DOI:
10.1039/C7RA01163F
(Paper)
RSC Adv., 2017,
7, 18793-18802
Bioremoval of Cu2+ from CMP wastewater by a novel copper-resistant bacterium Cupriavidus gilardii CR3: characteristics and mechanisms†
Received
26th January 2017
, Accepted 10th March 2017
First published on 28th March 2017
Abstract
Bacteria of the genus Cupriavidus are known for the ability of resistance to various heavy metals and metal-binding capability. Herein, we investigated the bioremoval of Cu2+ from synthesized chemical–mechanical polishing (CMP) wastewater by living cells of Cupriavidus gilardii CR3, a novel copper-resistant bacterium isolated in our previous study. The surface topography changes of strain CR3 were observed by SEM-EDX, where images showed that binding took place on the bacterial cell surface. FTIR spectra provided evidence that carboxyl, hydroxyl, amino, and phosphate groups on the surface of strain CR3 could be available for characteristic coordination bonding with Cu2+. Zeta potential confirmed that electrostatic interaction was involved in Cu2+ binding. The biosorption and bioaccumulation of Cu2+ by strain CR3 was highly pH-dependent, and the optimum pH value was 5.0. The maximum binding capacity for Cu2+ was 18.33 mg g−1 and the bioremoval efficiency was 27% under optimal conditions. The Cu2+ binding process obeyed the Langmuir isotherm (R2 = 0.99). Kinetic data were properly fitted with both pseudo-second order kinetic model (R2 = 0.99) and an intraparticle diffusion model (R2 = 0.98). It can be concluded that living cells of C. gilardii CR3 have the potential to be utilized for the removal of Cu2+ from CMP wastewater.
1. Introduction
Chemical–mechanical polishing/planarization (CMP) is a critical step in semiconductor circuits, which is employed after the metal deposition step to eliminate any topography on a wafer's surface with the combination of chemical reactions and mechanical forces.1 Copper metallization is rapidly replacing the aluminum-based processes in the CMP process. Copper is high resistance to electromigration effects and low electrical compared to early used aluminum. This transition, however, poses new challenges to wastewater treatment.2 Copper CMP effluents can account for 30–40% of the water discharge in the semiconductor manufacturing and contain high concentrations of soluble copper and a complex mixture of organic constituents.3 Copper is toxic to aquatic organisms, which has been reported to cause toxic effects on algae and fish or amphibians at as low as 5–10 ppb.4 The dissolved copper in CMP wastewater is in the range of 5–100 mg L−1.5 The 100 mg L−1 concentration of copper in CMP wastewater has also been reported.6 It is thus evident that the copper concentration range of CMP wastewater is by far higher than copper effective concentration towards aquatic faunas and its concentration is strictly regulated in many countries both for discharges to municipal wastewater treatment facilities and for direct discharge to receiving waters. In China, the integrated circuit has been designated as a pillar industry in the 10th Five Year Economic plan since 2000. This has certainly driven large scale domestic and foreign semiconductor manufacturing plants in the past decade. High production volumes of semiconductor circuits bring forth a large amount of water consumption and increase the amount of wastewater discharged into the local environment. Therefore, an appropriate treatment of copper CMP wastewater is indispensable to meet stringent discharge regulations.
Various technologies have been employed for the removal of copper form copper CMP wastewater.7 Flocculation/clarification is a conventional treatment for the removal of copper ions. However, this process requires large tanks and generates an undesirable copper-rich sludge and secondary environmental pollution. Other methods such as those employing membrane,8 ion exchange,9 and electrochemical technologies10 have also been considered. Drawbacks associated with the abovementioned methods including the addition of chemicals or high-cost limit their application. Accordingly, the need for the development of alternatives to copper CMP wastewater treatment has led to an increasing interest in sorption processes.5,11 Among various sorbents, bacterial biomass might be an efficient class of biosorbents for the removal of metal ions because of their high surface to volume ratios and presence of potentially active chemisorption sites.12,13 Both the living and non-living cells of bacteria can be used as biosorbents and have their own advantages and disadvantages.14 However, it is a controversial issue to choose living and non-living bacterial cells to remove heavy metals. Living cells might be preferential in the continuous treatment of heavy metal-bearing effluent15 and have been successful in the treatment of heavy metals in genuine industrial and municipal effluents.16 Biosorption and bioaccumulation often simultaneously participate in heavy metal removal by living bacterial cells.17 Hence, the use of heavy metal-resistant bacteria has gained extensive attention in the removal of heavy metals from aqueous solution because they can resist the toxicity of heavy metals even at high concentrations due to specific resistance systems, such as efflux and uptake mechanisms, and extracellular precipitation.18 Several studies have explored the copper removal form model copper CMP wastewater using metal tolerant bacteria from the genus Staphylococcus7 and Lactobacillus casei.19 However, bacterial mechanisms for the removal of various heavy metals are often highly specific for certain metals due to the heterogeneity of bacterial cell constituents.13 The high diversity and complex structure of bacteria imply that there are many ways for the metal to be bound by bacterial cells. Hence, there is much work that needs to be carried out in screening promising bacteria for their features and mechanisms for the bioremoval of heavy metal species.
The genus Cupriavidus is a Gram-negative β-proteobacteria that is well known for its resistance to heavy metals. Accordingly, Cupriavidus species is of great interest in the field of environmental biotechnology.20 Some Cupriavidus sp. such as C. metallidurans CH34,21 C. necator N-1,22 C. pinatubonensis JMP134,23 and C. taiwanensis24 have been reported to resist various heavy metals. In particular, C. metallidurans CH34 has been employed in the removal of heavy metals by biosorption and bioprecipitation in a bioreactor.25,26 To the best of our knowledge, the ability of C. gilardii in terms of removal of heavy metals has not been documented to date. In our previous study, we isolated a C. gilardii strain CR3 that was resistant to Cd2+, Zn2+, Co2+, Cu2+, Cu+, Hg2+, Pb2+, Ni2+, CrO42−, and Ag+ at concentrations up to 4 mM.27 Moreover, the whole genome of C. gilardii was first sequenced using strain CR3 as a reprehensive. Sequence analysis showed that the genome of the bacterium consists of two chromosomes and does not possess a plasmid.27 In addition, the genes responsible for heavy metal resistance in most bacteria, such as Pseudomonas syringae28 and Escherichia coli, are located on either plasmid(s) or dispersed between the chromosome and plasmids.29 There are several exceptions in which heavy metal resistant genes are only located in the bacterial chromosome, suggesting their heavy metal resistance ability.30 C. gilardii CR3 is such a distinctive bacterium. It can be inferred that C. gilardii CR3 has the ability to accumulate heavy metals at the outside of the cells and is thus bound to cell surface structures via binding heavy metals.
Herein, we further investigated the bioremoval of copper ions from synthetic Cu-CMP wastewater by living cells of C. gilardii CR3 as a biosorbent. The experiments were designed based on the hypothesis that biosorption and bioaccumulation were assumed to occur in the living biomass. The major objectives of this study are two fold: (1) to study the characteristics of the bioremoval of copper by living cells of C. gilardii CR3 and (2) to understand the mechanisms of biosorption and bioaccumulation of copper in Cupriavidus gilardii.
2. Materials and methods
2.1 Preparation of biosorbent and synthetic Cu-CMP wastewater
The bacterium strain of C. gilardii CR3 used in this study was isolated from a natural asphalt deposit in our previous study.27 The GenBank accession number of C. gilardii CR3 is JX945578. C. gilardii CR3 was grown in a Luria broth (LB) medium that contained 10.0 g tryptone, 5.0 g yeast extract, and 10.0 g NaCl per liter. Cells of C. gilardii CR3 were initially inoculated into sterile LB and placed on a shaker at 28 °C for 24 h. The live cells were harvested at an early stationary phase by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min. The cells were rinsed with phosphate buffer solution (PBS) three times and were subsequently stored in PBS at 4 °C. Cell concentrations in the suspension were determined by drying an aliquot onto a pre-weighed aluminum foil container to a constant weight at 60 °C.
000g for 5 min. The cells were rinsed with phosphate buffer solution (PBS) three times and were subsequently stored in PBS at 4 °C. Cell concentrations in the suspension were determined by drying an aliquot onto a pre-weighed aluminum foil container to a constant weight at 60 °C.
Synthetic Cu-CMP wastewater used in this study was similar to that described by Mosier et al. (2015).19 Briefly, the stock synthetic Cu-CMP wastewater containing 5 mM Cu2+ was prepared with CuCl2·2H2O and deionized water.
All chemicals used in this study were of analytical grade and were purchased from Sigma Aldrich Co., Ltd, USA. Biochemicals were obtained from Dingguo Biotechnology Co., Ltd., China.
2.2 Scanning electron microscopy and energy dispersive X-ray spectroscopy
The concentrations of Cu2+ in the supernatant were determined via atomic absorption spectrophotometer (AA6300, Shimadzu, Japan). Scanning electron microscopy equipped with energy dispersive X-ray spectroscopy (SEM-EDX) was used to study the chemical characteristics of C. gilardii CR3 before and after Cu2+ binding. The biosorbent samples were fixed using 2.5% glutaraldehyde for 12 h and dehydrated in graded concentrations by alcohol. Samples were freeze-dried for 24 h and the dried cells were coated with a thin layer of gold and examined using SEM-EDX.
2.3 Fourier transform infrared spectroscopy
The possible involvement in the functional groups of cells of strain CR3 during the removal of Cu2+ from aqueous solution was elucidated using the Fourier transform infrared spectroscopy (FTIR) analysis. FTIR spectra of the cells loaded with and without Cu2+ were obtained in KBr discs using an FTIR spectrometer (Perkin Elmer 2000). The FTIR samples were analyzed in the range of 4000–400 cm−1 with a resolution of 4 cm−1.
2.4 Zeta potential
The zeta potential for C. gilardii CR3 was measured before and after the Cu2+ binding using the Zeta Meter (Malvern, Nano ZS90). The concentration of Cu2+ was set at 64 mg L−1 in the measurement of zeta potential versus pH of cells (2.0–7.0).
2.5 Batch experiments
All the batch experiments were carried out by mixing 1 g L−1 biosorbent at ambient temperature (28 ± 2 °C) and the mixtures were agitated at the speed of 180 rpm for 24 h. Control experiments were carried out in the free-biosorbent in every test to investigate the removal of Cu2+, which might occur via chemical precipitation and sorption on to vessel walls.
The effects of pH, initial Cu2+ concentration of CMP wastewater, and contact time on biosorption and bioaccumulation capacity were studied. The effect of wastewater pH on the binding capacity of bacteria for Cu2+ was determined in the pH range from 2.0 to 7.0 with the intervals of 1.0 using 50 mL solutions containing 64 mg L−1 of Cu2+. The Cu-CMP wastewater was adjusted to required pH values using 0.1 mol L−1 HCl and 0.1 mol L−1 NaOH. Biosorbent was added to Cu-CMP wastewater. Similarly, the effect of initial Cu2+ concentration on biosorption and bioaccumulation was conducted by changing the initial Cu2+ concentration to 6.4, 19.2, 32, 44.8, 64, 83.2, 96, 108.8, and 128 mg L−1 (at optimum pH 5.0 for 24 h). In addition, the batch studies were carried out at room temperature (28 ± 2 °C) with 64 mg L−1 of Cu2+ using 1 g L−1 of biosorbent dosage at different time intervals between 5 and 400 min at pH 5.0. The Cu2+ binding capacity and removal efficiency of the tested bacterium were calculated according to the following equation:
| |
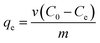 | (1) |
| |
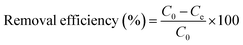 | (2) |
where
qe is the amount of Cu
2+ uptake by unit of sorbents in mg g
−1,
v is the volume of Cu-CMP wastewater in the reaction system in L,
m is the dosage of added biosorbents in g, and
C0 and
Ce are the initial and equilibrium concentration of Cu
2+ in the solution in mg L
−1, respectively.
2.6 Isotherms and kinetics
The experimental data were evaluated using a Langmuir isotherm and Freundlich isotherm equations. The linear form of the Langmuir isotherm and Freundlich isotherm equations can be expressed as follows:| |
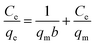 | (3) |
| |
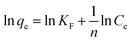 | (4) |
where qe is the amount of Cu2+ uptake by unit of sorbents in mg g−1, Ce is the equilibrium concentration of Cu2+ in the solution in mg L−1, qm is the theoretical monolayer sorption capacity in mg g−1, n and KF are Freundlich equilibrium constants, and b is the Langmuir equilibrium constant.
To analyze the kinetics of Cu2+ using C. gilardii CR3, the linear form of the pseudo-second order kinetics and the intraparticle diffusion model were applied to the experimental data:
| |
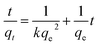 | (5) |
| |
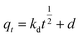 | (6) |
where
k,
kd, and
d are the rate constant of kinetic sorption,
qe is the amount of Cu
2+ uptake by unit of sorbents in mg g
−1, and
qt (mg g
−1) is the amount of Cu
2+ uptake at any given time
t (min).
2.7 Statistical analysis
All the experiments were performed in triplicate. The data is shown as mean values with the error bars representing the 95 percent confidence intervals around the average value.
3. Results and discussion
3.1 SEM-EDX analysis
Cell surface properties govern the metal–microorganism interactions to a large extent.31 The morphological characterization of C. gilardii CR3 was studied before and after Cu2+ was loaded. After Cu2+ was loaded, the morphology of the bacterial cells did not show a greater variation in cell size and surface roughness. Moreover, the structural integrity of the cell was clear, without appearance of cell damage. Interestingly, reticulo-filamentous substances were obviously observed in the outer cells after Cu2+ was loaded (Fig. 1b), which were mostly like those secreted by C. gilardii CR3 in the presence of ions from copper stress. This might reflect a self-protection of the strain CR3. Extracellular polymeric substances released by bacteria have been recommended as surface active agents and have an important role in binding heavy metals.32,33 The present results might be associated with the genetic characteristics of C. gilardii CR3. Strain CR3 was isolated from a natural asphalt seep located at the Rancho La Brea Tar Pits (Los Angeles, USA), in which these natural asphalt seeps have existed for at least 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 year.34 The distinctive echo niche caused bacterium CR3 to develop intrinsic heavy metal resistance mechanisms including self-detoxification through ion efflux, metal-complexation and metal-reduction, and a powerful DNA self-repair.27 Moreover, the EDX analysis confirmed that Cu2+ combined with the biomass of C. gilardii CR3 (Fig. 2). There was no copper peak appearing on the cells before Cu2+ was loaded (Fig. 2a). In contrast, two additional signals of copper existed in the EDX profile after Cu2+ was loaded on the cells (Fig. 2b).
000 year.34 The distinctive echo niche caused bacterium CR3 to develop intrinsic heavy metal resistance mechanisms including self-detoxification through ion efflux, metal-complexation and metal-reduction, and a powerful DNA self-repair.27 Moreover, the EDX analysis confirmed that Cu2+ combined with the biomass of C. gilardii CR3 (Fig. 2). There was no copper peak appearing on the cells before Cu2+ was loaded (Fig. 2a). In contrast, two additional signals of copper existed in the EDX profile after Cu2+ was loaded on the cells (Fig. 2b).
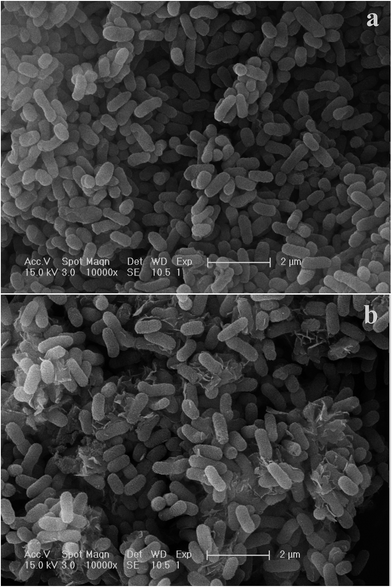 |
| | Fig. 1 Scanning electron microscopy (SEM) images of Cupriavidus gilardii CR3 in the absence (a) and presence (b) for Cu2+ concentration of 64 mg L−1 at pH 5.0 for 24 h. | |
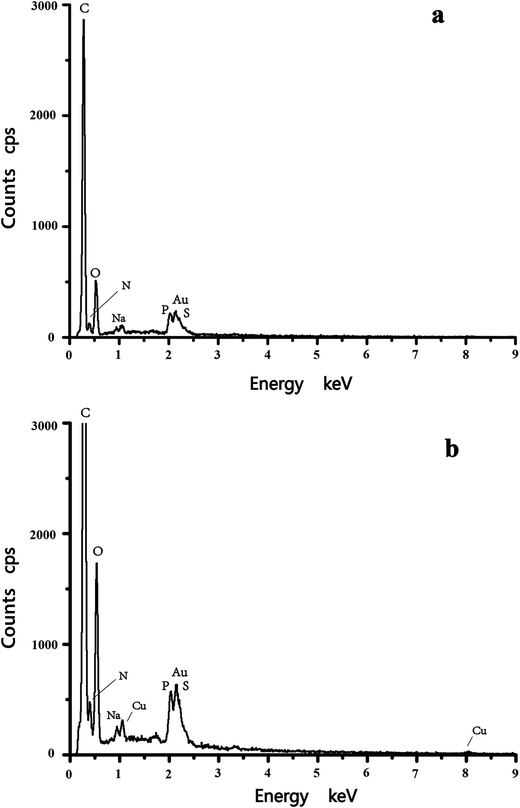 |
| | Fig. 2 Energy dispersive X-ray spectroscopy (EDX) of Cupriavidus gilardii CR3 in the absence (a) and presence (b) for Cu2+ concentration of 64 mg L−1 at pH 5.0 for 24 h. | |
3.2 FTIR analysis
A number of sorption peaks that represented various functional groups of the cell surface were observed in the FTIR spectra (Fig. 3). The major sorption bands of C. gilardii CR3 were as follows: 3307 cm−1, corresponding to –OH and –NH stretching associated with protein; 2927 cm−1, for asymmetric/symmetric stretching vibration of CH2; 1654, 1390, and 1072 cm−1, for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of amide I, the symmetric stretching C
O stretching of amide I, the symmetric stretching C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peptidic bond of carboxyl groups, and the stretching of PO2− in phosphodiesters and phosphates, respectively; and 545 cm−1, for phosphate or sulphate functional groups. In a comparison with the spectra of C. gilardii CR3 prior to being treated with Cu2+, a minute shift in the peak position was observed (<15 cm−1) after Cu2+ binding. The band shifts were from 3307 cm−1 to 3309 cm−1 for –OH and –NH, from 1390 cm−1 to 1391 cm−1 for –COOH, and from 1072 cm−1 to 1069 cm−1 for PO2− in the presence of Cu2+. Functional groups such as carboxyl, hydroxyl, amino, and phosphate groups played a key role during the metal ion binding process.35,36 The present FTIR analysis also provided evidence that these functional groups were involved in the binding of copper for C. gilardii CR3.
O peptidic bond of carboxyl groups, and the stretching of PO2− in phosphodiesters and phosphates, respectively; and 545 cm−1, for phosphate or sulphate functional groups. In a comparison with the spectra of C. gilardii CR3 prior to being treated with Cu2+, a minute shift in the peak position was observed (<15 cm−1) after Cu2+ binding. The band shifts were from 3307 cm−1 to 3309 cm−1 for –OH and –NH, from 1390 cm−1 to 1391 cm−1 for –COOH, and from 1072 cm−1 to 1069 cm−1 for PO2− in the presence of Cu2+. Functional groups such as carboxyl, hydroxyl, amino, and phosphate groups played a key role during the metal ion binding process.35,36 The present FTIR analysis also provided evidence that these functional groups were involved in the binding of copper for C. gilardii CR3.
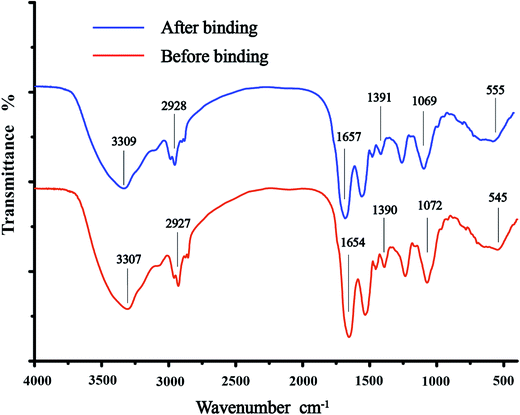 |
| | Fig. 3 Fourier transform infrared spectroscopy (FTIR) of Cupriavidus gilardii CR3 before and after binding the initial Cu2+ concentration of 64 mg L−1 at pH 5.0 for 24 h. | |
3.3 Zeta potential analysis
Bacterial surface charge is usually characterized by the zeta potential.37 To elucidate the mechanism that C. gilardii CR3 sorbs Cu2+, the zeta potentials of C. gilardii CR3 were measured at different pH values with and without Cu2+ binding (Fig. 4). The isoelectric point of C. gilardii CR3 was around 2.2. The zeta potential of cells changed from positive to negative once the pH exceeded 2.2, and the negative values of zeta potential increased with continuously increasing pH. The values of the zeta potential of the cell ranged from about +3 mV to about −15 mV. The variations of zeta potential in strain CR3 with pH revealed the electrostatic interaction between bacteria and Cu2+ in an aqueous system, which is consistent with the previous observations that most bacteria carry a net negative charge.35 Additionally, the obtained results indicated that the zeta potentials of the cell with and without binding Cu2+ exhibited similar trends over the pH range from 2.0 to 7.0. The absolute values of the zeta potential of the cells after binding with Cu2+ were lower than those prior to binding at the pH range from 2.2 to 7.0.
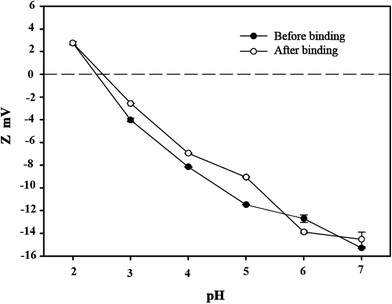 |
| | Fig. 4 Zeta potential of Cupriavidus gilardii CR3 before and after binding initial Cu2+ concentration of 64 mg L−1 at different pH values. | |
3.4 Effect of pH on copper bioremoval
The pH value of the aqueous solution is an important variable in the binding of metals on the biosorbents. The influence of the initial pH value of the solution on the removal of Cu2+ was examined in the pH range from 2 to 7. The effects of pH on the binding of Cu2+ at various pH values (2.0–7.0) are shown in Fig. 5. The results showed that pH values significantly affect bacterial binding capacity to copper in the solution. The capacity of C. gilardii CR3 and removal efficiency of Cu2+ increased with the elevation of the pH value. At the pH range of 2.0–5.0, the binding capacity sharply increased from 2.08 mg g−1 (removal 2% Cu2+) to 18.33 mg g−1 (removal 27% Cu2+). A 30% removal of Cu2+ occurred at pH between 6.0 and 7.0.
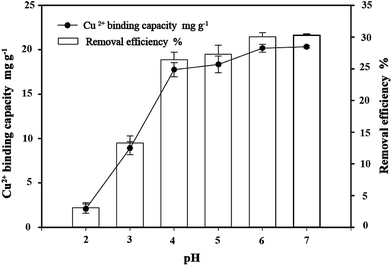 |
| | Fig. 5 Effect of pH on copper bioremoval of Cupriavidus gilardii CR3 with an initial Cu2+ concentration of 64 mg L−1 for 24 h. | |
The pH value affects the solution chemistry of metal ions and the activity of the functional groups of the cell wall.38 Copper can be present in solution as three different species: Cu2+, CuOH+, and Cu(OH)2.39 Under lower pH conditions, Cu2+ and CuOH+ were more favorable copper species. The competition of Cu2+ and protons in solution to the binding sites on the bacterium cell wall can explain the relationship between pH values and the uptake of metal ions.38 In this study, the binding capacities of copper were greatly influenced by the pH value of the solution. At pH 2.0, the lowest binding capacity was obtained. The reason for this is that H3O+ in the solution may more easily occupy the binding sites on the surface of C. gilardii CR3 than copper. With an increase in pH (2.0–5.0), the amount of negative charges on the surface of the strain CR3 increased (Fig. 4), which led to the enhancement in binding capacity. When the pH value was over 5.3, the precipitation of Cu2+ hydroxide occurred. The formation of metal hydroxide and other metal–ligand complexes significantly reduced the amount of metal ions bound at higher pH.40 Therefore, the optimum pH condition was 5.0 in this study, and all the subsequent experiments were conducted at pH 5.0.
3.5 Effect of initial Cu2+ concentration on copper bioremoval
As depicted in Fig. 6, the Cu2+ binding capacity of C. gilardii CR3 increased with the increase of initial Cu2+ concentration from 6.4 mg L−1 to 128 mg L−1, and the removal efficiency inversely changed. The rapid removal of Cu2+ was observed under the lower Cu2+ condition (<64 mg L−1). With an increase in Cu2+ concentration, the removal of Cu2+ became gradually slow. The lowest removal ratio was achieved at the highest initial Cu2+ concentration, where the equilibrium binding capacity for the cells was 22 mg g−1. Therefore, the intersecting point of the two curves was the best ratio of the Cu2+ concentration and bacterial concentration.
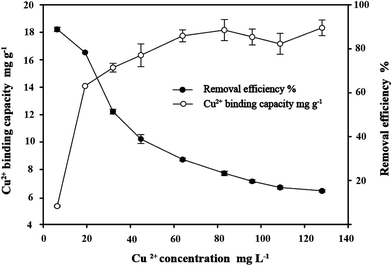 |
| | Fig. 6 Effect of initial Cu2+ concentration on copper bioremoval of Cupriavidus gilardii CR3 at pH 5.0 for 24 h. | |
3.6 Effect of contact time on copper bioremoval
The equilibrium time is of crucial importance because it is one of the parameters for economical wastewater treatment plant application. The effect of contact time on bioremoval of C. gilardii CR3 at the initial concentration of 64 mg L−1 is shown in Fig. 7. The results revealed that the binding process as a function of contact time occurs in two steps. The first step involves a rapid metal uptake within the first 50 min of contact time, suggesting that this step was a rapid process. Within the first 50 minutes of binding, 76% of the total binding capacity was achieved. Rapid binding during the initial stage of the process was attributed to the larger surface area that was available for the binding of metal ions.41 The second step was characterized by the subsequent slower binding capacity of the metal. The exhaustion of binding sites during the process resulted in lower sorption rates during the second stage.42
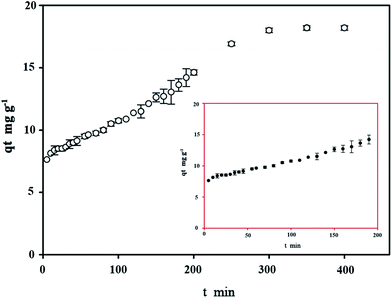 |
| | Fig. 7 Effect of contact time on copper bioremoval of Cupriavidus gilardii CR3 with an initial Cu2+ concentration of 64 mg L−1 at pH 5.0. | |
3.7 Isotherms study
Several sorption isotherms can be used to describe the equilibrium concentration of the sorbed species as a function of the fluid phase concentration. The Langmuir isotherm assumes monolayer sorption and the Freundlich isotherm is an empirical equation based on sorption for a heterogeneous surface.43,44 In the present study, the linearized Langmuir and Freundlich isotherms at various initial concentrations of copper ions were employed to determine the sorption isotherm data (Fig. 8). The correlation coefficient and other parameters obtained from Langmuir and Freundlich sorption isotherms for the sorption of Cu2+ are shown in Table 1. The Langmuir model (R2 = 0.99) exhibited a better fit to the data of copper than the Freundlich model (R2 = 0.49) in the studied initial Cu2+ concentration range. It could be inferred that the binding of the cells of C. gilardii CR3 to Cu2+ is monolayer sorption. Most of bacterial binding for metals follow the Langmuir isotherm.45–47 According to Langmuir isotherm, the predicted maximum biosorption capacity for C. gilardii CR3 was 18.25 mg g−1, which is very close to the experimental binding capacity qe value (18.33 mg g−1). These results suggest that the binding of copper by strain CR3 principally occurred at cell sorption rather than accumulation because the maximum binding capacity calculated by the Langmuir isotherm did not consider the amount of metal uptake by bacteria.48 Although both biosorption and bioaccumulation are involved in living cells for the removal of various heavy metals from aqueous solution, biosorption usually accounts for the main part in the binding capacity of heavy metals.49–51 For instance, the cell surface of a Lactococcus lactis strain accounted for over 95% of the amount of cadmium binding.52
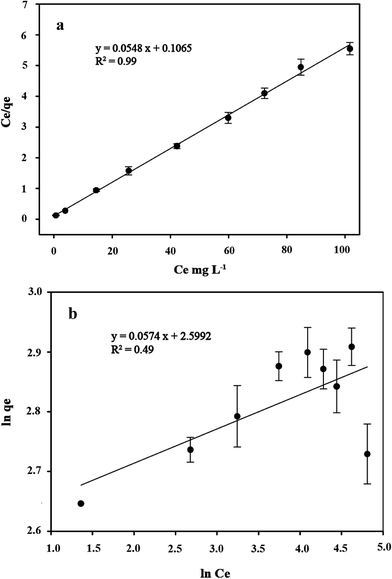 |
| | Fig. 8 The linearized form of a Langmuir isotherm (a) and Freundlich isotherm (b) of Cupriavidus gilardii CR3 for Cu2+ at pH 5.0 for 24 h. | |
Table 1 Isotherms of Cupriavidus gilardii CR3 for Cu2+ at pH 5.0 for 24 h
| Equilibrium model |
Langmuir |
Freundlich |
| Parameter |
qma |
bb |
R2 |
KFc |
1/nc |
R2 |
| The academic maximum amount of Cu2+ uptake per gram sorbents in mg g−1. Langmuir equilibrium constant. Freundlich equilibrium constant. |
| Value |
18.25 |
0.51 |
0.99 |
13.45 |
0.05 |
0.49 |
Additionally, a comparison within Cupriavidus strains showed that the copper biosorption capacity of C. gilardii CR3 was almost the same as that of the C. taiwanensis TJ208 (19.0 mg g−1), whereas lower than that of C. metallidurans CH34 (26.69 mg g−1) under the same experiment conditions.53 Therefore, it is still efficient compared to other bacteria in terms of copper biosorption capacity: the copper biosorption capacity of Bacillus sp. is 16.25 mg g−1 (ref. 54) and Sphaerotilus natans is 5.4 mg g−1.55 Thus, C. gilardii CR3 can be an interesting alternative for the bioremoval of Cu2+ from Cu-CMP wastewater.
3.8 Kinetics study
Kinetic studies are important for the practical application of binding process because they can provide insight into the reaction pathways and the mechanisms of the sorption reaction.56 To investigate the kinetic patterns of copper on C. gilardii CR3, the pseudo-second order kinetic model and intraparticle diffusion model were applied in this study (Fig. 9). The kinetics of copper binding by strain CR3 was well described by the pseudo-second order kinetic model (R2 = 0.99) and intraparticle diffusion model (R2 = 0.98) (Table 2). The experimental data fitting the pseudo-second order model demonstrated that the mechanism of the sorption process was controlled by chemical sorption involved in exchange sorption of metal ions and a surface proton.57,58 Moreover, the fit line with an intraparticle diffusion model did not pass through the origin, suggesting that intraparticle diffusion occurred in the binding process, but it was not the sole rate-controlling step.40 Furthermore, the amount of negative charges on the surface of strain CR3 increased with an increase in pH value based on the measurement of the zeta potential (Fig. 4), which confirmed that the electrostatic interaction contributed to the binding process. Therefore, chemisorption and physisorption might be simultaneously involved in the binding process by means of ion exchange and electrostatic interactions between copper ions and functional groups in the cells of strain CR3.
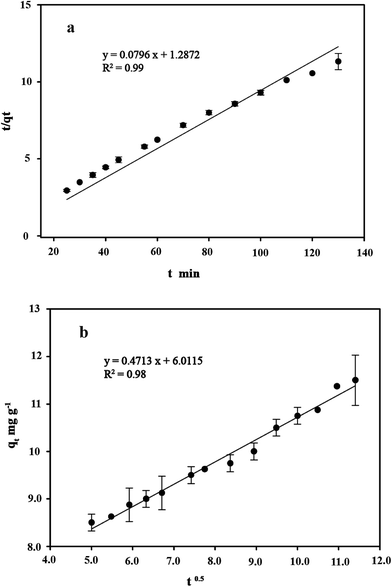 |
| | Fig. 9 The linearized form of the pseudo-second order kinetic model (a) and the intraparticle diffusion model (b) for the Cu2+ concentration of 64 mg L−1 bound by Cupriavidus gilardii CR3 at pH 5.0. | |
Table 2 Kinetics for Cu2+ concentration of 64 mg L−1 by Cupriavidus gilardii CR3 at pH 5.0
| Kinetics model |
The pseudo-second order kinetics |
The intraparticle diffusion |
| Parameter |
qea |
kb |
R2 |
kdb |
db |
R2 |
| The amount of Cu2+ uptake by unit sorbents in mg g−1. The rate constant of kinetic sorption. |
| Value |
12.56 |
0.004 |
0.99 |
0.47 |
6.01 |
0.98 |
4. Conclusions
The present study was carried out to use living cells of C. gilardii CR3 as a biosorbent for the removal Cu2+ from Cu-CMP wastewater. The binding ability was pH-dependent, and the highest binding capacity of copper ions (18.33 mg g−1) was achieved at pH 5.0. The biosorption equilibrium was well described by the Langmuir isotherm. Both the pseudo-second order kinetic model and the intraparticle diffusion model were in good agreement with all the experimental data. The obtained results may provide pre-stage for an initial evaluation of binding mechanisms and operational conditions for the continuous removal of Cu2+ from CMP wastewater utilizing living cells of C. gilardii CR3 in a fixed-bed reactor.
Authors' contributions
XW and FW conceived and designed the experiments. YY and XW performed the experiments. MH and DZ analyzed the data. MH and MH contributed reagents, materials, and analysis tools. XW and YY wrote the paper. XW and FW revised the paper.
Ethics approval and consent to participate
Not applicable.
Conflict of interest
The authors declare that they have no competing interests.
Abbreviations
| CMP | Chemical–mechanical polishing/planarization |
| LB | Luria broth |
| PBS | Phosphate buffer solution |
| SEM-EDX | Scanning electron microscopy equipped with energy dispersive X-ray spectroscopy |
| FTIR | Fourier transform infrared spectroscopy |
Acknowledgements
Analyses of SEM-EDX, FTIR, and zeta potential were carried out by the Analysis and Test Center, Northeast Normal University. The skillful technical assistance of Li Chen is gratefully acknowledged. We thank Prof. Fayek Negm, Dept. of Botany and Plant Sci. University California Riverside, USA, kindly helped us to improve the quality of our manuscript. This study was funded by the National Natural Science Foundation of China (No. 51678122), and Jilin Provincial Science and Technology Department Project, China (No. 20150414046GH).
References
- G. C. C. Yang, Environ. Prog. Sustainable Energy, 2002, 21, 57–62 CAS.
- B. Maag, Global Environ. Polit., 2000, 2, 134–136 Search PubMed.
- J. Hollingsworth, R. Sierra-Alvarez, M. Zhou, K. L. Ogden and J. A. Field, Chemosphere, 2005, 59, 1219–1228 CrossRef CAS PubMed.
- H. Chua, P. H. Yu, S. N. Sin and M. W. Cheung, Chemosphere, 1999, 39, 2681–2692 CrossRef CAS PubMed.
- W. Maketon, PhD thesis, University of Arizona, 2007.
- G. C. C. Yang and C. M. Tsai, J. Membr. Sci., 2006, 286, 36–44 CrossRef CAS.
- L. C. Stanley and K. L. Ogden, J. Environ. Manage., 2003, 69, 289–297 CrossRef PubMed.
- Y. N. Su, W. S. Lin, C. H. Hou and W. Den, J. Water Process Eng., 2014, 4, 149–158 CrossRef.
- W. Maketon and K. L. Ogden, Chemosphere, 2009, 75, 206–211 CrossRef CAS PubMed.
- G. C. C. Yang and C. M. Tsai, J. Membr. Sci., 2008, 321, 232–239 CrossRef CAS.
- W. Maketon, C. Z. Zenner and K. L. Ogden, Environ. Sci. Technol., 2008, 42, 2124–2129 CrossRef CAS PubMed.
- P. Kotrba, T. Macek, M. Mackova, G. Naja, B. Volesky and F. Pagnanelli, Bacterial biosorption and biosorbents, in Microbial Biosorption of Metals, ed. Y. S. Yun, K. Vijayaraghavan and S. W. Won, Springer Science and Business Media, New York, 2011, pp. 121–141 Search PubMed.
- P. S. Li and H. C. Tao, Crit. Rev. Microbiol., 2013, 41, 140–149 CrossRef PubMed.
- A. Malik, Environ. Int., 2004, 30, 261–278 CrossRef CAS PubMed.
- G. M. Gadd, J. Chem. Technol. Biotechnol., 2009, 84, 13–28 CrossRef CAS.
- P. Kotrba, T. Macek, M. Mackova, G. Naja, B. Volesky and F. Pagnanelli, Biosorption and metal removal through living cells, in Microbial Biosorption of Metals, ed. P. Kotrba, M. Mackova, J. Fišer and T. Macek, Springer Science and Business Media, New York, 2011, pp. 197–233 Search PubMed.
- K. Chojnacka, Environ. Int., 2010, 36, 299–307 CrossRef CAS PubMed.
- I. Kamika and M. N. Momba, BMC Microbiol., 2013, 13, 1–14 CrossRef PubMed.
- A. P. Mosier, J. Behnke, E. T. Jin and N. C. Cady, J. Environ. Manage., 2015, 160, 67–72 CrossRef CAS PubMed.
- W. M. Chen, C. H. Wu, E. K. James and J. X. Chang, J. Hazard. Mater., 2008, 151, 364–371 CrossRef CAS PubMed.
- P. J. Janssen, R. Van Houdt, H. Moors, P. Monsieurs, N. Morin, A. Michaux, M. A. Benotmane, N. Leys, T. Vallaeys, A. Lapidus, S. Monchy, C. Médigue, S. Taghavi, S. McCorkle, J. Dunn, D. Van der Lelie and M. Mergeay, PLoS One, 2010, 5, e10433 Search PubMed.
- A. Poehlein, B. Kusian, B. Friedrich, R. Daniel and B. Bowien, J. Bacteriol., 2011, 193, 5017 CrossRef CAS PubMed.
- A. Lykidis, D. Perez-Pantoja, T. Ledger, K. Mavromatis, I. J. Anderson, N. N. Ivanova, S. D. Hooper, A. Lapidus, S. Lucas, B. González and N. C. Kyrpides, PLoS One, 2010, 5, e9729 Search PubMed.
- C. Amadou, G. Pascal, S. Mangenot, M. Glew, B. Contemps, D. Capela, S. Carrère, S. Cruveiller, C. Dossat, A. Lajus, M. Marchetti, V. Poinsot, Z. Rouy, B. Servin, M. Saa, C. Schenowitz, V. Barbe, J. Batut, C. Médigue and C. Masson-Boivin, Genome Res., 2008, 18, 1472–1483 CrossRef CAS PubMed.
- R. Paterson, Immobilization of bacteria in composite membranes and development of tubular membrane reactors for heavy metal recuperation, in Effective membrane processes: new perspectivesed, ed. L. Diels, S. V. Roy, M. Mergeay, W. Doyen, S. Taghavi and R. Leysen, Kluwer Academic Publishers, The Netherlands, 1993, pp. 275–293 Search PubMed.
- A. E. Torma, M. L. Apel and C. L. Brieley, The use of Alcaligenes eutrophus immobilized in a tubular membrane reactor for heavy metal recuperation, in Biohydrometallurgical technologies, ed. L. Diels, S. V. Roy, S. Taghavi, W. Doyen, R. Leysen and M. Mergeay, The Minerals, Metals and Materials Society, Warrendale, 1993, pp. 133–144 Search PubMed.
- X. Y. Wang, M. L. Chen, J. F. Xiao, L. R. Hao, E. David Crowley, Z. W. Zhang, J. Yu, N. Huang, M. X. Huo and J. Y. Wu, PLoS One, 2015, 10, 1–21 Search PubMed.
- S. Puig, E. M. Rees and D. J. Thiele, Structure, 2002, 10, 1292–1295 CrossRef CAS PubMed.
- D. H. Nies, FEMS Microbiol. Rev., 2003, 27, 313–339 CrossRef CAS PubMed.
- J. Valdés, I. Pedroso, R. Quatrini, R. J. Dodson, H. Tettelin, R. Blake II, J. A. Eisen and D. S. Holme, BMC Genomics, 2008, 9, 82–86 CrossRef PubMed.
- E. Kordialik-Bogacka, Cent. Eur. J. Chem., 2011, 9, 348–351 CrossRef CAS.
- M. Watanabe, K. Kawahara, K. Sasaki and N. Noparatnaraporn, J. Biosci. Bioeng., 2003, 95, 374–378 CrossRef CAS PubMed.
- A. Iyer, K. Mody and B. Jha, Mar. Pollut. Bull., 2005, 50, 340–343 CrossRef CAS PubMed.
- J. S. Kim and D. E. Crowley, Appl. Environ. Microbiol., 2007, 73, 4579–4591 CrossRef CAS PubMed.
- F. Huang, Z. Dang, C. L. Guo, G. N. Lu, R. R. Gu, H. J. Liu and H. Zhang, Colloids Surf., B, 2013, 107, 11–18 CrossRef CAS PubMed.
- L. Xia, X. J. Xu, W. Zhu, Q. Y. Huang and W. L. Chen, Int. J. Mol. Sci., 2015, 16, 15670–15687 CrossRef CAS PubMed.
- V. D. M. Hc and H. J. Busscher, Appl. Environ. Microbiol., 2001, 67, 491–494 CrossRef.
- A. Esposito, F. Pagnanelli and F. Vegliò, Chem. Eng. Sci., 2002, 57, 307–313 CrossRef CAS.
- X. N. Li, A. R. Li, M. Z. Long and X. J. Tian, J. Environ. Health Sci. Eng., 2015, 13, 1–8 CrossRef PubMed.
- H. Xie, Q. Zhao, Z. Zhou, Y. Wu, H. Wang and H. Xu, RSC Adv., 2015, 5, 33478–33488 RSC.
- Y. Liu, G. Cui, C. Luo, L. Zhang, Y. Guo and S. Yan, RSC Adv., 2014, 4, 55162–55172 RSC.
- M. Torab-Mostaedi, M. Asadollahzadeh, A. Hemmati and A. Khosravi, J. Taiwan Inst. Chem. Eng., 2013, 44, 295–302 CrossRef CAS.
- G. Bayramoğlu, S. Bektaş and M. Y. Arica, J. Hazard. Mater., 2003, 101, 285–300 CrossRef.
- M. T. Veit, C. R. G. Tavares, S. M. Gomes-Da-Costa and T. A. Guedes, Process Biochem., 2005, 40, 3303–3308 CrossRef CAS.
- X. C. Chen, Y. P. Wang, Q. Lin, J. Y. Shi, W. X. Wu and Y. X. Chen, Colloids Surf., B, 2005, 46, 101–107 CrossRef CAS PubMed.
- R. M. Gabr, S. H. A. Hassan and A. A. M. Shoreit, Int. Biodeterior. Biodegrad., 2008, 62, 195–203 CrossRef CAS.
- Z. Khan, A. Rehman, S. Z. Hussain, M. A. Nisar, S. Zulfiqar and A. R. Shakoori, AMB Express, 2016, 6, 54 CrossRef PubMed.
- S. S. Shaukat, Biosorption of metals: state of the art, general features, and potential applications for environmental and technological processes, in Progress in Biomass and Bioenergy Production, ed. R. C. Oliveira, M. C. Palmieri and O. Garcia Jr, In Tech, Rijeka, 2011, pp. 151–176 Search PubMed.
- L. Velásquez and J. Dussan, J. Hazard. Mater., 2009, 167, 713–716 CrossRef PubMed.
- R. J. Yin, Q. X. Zhai, L. L. Yu, Y. Xiao, G. Wang, R. P. Yu, F. W. Tian and W. Chen, Eur. Food Res. Technol., 2016, 242, 1–9 CrossRef.
- T. Srinath, T. Verma, P. W. Ramteke and S. K. Garg, Chemosphere, 2002, 48, 427–435 CrossRef CAS PubMed.
- Y. Sheng, Y. Wang, X. Yang, B. Zhang, X. He, W. Xu and K. Huang, Environ. Toxicol. Pharmacol., 2016, 48, 183 CrossRef CAS PubMed.
- R. Biondo, S. F. Da, E. J. Vicente, J. E. Souza Sarkis and A. C. Schenberg, Environ. Sci. Technol., 2012, 46, 8325–8332 CrossRef CAS PubMed.
- S. Tunali, A. Çabuk and T. Akar, Chem. Eng. J., 2006, 115, 203–211 CrossRef CAS.
- F. Beolchini, R. Pagnanelli, L. Toro and F. Vegliò, Water Res., 2006, 40, 144–152 CrossRef CAS PubMed.
- Y. S. Ho and G. Mckay, Water Res., 2000, 34, 735–742 CrossRef CAS.
- G. Padmapriya and A. G. Murugesan, Desalin. Water Treat., 2015, 53, 3513–3520 CrossRef CAS.
- S. Khosravihaftkhany, N. Morad, A. Z. Abdullah, T. T. Teng and N. Ismail, RSC Adv., 2015, 5, 106498–106508 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra01163f |
|
| This journal is © The Royal Society of Chemistry 2017 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *ab and
Mingxin Huob
*ab and
Mingxin Huob
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min. The cells were rinsed with phosphate buffer solution (PBS) three times and were subsequently stored in PBS at 4 °C. Cell concentrations in the suspension were determined by drying an aliquot onto a pre-weighed aluminum foil container to a constant weight at 60 °C.
000g for 5 min. The cells were rinsed with phosphate buffer solution (PBS) three times and were subsequently stored in PBS at 4 °C. Cell concentrations in the suspension were determined by drying an aliquot onto a pre-weighed aluminum foil container to a constant weight at 60 °C.
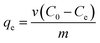
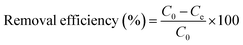
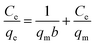
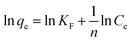
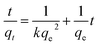
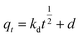
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 year.34 The distinctive echo niche caused bacterium CR3 to develop intrinsic heavy metal resistance mechanisms including self-detoxification through ion efflux, metal-complexation and metal-reduction, and a powerful DNA self-repair.27 Moreover, the EDX analysis confirmed that Cu2+ combined with the biomass of C. gilardii CR3 (Fig. 2). There was no copper peak appearing on the cells before Cu2+ was loaded (Fig. 2a). In contrast, two additional signals of copper existed in the EDX profile after Cu2+ was loaded on the cells (Fig. 2b).
000 year.34 The distinctive echo niche caused bacterium CR3 to develop intrinsic heavy metal resistance mechanisms including self-detoxification through ion efflux, metal-complexation and metal-reduction, and a powerful DNA self-repair.27 Moreover, the EDX analysis confirmed that Cu2+ combined with the biomass of C. gilardii CR3 (Fig. 2). There was no copper peak appearing on the cells before Cu2+ was loaded (Fig. 2a). In contrast, two additional signals of copper existed in the EDX profile after Cu2+ was loaded on the cells (Fig. 2b).


![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of amide I, the symmetric stretching C
O stretching of amide I, the symmetric stretching C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peptidic bond of carboxyl groups, and the stretching of PO2− in phosphodiesters and phosphates, respectively; and 545 cm−1, for phosphate or sulphate functional groups. In a comparison with the spectra of C. gilardii CR3 prior to being treated with Cu2+, a minute shift in the peak position was observed (<15 cm−1) after Cu2+ binding. The band shifts were from 3307 cm−1 to 3309 cm−1 for –OH and –NH, from 1390 cm−1 to 1391 cm−1 for –COOH, and from 1072 cm−1 to 1069 cm−1 for PO2− in the presence of Cu2+. Functional groups such as carboxyl, hydroxyl, amino, and phosphate groups played a key role during the metal ion binding process.35,36 The present FTIR analysis also provided evidence that these functional groups were involved in the binding of copper for C. gilardii CR3.
O peptidic bond of carboxyl groups, and the stretching of PO2− in phosphodiesters and phosphates, respectively; and 545 cm−1, for phosphate or sulphate functional groups. In a comparison with the spectra of C. gilardii CR3 prior to being treated with Cu2+, a minute shift in the peak position was observed (<15 cm−1) after Cu2+ binding. The band shifts were from 3307 cm−1 to 3309 cm−1 for –OH and –NH, from 1390 cm−1 to 1391 cm−1 for –COOH, and from 1072 cm−1 to 1069 cm−1 for PO2− in the presence of Cu2+. Functional groups such as carboxyl, hydroxyl, amino, and phosphate groups played a key role during the metal ion binding process.35,36 The present FTIR analysis also provided evidence that these functional groups were involved in the binding of copper for C. gilardii CR3.







