates and then incubated for 24 h at 37 °C. After 1–2 h at room temperature (23–25 °C), each of the plates was seeded with 25 adult worms. For the biosurfactant treatment the plates were spread with 100 μL of biosurfactant (10 μg mL−1) and air dried. Then the culture (10 μL) of Vibrio cholerae was spread above the surfactant. The plates were then incubated at 25 °C and mortality rate was observed every 24 h up to 240 h. Five replicates were performed on every trial. E. coli OP50 was employed as a negative control. A worm was considered dead when it no longer responded to touch. Any worms that died as the result of being stuck to the walls of the plate were excluded from this analysis.Statistical analysis
All experiments were replicated three times independently. Results were expressed as mean ± standard deviation. Differences in activity were analysed by ANOVA (p < 0.05) using GraphPad prism 5. To evaluate differences between C. elegans growth challenge conditions, a log rank test was carried out and Kaplan–Meier curves were generated using survival duration in days for each worm.Results and discussion
Isolation and identification of dominant biosurfactant producers
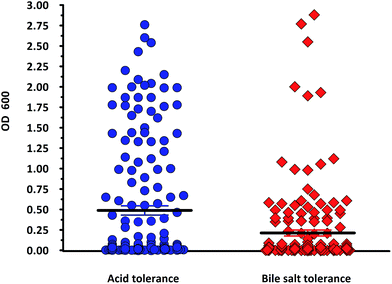
In the present study, a total of 170 morphologically distinct bacterial isolates were obtained from the rectal swabs of 25 healthy individuals. In general, tolerance to acid and bile salts has been considered a favourable condition for colonization and metabolic activity of bacteria in the host's intestine. Therefore, being tolerant to bile salt and acidic environment is an important criterion for the selection of intestinal isolates to assure their viability and functionality. Survival at pH 3.0 and at a bile concentration of 0.3% are optimal factors for intestinal bacterial strains.32 Based on the rapid growth in the media containing acid and bile salts, only 54 isolates were selected and used for the screening of biosurfactant production (Fig. 2). Among the 54 stable isolates the strains (n = 12) DMTMMK1, DMTMMK4, DMTMMK10, DMTMMK12, DMTMMK18, DMTMMK19, DMTMMK20, DMTMMK24, DMTMMK27, DMTMMK31, DMTMMK34 and DMTMMK47 were considered as potential biosurfactant producers. A clear halo zone was observed around the colonies on a light blue mineral salts agar plate containing the cationic surfactant CTAB and the basic dye methylene blue. On the agar plates containing tributyrin, the positive bacterial isolates produced a clear zone around the colonies, demonstrating lipase production by the strains. It was noted that some of the isolates that showed surfactant activity with the above-mentioned screening techniques failed to show haemolysis. Of 12 isolates, 7 showed β-haemolysis with complete zone of clearance and the remaining 5 isolates did not show any haemolysis. The ability of the biosurfactant to emulsify liquid hydrocarbons, such as coconut oil, corn oil, sesame oil, olive oil and cooked oil, was determined. Coconut oil proved better to work with than any other oil tested. Emulsification index of the leading isolates is illustrated in Fig. 3. The oil collapse pattern was recorded (Fig. 4). All the 12 isolates tested were able to form emulsions, with various emulsification index values. Three isolates, DMTMMK18, DMTMMK34, and DMTMMK47, showed very low emulsification index with slight emulsion stability, with the emulsions formed breaking up after only a few minutes. The strain DMTMMK1 was designated the potential biosurfactant producer based on the results of screening tests including oil displacement (8 mm), lipase activity (14 mm), positive drop collapsing test and emulsification activity (78.6%). The isolate DMTMMK1 was characterized as motile, Gram-negative small rods; and hydrolysed cellulose and tributyrin but not casein and starch. The strain utilized glucose and did not utilize lactose. This preliminary identification is supported by sequence analysis of the 16S rDNA gene of the selected strain. Taxonomic affiliation of the isolate was retrieved and representatives of maximally homologous (96–99%) sequences of the isolate were obtained from BLAST program of NCBI (https://blast.ncbi.nlm.nih.gov/) and were used for the construction of phylogenetic affiliation by MEGA 6.0. The phylogenetic analysis and BLAST search showed that the isolate belongs to the genus Proteus.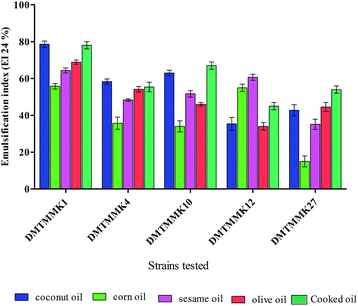 | ||
| Fig. 3 % Effect of various oils on the emulsification index of the biosurfactant. Mean values of triplicate independent experiments and SD are shown. | ||
Finally, based on morphology, biochemical characteristics (ESI Table S1†), and phylogenetic analysis (ESI Fig. S1†), the isolate DMTMMK1 was identified as Proteus mirabilis DMTMMK1. The 16S rRNA sequence of DMTMMK1 was deposited in NCBI with accession number KU311145.1.
Tolerance of the strain DMTMMK1 to simulated human GI (gastrointestinal) tract
The linear regression of the loss of colony forming units (CFU) showed that P. mirabilis DMTMMK1 had acceptably high resistance to the simulated human GI tract. The strain survived well in 0.3% w/v bile salts, with more than 3.5 CFU per mL present after 3 h. This potential to survive high bile concentrations indicates the existence of tolerance mechanisms and confirms that strains possessing strong antagonistic effect against enteric pathogens should be able to compete effectively with the pathogens in the GI tract.Characterization of biosurfactant produced from Proteus mirabilis DMTMMK1
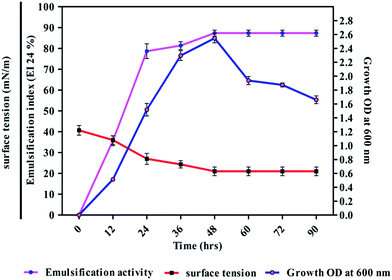
The strain DMTMMK1 was grown in MRS medium for 96 h and cell biomass (growth, OD at 600 nm), surface tension and emulsification activity were evaluated (Fig. 5). The maximum emulsification index was observed at a growth of 1.98 × 109 cells per mL with reduction of surface tension 24 mN m−1. An emulsion is formed when one liquid phase is distributed as microscopic droplets with continuous liquid phase. Stability of such emulsions is therefore an important criterion to study when considering the potential application of such biosurfactants. The stability of the biosurfactant from DMTMMK1 was verified over a wide variety of oils up to 64 h. An emulsion formed from olive oil was found to be stable over a period of up to 18 h although a considerable loss of emulsion stability was observed beyond 18 h. The emulsion formed from coconut oil maintains stability up to 77% even after 60 h incubation. A very important characteristic of surfactant is the critical micelle concentration (CMC), which is defined as the surfactant concentration requisite to form micelles. When concentration of the surfactant attains CMC, the surface tension remains relatively constant due to the interface saturation with the surfactant. Hence, it is important to obtain a surfactant that has not only low CMC but also high surface activity. As shown in Fig. 5 the CMC of the surfactant was found to be 0.75 mg mL−1 and the surface tension was reduced from 45 mN m−1 to 19 mN m−1. The biosurfactant extracted from the acid precipitant was initially characterized by TLC, which revealed a single spot with an Rf value of 0.79 while visualized under UV light, confirming the presence of glycolipid (ESI Fig. S2†).27 The FTIR spectrum revealed the presence of polysaccharide and lipid component in the biosurfactant (Fig. 6a). The presence of hydroxyl group was confirmed by infrared spectrum of the purified biosurfactant, which showed broad stretching at 3390 cm−1. Absorption around 2947 cm−1 and 2835 cm−1 was assigned to the symmetric stretch –CH of CH2 and CH3 groups of aliphatic chains.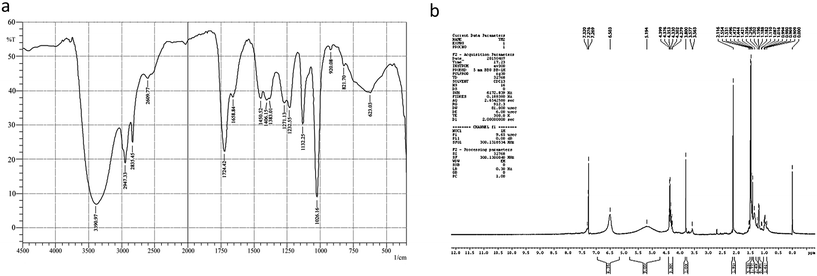 | ||
| Fig. 6 (a) FTIR spectrum of purified biosurfactant from Proteus mirabilis DMTMMK1. (b) 1H NMR spectrum for biosurfactant from Proteus mirabilis DMTMMK1. | ||
Also, an intense absorption band at 1724 cm−1 and a weak symmetric stretching peak around 1658 cm−1 indicated the occurrence of ester carbonyl groups (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in COOH) in the biosurfactant. Peaks at 1271 cm−1 and 1232 cm−1 were found to be ether and C–O stretching vibration in sugars. Peaks at 1026 cm−1 and 623 cm−1 were due to stretching vibrations of polysaccharides and CH2 group, respectively, confirming the presence of glycolipid moieties.
O in COOH) in the biosurfactant. Peaks at 1271 cm−1 and 1232 cm−1 were found to be ether and C–O stretching vibration in sugars. Peaks at 1026 cm−1 and 623 cm−1 were due to stretching vibrations of polysaccharides and CH2 group, respectively, confirming the presence of glycolipid moieties.
The FTIR spectrum of the biosurfactant showed that the purified biosurfactant was glycolipid in nature. The 1H NMR spectrum of the purified biosurfactant in CDCl3 further confirmed a typical glycolipid type structure and characteristic proton chemical shift peaks were observed. Multiple signals of protons of (–CH3) below 1.0 ppm and (–CH2) between 1.0 and 1.4 ppm showed the presence of a linear alkane (Fig. 6b). Proton NMR confirmed the presence of α-carbon protons with the corresponding signals between 3.564 to 5.194 and the side chain protons were confirmed with the peaks at 0.25–3.0 ppm. The presence of an intense singlet at 3.57 ppm observed in the biosurfactant suggested the presence of one methyl ester group in the biosurfactant. The presence of the methyl ester group in the structure of a biosurfactant has been related with an increase of its hydrophobicity and, consequently an increment of surfactant power, and antimicrobial activities. From TLC, FTIR and NMR analysis, the biosurfactant produced by Proteus mirabilis strain DMTMMK1 was characterized as a glycolipid.
Antimicrobial activity of biosurfactant
Biosurfactants with antimicrobial properties found to date are produced by bacteria of terrestrial origin. The number of reports on antimicrobial biosurfactant molecules from human intestinal microflora is limited. The antimicrobial activity of the crude biosurfactant from the intestinal isolate Proteus mirabilis DMTMMK1 was determined by the zone of growth inhibition in well diffusion assay. The solvent-extracted as crude biosurfactant showed antimicrobial activity against the majority of the tested strains. Minimum inhibitory concentration (MIC) values were determined and are summarized in Fig. 7. The largest inhibition zones due to the biosurfactant were observed against Vibrio cholerae, Shigella flexneri and Bacillus cereus. The biosurfactant was also found to have significant inhibitory effect against E. coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae. Vibrio cholerae and Shigella flexneri were seen to be inhibited at relatively lower biosurfactant concentrations of 15 μg mL−1 and 25 μg mL−1 respectively. However the MIC values for other tested bacteria such as Bacillus cereus, Klebsiella pneumoniae, and Pseudomonas aeruginosa ranged from 40 to 95 μg mL−1. The MIC value of biosurfactant was higher for Salmonella enterica at 175 μg mL−1. Experiments were performed with penicillin, streptomycin, doxycycline and azithromycin to study the relative efficacy of the Proteus mirabilis DMTMMK1 biosurfactant as an antibacterial agent. As a result the MIC values for the crude biosurfactant against the tested organisms were found to be less than those of penicillin and slightly higher than those of streptomycin, azithromycin and doxycycline. Resistance to penicillin was observed in the case of Klebsiella pneumonia, which was inhibited by biosurfactant at 65 μg mL−1 concentration. The isolated biosurfactant non-selectively showed activity against both Gram-positive and Gram-negative bacterial strains. The antimicrobial results obtained from our studies were similar to the results of biosurfactant studies using Bacillus circulans and rhamnolipids produced by Pseudomonas aeruginosa which have been reported to be active against Gram-negative bacteria.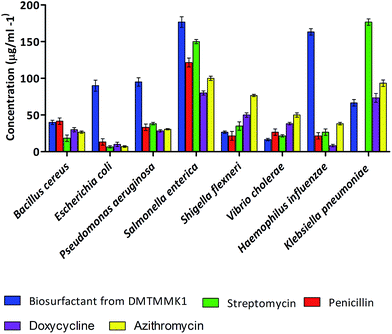 | ||
| Fig. 7 Distribution of MIC values for the biosurfactants tested against eight different bacterial pathogens. | ||
Vibrio cholerae is a non-spore-forming pathogen causing a severe diarrhoeal disease. In its life-threatening manifestation, cholera is one of the most rapidly fatal illnesses known. Most antibiotics and chemotherapeutic agents have no value in cholera therapy, but the biosurfactant from P. mirabilis DMTMMK1 was found to be effective against Vibrio cholerae at a dosage as low as 15 μg mL−1, showing its effectiveness and potential use in treating infections.
Evaluation of PI uptake with biosurfactant treatment using cell sorter
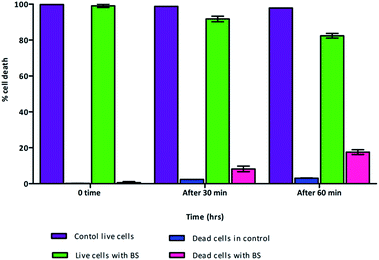
Mostly the viable cell fraction is determined by the addition of dyes, such as resazurin, XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide), and FDA (fluorescein diacetate), which are metabolically transformed to a quantifiable end product. We adapted flow cytometric enumeration of Vibrio cholera, which showed significant inhibition of bacterial growth following incubation with biosurfactant. Flow cytometry results supported that biosurfactant had a bactericidal effect against the Vibrio cholerae strains by damaging the cytoplasmic membrane, resulting in cellular leakage. The cell membrane plays a chief self-protective role in defending the pathogen against a number of drug targets and thereby plays a substantial role in drug resistance mechanisms. Nucleic acid staining with propidium iodide (PI), which is an impermeant dye, stains only membrane-damaged cells, helping to distinguish intact bacteria from membrane-compromised ones. Fig. 8 shows the distribution of fluorescence intensities for cells of Vibrio cholerae after exposure to biosurfactant from 0 min and 60 min compared with a control without biosurfactant treatment for which the bacteria were 99.9% viable. Around 9.8% of the Vibrio cholerae cells incubated with biosurfactant were membrane-compromised and 90.2% of cells were viable after 1 h of treatment (Fig. 9).However, after 2 h of treatment, more cells were permeabilized, indicating a substantial increase in the ratio of dead cells with a distribution of 18.9% dead and 81.1% live. Clear discrimination between viable intact and permeabilized cells was noted by monitoring the extents to which cells accumulated PI during the biosurfactant treatment. This is the first study using flow cytometry to verify the cell permeability and membrane damage in Vibrio cholerae biofilm cells treated with biosurfactant.
Inhibition and disruption of Vibrio cholerae biofilm
Biofilm formation by Vibrio cholerae is a key factor that greatly contributes to virulence and disease dissemination, which makes it an ideal target for disease control. Much growing evidence has suggesting that Vibrio cholerae can form biofilm-like aggregates during infection, which could play a critical role in pathogenesis and disease transmission.31The effect of biosurfactants on microbial adhesion and biofilm disruption has been widely described, and has become an effective strategy to diminish biofilm formation and combat colonization by pathogenic communities. The biosurfactant from Proteus mirabilis DMTMMK1 was applied as initial attachment inhibitor and post-treatment agent for biofilm reduction.
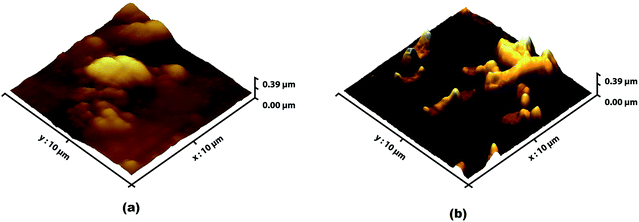
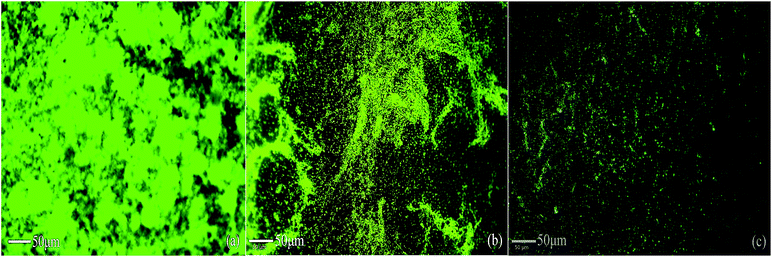
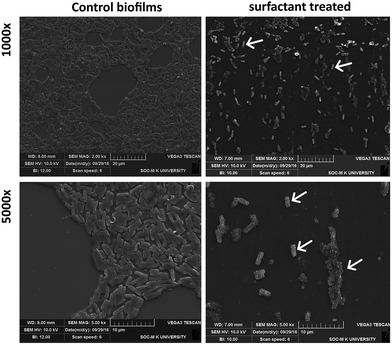
Apart from inhibiting biofilm formation, it disrupted pre-formed mature biofilms by 68.62 ± 5.17% even at the shortest incubation time of 30 min. The minimum concentration of biosurfactant that showed an anti-biofilm effect against Vibrio cholerae was 5 μg mL−1. Compared with the control sample, biofilm formation was reduced by 89.5 ± 1.9% (p < 0.05) and 77.76 ± 2.1% when 15 μg mL−1 and 10 μg mL−1 of biosurfactant were added, respectively. When 15 μg mL−1 of biosurfactant was applied for 2 h, 78.2 ± 3.1% (p < 0.05) of the Vibrio cholerae cells became detached from the surface. The observations on disruption of pre-formed biofilm were supported by AFM images, which showed unstructured biofilm formation with the incubation of biosurfactant. The AFM images clearly differentiate the control biofilm with strong extracellular polymeric substances from that with distributed microcolonies after biosurfactant treatment (Fig. 10). HCS analysis revealed a significant reduction in surface area occupied by biosurfactant-treated Vibrio cholerae (Fig. 11 and 12). Dispersion of mature biofilms was observed in the disruption assay and uneven scattered microcolonies were observed in the inhibition assay, as compared with respective untreated controls. SEM imaging of Vibrio cholerae grown in the presence of biosurfactant from DMTMMK1 showed noticeable damage in the cell membrane accompanied by alteration in morphology (Fig. 13).
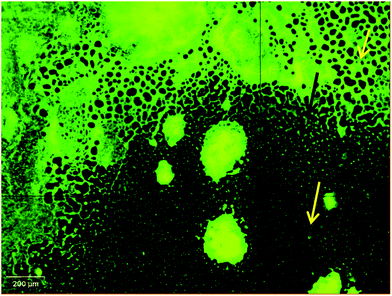 | ||
| Fig. 12 Disruption pattern of mature biofilm formed after 48 h incubation. The damaged part of the biofilm is indicated with arrows. | ||
Surface roughness was clearly observed for cells with biosurfactant treatment. The morphological alteration plays a significant role in virulence, host cell adhesion and evasion of the immune system. From SEM observations, control cells showed an increased number of attachments in the biofilm state. Further, microscopic analysis clearly revealed alteration in the shape of the Vibrio cells from normal elongated rods to shorter ones in the treated groups compared with the control.
Inhibition of Vibrio cholerae proteases and motility by biosurfactant
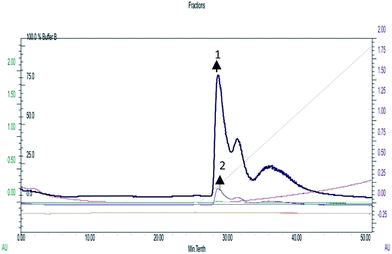
A recent study conducted by Smith et al. has provided a new insight into the role of secreted proteases in the dynamics and assembly of biofilm formation by Vibrio cholerae.33 They hypothesized that proteases from Vibrio cholerae could have an active role in biofilm recruitment/reinforcement. Considering the key roles of Vibrio cholerae proteases for bacterial colonization and growth of biofilms, this factor could be a potential target to tackle bacterial colonization and biofilm formation. In secreted protease assay, biosurfactant exhibited potential inhibition of proteases which are the vital virulence factors of Vibrio cholerae. After incubation with biosurfactant the percentage of secreted proteases decreased by up to 58 ± 3.6% (p < 0.005) (Fig. 14). FPLC results clearly support that only control biofilm without any surfactant treatment exhibited a prominent absorbance peak but in the presence of biosurfactant the peak was reduced. The individual peak exhibited strong proteolytic activity on azocasein in the case of control biofilm (Fig. 15).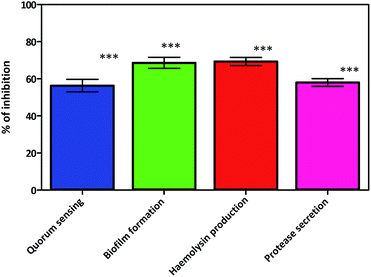 | ||
| Fig. 14 Virulence factor inhibition of biosurfactant against Vibrio cholerae. Mean values of triplicate independent experiments and SD are shown. ***Indicates statistical significance (p > 0.005). | ||
Motility is thought to contribute to virulence by increasing the chance for association of the Vibrio cholerae with the intestinal mucosa. Biosurfactant produced by Proteus mirabilis DMTMMK1 triggered a significant antimotility effect against Vibrio cholerae and effectively slowed the growth at the centre of the soft agar stabs (ESI Fig. S3†). The antimotility effect limits the size of the bacterial movement in stabs with a limited number of cells and consequently reduces the biofilm formation. The bacterial motility serves as a potential resistance mechanism towards antibiotics and leads to virulence which is arrested by the biosurfactant.
The haemolysin activity was reduced in tubes to which CFS (cell-free supernatant) collected from 24 h cultures of Vibrio cholerae treated with biosurfactant was added. Inhibition of haemolytic activity, provides direct evidence that biosurfactant controls the phenotypic expression of this virulence factor in the strain.
A significant reduction (69.33 ± 3.76%) (Fig. 14) of haemolysin was observed. Inhibition or reduction in biofilm formation and motility would be predicted to be an outcome of biosurfactant inhibiting the QS pathways of Vibrio cholerae. This was proved in the preliminary study in which lactone compounds were quantified, where biosurfactant inhibited the production of acyl homoserine lactones (AHLs), whose production is controlled by a QS mechanism.34,35 During lactone quantification 56.3 ± 5.85% lactone secretion inhibition was observed in Vibrio cholerae (Fig. 14). The loss of lactone absorbance in treated cultures is indicative of AHL inhibition by biosurfactant which accompanied the loss in QS signalling which is responsible for biofilm formation and virulence.
Cytotoxicity assays for biosurfactant in eukaryotic cells
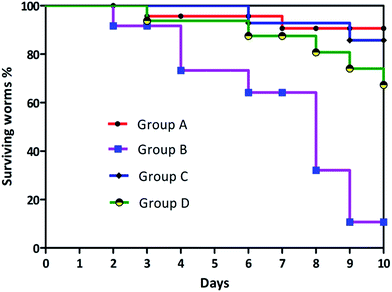
Cytotoxicity of biosurfactant was evaluated using the Vero cell line. Vero cells were treated with biosurfactant from Proteus mirabilis at various concentrations (5 μg to 700 μg) for 48 h. The results obtained clearly show a decrease in the percentage of viable cells with increasing biosurfactant concentrations and exposure times, thus suggesting a cytostatic/cytotoxic effect of the biosurfactant against the studied cell line. Based on these findings, 600 μg mL−1 biosurfactant was found to be effective in decreasing Vero cell viability to values between 45 and 50%. The differences obtained for the range of biosurfactant concentrations evaluated as compared with the control were found to be statistically significant (p-value < 0.005). As a result the biosurfactant seems to have a safer profile when exposed to normal cells at appropriate concentrations. Survival assay was conducted to determine the effect of Vibrio cholerae with biosurfactant treatment on the lifespan of C. elegans. A log rank test was adapted to search for significant differences in survival between the two different conditions. The Kaplan–Meier plots for challenge experiments using C. elegans feeding are shown in Fig. 16. Vibrio cholerae displays a great negative impact on the survival of C. elegans, and when the bacteria were grown in NGM medium amended with biosurfactant, a great increase in the lifespan and survival rate of C. elegans was observed. Vibrio cholerae killed all of the exposed worms within 6 days, with a toxic dose (TD50) at 3 days. Vibrio cholerae appears to belong to the “slow killer” group in which slow killing has been observed to occur within a 2.5–3 day period.A log rank (Mantel–Cox) test was performed to examine the significant differences in survival between the four different conditions. The Kaplan–Meier plots for C. elegans fed with E. coli OP50 and Vibrio cholerae with and without surfactant treatment are shown in Fig. 16. χ2-Statistics and p-value were found to be 25.70 and <0.0001 respectively. The survival proportion was only 10.69444 in the case of Vibrio cholerae-infected worms, which was increased to 67.308 when the plates were overlayered with surfactant. As mentioned before, biosurfactant significantly inhibits the virulence of Vibrio cholerae which is required for the colonization and disease progression in C. elegans.
Conclusion
Diversity of the gut microbiota was evident from the isolation process from a small population of individuals, which yielded a total number of 170 isolates. A further 54 strains showed both acid and bile salt tolerance, indicating the prime characteristics of intestinal inhabitants. The biosurfactant-producing nature of the strains was confirmed through various screening assays. The intestinal bacterial strain Proteus mirabilis DMTMMK1 was successfully found to be a potent producer of biosurfactant with the ability to emulsify a wide range of hydrocarbons in an effective way. In this work we have demonstrated the antimicrobial and anti-biofilm properties of the biosurfactant isolated from Proteus mirabilis DMTMMK1 against several pathogenic organisms. Besides this, distortion of biofilm and enhanced dead cell population in the presence of biosurfactants was seen with the AFM, HCS and FACS analysis. The ability of the biosurfactant from Proteus mirabilis DMTMMK1 to disrupt mature biofilms, inhibit biofilm formation and diminish biofilm viability makes it a possible innovative agent to treat biofilm-associated infections of Vibrio cholerae. The results obtained indicate the potential application of this biosurfactant as an alternative antimicrobial and anti-adhesive agent in the field of medicine.Acknowledgements
The authors acknowledge the Department of Biotechnology – Interdisciplinary Programme for Life Sciences (DBT-IPLS, India) and the Department of Microbial Technology for the facilities provided. We are very thankful to Dr B. Ashock Kumar for helping in C. elegans cytotoxicity analysis. We thank Miss M. Niraimathi and Mr Balaji for their help in HCS and SEM imaging.References
- C. A. Lozupone, J. I. Stombaugh, J. I. Gordon, J. K. Jansson and R. Knight, Nature, 2012, 489, 220–230 CrossRef CAS PubMed.
- J. Qin, R. Li, J. Raes, M. Arumugam, S. Burgdorf, C. Manichanh, T. Nielsen, N. Pons, T. Yamada, D. R. Mende, J. Li, J. Xu, S. Li, D. Li, J. Cao, B. Wang, H. Liang, H. Zheng, Y. Xie, J. Tap, P. Lepage, M. Bertalan, J. Batto, T. Hansen, D. Le Paslier, A. Linneberg, H. B. Nielsen, E. Pelletier, P. Renault, Y. Zhou, Y. Li, X. Zhang, S. Li, N. Qin and H. Yang, Nature, 2010, 464, 59–65 CrossRef CAS PubMed.
- C. G. Buffie and E. G. Pamer, Nat. Rev. Immunol., 2013, 13, 790–801 CrossRef CAS PubMed.
- D. Kelly, S. Conway and R. Aminov, Trends Immunol., 2005, 26, 326–333 CrossRef CAS PubMed.
- F. Sommer and F. Bäckhed, Nat. Rev. Microbiol., 2013, 11, 227–238 CrossRef CAS PubMed.
- S. Caballero and E. G. Pamer, Annu. Rev. Immunol., 2015, 33, 227–256 CrossRef CAS PubMed.
- J. Bérdy, J. Antibiot., 2005, 58, 1–26 CrossRef PubMed.
- S. Mukherjee, P. Das and R. Sen, Trends Biotechnol., 2006, 24, 509–515 CrossRef CAS PubMed.
- J. D. Van Hamme, A. Singh and O. P. Ward, Biotechnol. Adv., 2006, 24, 604–620 CrossRef CAS PubMed.
- M. G. Healy, C. M. Devine and R. Murphy, Stud. Environ. Sci., 1997, 66, 179–195 CAS.
- E. Z. Ron and E. Rosenberg, Environ. Microbiol., 2001, 3, 229–236 CrossRef CAS PubMed.
- P. Singh and S. S. Cameotra, Trends Biotechnol., 2004, 22, 142–146 CrossRef CAS PubMed.
- F. Ahimou, P. Jacques and M. Deleu, in Enzyme and Microbial Technology, 2000, vol. 27, pp. 749–754 Search PubMed.
- A. Daverey, K. Pakshirajan and S. Sumalatha, Clean Technol. Environ. Policy, 2011, 13, 481–488 CrossRef CAS.
- M. Nitschke, S. G. V. A. O. Costa, R. Haddad, L. A. G. Gonçalves, M. N. Eberlin and J. Contiero, Biotechnol. Prog., 2005, 21, 1562–1566 CrossRef CAS PubMed.
- L. Rodrigues, H. Van Der Mei, I. M. Banat, J. Teixeira and R. Oliveira, FEMS Immunol. Med. Microbiol., 2006, 46, 107–112 CrossRef CAS PubMed.
- L. Rodrigues, I. M. Banat, J. Teixeira and R. Oliveira, J. Antimicrob. Chemother., 2006, 57, 609–618 CrossRef CAS PubMed.
- I. M. Banat, R. S. Makkar and S. S. Cameotra, Appl. Microbiol. Biotechnol., 2000, 53, 495–508 CrossRef CAS PubMed.
- M. M. C. Velraeds, H. C. Van Der Mei, G. Reid and H. J. Busscher, Appl. Environ. Microbiol., 1996, 62, 1958–1963 CAS.
- T. Meylheuc, C. J. Van Oss and M. N. Bellon-Fontaine, J. Appl. Microbio., 2001, 91, 822–832 CrossRef CAS.
- B. Dodge, B. Van Der Pol, M. Reece, D. Malebranche, O. Martinez, G. Goncalves, P. Schnarrs, R. Nix and J. D. Fortenberry, Sex. Health, 2012, 9, 190–191 Search PubMed.
- M. T. Liong and N. P. Shah, J. Dairy Sci., 2005, 88, 55–66 CrossRef CAS PubMed.
- P. G. Carrillo, C. Mardaraz, S. I. Pitta-Alvarez and A. M. Giulietti, World J. Microbiol. Biotechnol., 1996, 12, 82–84 CrossRef CAS PubMed.
- N. M. Pinzon and L. K. Ju, Biotechnol. Lett., 2009, 31, 1583–1588 CrossRef CAS PubMed.
- M. Abouseoud, R. Maachi, A. Amrane, S. Boudergua and A. Nabi, Desalination, 2008, 223, 143–151 CrossRef CAS.
- J. G. Holt, N. Krieg, P. Sneath, J. Staley and S. Williams, Williams Wilkins, 1994 Search PubMed.
- L. V. Andreou, Methods Enzymol., 2013, 529, 143–151 CAS.
- P. Vandana and J. K. Peter, Int. J. Adv. Technol. Eng. Sci., 2014, 2348–7550 Search PubMed , https://www.ijates.com.
- G. S. Kiran, B. Sabarathnam and J. Selvin, FEMS Immunol. Med. Microbiol., 2010, 59, 432–438 CrossRef CAS PubMed.
- R. D. Rufino, J. M. de Luna, G. M. de Campos Takaki and L. A. Sarubbo, Electron. J. Biotechnol., 2014, 17, 34–38 CrossRef.
- S. L. Iversen and M. H. Jörgensen, Biotechnol. Tech., 1995, 9, 573–576 CrossRef CAS.
- A. Hosono, J. Dairy Sci., 1999, 82, 243–248 CrossRef.
- D. R. Smith, M. Maestre-Reyna, G. Lee, H. Gerard, A. H.-J. Wang and P. I. Watnick, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 10491–10496 CrossRef CAS PubMed.
- M. R. Parsek and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8789–8793 CrossRef CAS.
- M. R. Parsek, D. L. Val, B. L. Hanzelka, J. E. Cronan and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 4360–4365 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra01934c |
| This journal is © The Royal Society of Chemistry 2017 |