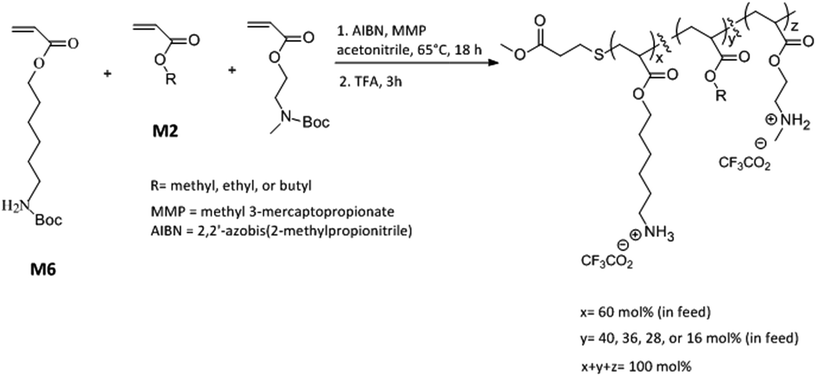
Open Access Article
Open Access ArticleRapid bactericidal activity of an amphiphilic polyacrylate terpolymer system comprised of same-centered comonomers with 2-carbon and 6-carbon spacer arms and an uncharged repeat unit†
Kamia Punia‡
a,
Ashish Punia‡a,
Kaushiki Chatterjeeb,
Sumit Mukherjee c,
Jimmie Fatab,
Probal Banerjeec,
Krishnaswami Rajaa and
Nan-Loh Yang
c,
Jimmie Fatab,
Probal Banerjeec,
Krishnaswami Rajaa and
Nan-Loh Yang *a
*a
aPh.D. Program in Chemistry at the Graduate Center of the City University of New York, New York, NY 10016, USA. E-mail: Nanloh.yang-cepm@csi.cuny.edu
bPh.D. Program in Biology at the Graduate Center of the City University of New York, New York, NY 10016, USA
cPh.D. Program in Biochemistry at the Graduate Center of the City University of New York, New York, NY 10016, USA
First published on 6th February 2017
Abstract
The global health threat of antimicrobial resistance has created a pressing need to develop practical alternatives to conventional antibiotic agents. Peptide mimetic synthetic amphiphilic polymers are known to non-specifically disrupt the bacterial cell surface thus leading to highly hindered bacterial resistance development. We investigated the antibacterial activities of a terpolymer macromolecular architecture with a combination of 6-carbon and 2-carbon spacer arms (distance from polymer backbone to pendent cationic center) interspersed with counits with hydrophobic side groups. A random copolymer with a combination of 6-carbon spacer arm repeat units (60 mol%) and 2-carbon spacer arm (40 mol%) units is moderately active against bacteria and shows very low hemolytic activity. Incorporation of comonomer units with alkyl side groups, by replacing different levels of 2-carbon spacer arm counit, led to substantial increments in antibacterial activities without detrimental effects on hemolytic activities leading to highly selective (bacteria over red blood cells) antibacterial activity. Time-kill studies revealed rapid bactericidal activity of the terpolymer against both Staphylococcus aureus and Escherichia coli with 100% killing efficiency achieved within 1 h of polymer treatment, corresponding to a 5-log reduction of bacterial colony forming units. These results indicate the high potential of this amphiphilic terpolymer architecture in the development of alternatives to antibiotics.
Introduction
The global threat of antimicrobial resistance has created a pressing need for alternatives to conventional antibiotics.1–5 A recent report has estimated more than 10 million annual deaths by 2050 as a result of antimicrobial resistance, which would surpass the number of fatalities caused by cancer.6 A steep decline in the introduction of new antibiotics has further exacerbated the threat from superbugs, a trend resulting in part from the huge costs involved in the development through clinical approval of new antibiotics followed by rapid development of antimicrobial resistance resulting in limited therapeutic utilization.1 Synthetic amphiphilic polymers which mimic the design and activity of natural antimicrobial peptides (AMPs) can provide the urgently needed solution to the superbug threat.7–9 The non-specific mode of antibacterial action (membrane rupture through electrostatic and lipophilic interactions) of AMPs and their synthetic polymeric mimics hinders the development of bacterial resistance.10,11 High activity of synthetic amphiphilic polymers against antibiotic resistant bacteria and the inability of bacteria to develop resistance against synthetic amphiphilic polymers, in contrast to conventional antibiotic, has been documented.12Understanding the role that polymeric design features plays in their antibacterial activities and toxicities towards mammalian cells is critical for their successful wide scale application as therapeutic agents. For biomedical applications, selective activity of amphiphilic polymers against bacteria over mammalian cells is highly desired. However, the macromolecular design features affecting the bacterial cell versus mammalian cell toxicity have not been well understood, and a number of studies in recent years have focused on the development of structure–activity relationships for synthetic amphiphilic polymers.13–44
We have recently shown that control of spatial cationic charge density through variation in spacer arm (distance from polymer backbone to cationic center) design can lead to highly selective (bacteria over red blood cells) antibacterial activity.41 The homopolymer of 6-carbon spacer arm is highly antibacterial and hemolytic, whereas the homopolymer of 2-carbon spacer arm monomer has low activity against Escherichia coli (E. coli) but displayed non-hemolytic activity.41 Interestingly, copolymerization of just a small mol% of 2-carbon spacer arm monomer with 6-carbon spacer arm monomer led to three orders of magnitude reduction in hemolytic activity without significant deterioration of antibacterial activities.41 On the other hand, it has been shown in earlier reports that “separate center” copolymer architecture with cationic and hydrophobic groups on separate repeat units shows higher antibacterial and hemolytic activity in comparison with “same center” homopolymer with cationic and hydrophobic groups on same repeat unit.44
Here we report a new terpolymer architecture with a combination of “same center” 6-carbon and 2-carbon spacer arm design with “separate center” architecture by interspersing cationic repeat units with hydrophobic repeat units. The mol% content of 6-carbon spacer arm repeat units was kept approximately constant while the effects of variation in mol% of 2-carbon spacer arm with respect to alkyl acrylates and the length of alkyl side group were investigated on the antibacterial and hemolytic activities of these terpolymers. Our investigations found that the combination of spacer arm and separate center design can have a synergistic effect leading to increment in antibacterial activity and the optimization of amphiphilic balance and lipophilicity can result in concomitant lower hemolytic activity. Moreover, the resultant copolymer displayed rapid bactericidal activity against both Gram negative and Gram positive bacteria underlining the anti-infective potential of these amphiphilic macromolecules.
Experimental
Materials and instrumentation
6-Amino-1-hexanol, 2-(methylamino)ethanol, 2,2′-azobis(2-methylpropionitrile) (AIBN), methyl 3-mercaptopropionate (MMP), N,N-diisopropylethylamine, acetonitrile (anhydrous), hexane, diethyl ether, and methanol were purchased from Sigma-Aldrich and were used without further purification. Acryloyl chloride was purchased from Sigma-Aldrich and was purified by distillation prior to use. Ethyl acrylate, methyl acrylate, and butyl acrylate were obtained from Sigma-Aldrich and were treated with inhibitor remover prior to use. Trifluoroacetic acid and di-tert-butyl dicarbonate (t-Boc) were purchased from Alfa Aesar and were used without further purification.1H NMR spectra were obtained on a Varian Unity NMR spectrometer (600 MHz) using CDCl3 or D2O as solvents. Molecular weights of polymers were estimated against linear polystyrene standards via gel permeation chromatography using an EcoSec HLC-83220 gel permeation chromatography instrument (RI detector, TSKgel SuperHZ-N (3 μm 4.6 mm ID) and TSKgel SuperHZ-M (3 μm 4.6 mm ID) columns). Tetrahydrofuran was used a solvent for GPC at a flow rate of 0.35 mL min−1.
Synthesis of monomers and polymers
N-Boc protected amine functionalized acrylate monomers (M6 and M2, Scheme 1) were synthesized as described in literature.41 Copolymers were synthesized via free radical polymerization and N-Boc protecting groups were subsequently cleaved by treatment with excess trifluoroacetic acid.40–44 A representative synthesis procedure is as follows. 1.30 g (4.8 mmol) of monomer M6, 0.51 g (2.24 mmol) of monomer M2, and 0.096 g (0.96 mmol) of ethyl acrylate were added into a 100 mL round bottom flask already charged with 0.013 g (0.08 mmol) of AIBN. In this reaction mixture, 0.048 g (0.4 mmol) of methyl 3-mercaptopropionate was added as a chain transfer agent. Acetonitrile (8 mL) was added as a solvent and the reaction mixture was subsequently degassed with nitrogen for 15 minutes using a stainless steel needle. The mixture was stirred at 65 °C for 18 h followed by evaporation of solvent using rotavapor. The polymer was then repeatedly precipitated in hexane by first dissolving in small quantity of tetrahydrofuran. To cleave N-Boc protecting groups, polymer was treated with excess trifluoroacetic acid (TFA) for 3 h. After the completion of reaction, excess TFA was removed using rotavapor and polymer was dissolved in small quantity of methanol and repeatedly precipitated into diethyl ether. The resultant polymer was dried under vacuum for 2 days and lyophilized. Yield 0.58 g.Determination of antibacterial activity
Polymer stock solutions were prepared in dimethyl sulfoxide and two-fold dilutions were prepared using distilled water. All biological activity experiments were performed as per modifications of our previous publications.40–44 An aliquot of E. coli (TOP 10, ampicillin resistant) was inoculated into freshly prepared Luria–Bertani (LB) broth. After overnight incubation of E. coli at 37 °C (under stirring); 1 mL of bacterial suspension was diluted with 9 mL of freshly prepared LB broth to enable log phase growth. Log phase growth of E. coli was confirmed by an increase in optical density from OD600 ∼ 0.1 to OD600 ∼ 0.45–0.5. This bacterial cell suspension in log phase was diluted with fresh LB broth to prepare a final stock cell suspension with OD600 = 0.001. 90 μL of this stock cell suspension was added into each well of the 96 well tissue culture plate followed by the addition of 10 μL of polymer concentrations. The 96 well plates were incubated at 37 °C (under stirring) for 18 h and bacterial cell growth was measured as turbidity at optical density at λ = 600 nm (OD600). Minimum inhibitory concentration (MIC) is defined as the lowest polymer concentration required to inhibit 100% bacterial cell growth. MICs were similarly determined against Staphylococcus aureus (S. aureus) (ATCC 25923) with the exception that Muller–Hinton broth was used in place of Luria–Bertani broth. The MIC values shown here are the averages of three separate experiments performed on different days.Determination of hemolytic activity
Freshly drawn mice blood was centrifuged at 3000 rpm for 15 minutes, and white blood cells and plasma were removed as supernatant followed by washing (2×) red blood cells (RBCs) with Tris-buffered saline (TBS). 1 mL of RBCs were diluted with TBS to obtain 2.5% RBC stock suspension. 130 μL of this RBC suspension, 15 μL of TBS, and 15 μL of polymer concentration were added to each 0.6 mL microcentrifuge tubes. The tubes were incubated under shaking at 37 °C for 1 h. The suspension was centrifuged at 4000 rpm for 5 minutes and 30 μL supernatant was added (in triplicate) into each well of the 96 well tissue culture plate. The lysing ability of polymers towards RBCs was determined in terms of hemoglobin concentration by measuring optical density at λ = 414 nm (OD414). Hemolytic concentration-50% (HC50) value is defined as the minimum polymer concentration required to lyse 50% of the RBCs within an incubation period of 1 h. For 100% hemolysis, 1% triton was used as a positive control. The HC50 values reported here are the averages of three separate experiments. Following formula was used to calculate HC50:All experiments were performed in compliance with the relevant laws and institutional guidelines of Institutional Animal Care and Use Committee (IACUC). The experiments have been approved by IACUC at College of Staten Island.
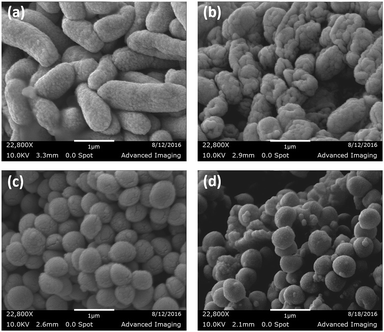
Field emission scanning electron microscopy (SEM) analysis of bacterial membrane rupture
E. coli and S. aureus cells were incubated (at 37 °C under shaking) overnight in Luria–Bertani broth and Muller–Hinton broth respectively. Bacterial cells from the overnight cell culture were bought to log phase growth by following the similar procedure as described above in determination of antibacterial activity. This cell suspension in log phase growth was subsequently diluted with freshly prepared nutrient broth to obtain a final stock cell suspension of approximately 108 CFU mL−1 (OD600 ∼ 0.1 for E. coli; and OD600 ∼ 0.2 for S. aureus). 900 μL of this cell suspension was mixed with 100 μL of polymer concentration in a 1.5 mL centrifugation tube and incubated for 2 h at 37 °C under shaking. After 2 h, the cell suspension was centrifuged at 4000 rpm for 5 minutes and supernatant was decanted. Bacterial cells were washed twice with phosphate-buffered saline and subsequently fixed using glutaraldehyde (2.5%) treatment for 1 h. Cells were then washed with distilled water and dehydrated sequentially with 30%, 50%, 70%, 80%, 90%, and 100% (v/v%) aqueous ethanol solutions. E. coli and S. aureus cell suspensions without polymer treatment were used as control. The resultant dehydrated cells were dried under vacuum (room temperature) for 2 days and were then mounted on SEM sample holder using carbon tape. For SEM analysis, gold/palladium (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) alloy coating was applied on samples via sputter coater and samples were analyzed at an operating voltage of 5 kV on an AMRAY 1910 field emission scanning electron microscope.
40) alloy coating was applied on samples via sputter coater and samples were analyzed at an operating voltage of 5 kV on an AMRAY 1910 field emission scanning electron microscope.
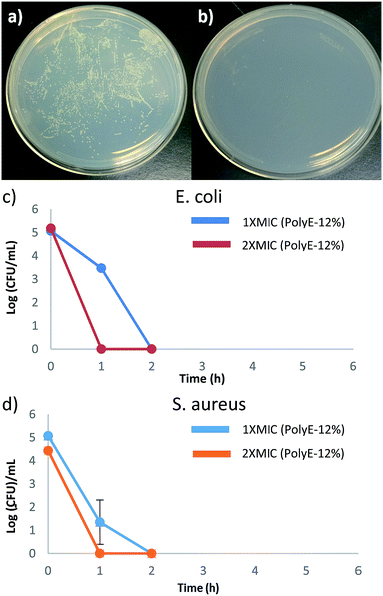
Determination of time dependent killing efficiency
E. coli and S. aureus cell suspensions were prepared at log phase growth as described above. Bacterial cell suspensions with approximately 105 CFUs mL−1 were treated with polymer concentrations (1× MIC and 2× MIC) at 37 °C under shaking. At time intervals of 0 h, 1 h, and 2 h; the cell suspension was taken out and serially diluted (in ten folds). 20 μL of final cell suspension was streaked on agar plate and incubated for 24 h (at 37 °C). The CFUs were counted using the Image J processing and analysis software. Bacterial cell suspension without polymer treatment was used control. Results reported here are the averages of two separate experiments and the detection limits of experiments were in the range of 228–320 CFUs in agar plates.Results and discussion
Design and synthesis of copolymers
Copolymers were synthesized via free radical polymerization as shown in Scheme 1. The feed mol% of M6 monomer was kept constant at 60 mol% and the relative mol% of M2 monomer and alkyl acrylate was gradually varied as the mol% of alkyl acrylate was changed from 0 mol% to 24 mol% (Scheme 1 and Table 1). The purity and composition of copolymers were determined by 1H NMR and the actual mole percentages of comonomers were found to be in close approximation to corresponding feed mole percentages. Molecular weights of N-Boc protected polymers were estimated by gel permeation chromatography (GPC) using linear polystyrene standards. All polymers were synthesized at similar molecular weights as confirmed by GPC (Table 1 and ESI†). “PolyM6–M2” represents the polymer with 60 mol% of M6 monomer and 40 mol% M2 monomer and other copolymers in this series are represented by the representative nomenclature “PolyX–Y%” where X indicates the identity of alkyl acrylate (X = M (methyl acrylate); E (ethyl acrylate); and B (butyl acrylate)) and Y% is the feed mol% of corresponding alkyl acrylate.| Polymer | Feed mol% of alkyl acrylate | Actual mol% of alkyl acrylatea | Mnb (kDa) | PDIc | E. coli MIC (μg mL−1) | S. aureus MIC (μg mL−1) | HC50 (μg mL−1) | Selectivity (HC50/MIC) | |
|---|---|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | ||||||||
| a Calculated from 1H NMR.b Number average molecular weight estimated from gel permeation chromatography (GPC).c Polydispersity index calculated from GPC. | |||||||||
| PolyM6–M2 | 0 | 0 | 3.8 | 1.4 | 125 | 62 | >2000 | >16 | >32 |
| PolyM-4% | 4 | 3 | 3.7 | 1.4 | 125 | 62 | >2000 | >16 | >32 |
| PolyM-12% | 12 | 10 | 3.7 | 1.4 | 62 | 62 | >2000 | >32 | >32 |
| PolyM-24% | 24 | 21 | 3.7 | 1.4 | 30 | 62 | 423 | 14 | 7 |
| PolyE-4% | 4 | 4 | 3.7 | 1.4 | 93 | 62 | >2000 | >21 | >32 |
| PolyE-12% | 12 | 12 | 3.7 | 1.4 | 62 | 62 | >2000 | >32 | >32 |
| PolyE-24% | 24 | 21 | 3.8 | 1.4 | 22 | 46 | 384 | 17 | 8 |
| PolyB-4% | 4 | 4 | 4.1 | 1.4 | 62 | 62 | 1428 | 23 | 23 |
| PolyB-12% | 12 | 11 | 3.9 | 1.3 | 15 | 38 | 250 | 16 | 7 |
| PolyB-24% | 24 | 24 | 4.4 | 1.3 | 7.8 | 22.5 | 12.6 | 1.6 | 0.56 |
Antibacterial activity
Antibacterial activities of polymers were determined against Gram negative E. coli and Gram positive S. aureus in terms of MICs and are as shown in Fig. 1 and Table 1. It has been shown in earlier reports that homopolymer with 2-carbon spacer arm repeat units display low activity against E. coli.41,44,45 We have earlier reported that incorporation of a longer 6-carbon spacer arm repeat unit with 2-carbon spacer arm counits in a random copolymer led to significant increase in antibacterial activity.41 Incorporation of longer spacer arm can reduce the local cationic charge density and may also result in higher membrane rupture ability due to “snorkel effect” in which the cationic centers attach to negatively charged bacterial cell surface and longer alkyl spacer arms can then permeabilize through the hydrophobic core of bacterial lipid bilayer.41,45 Furthermore, inclusion of repeat unit with longer spacer arms increases the spatial distance between cationic groups. Increase of backbone spacer distance has been reported to result in higher antibacterial activity in cationic polymers obtained from alternating ring-opening metathesis polymerization.22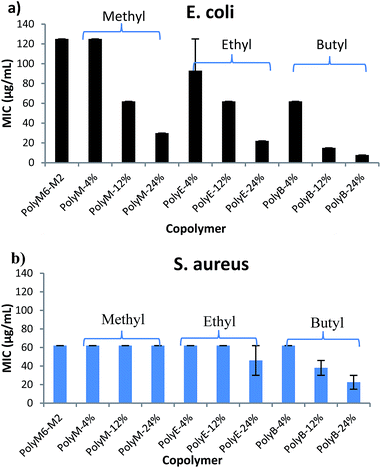 | ||
| Fig. 1 Antibacterial activities (MIC) of copolymers towards (a) E. coli and (b) S. aureus. Error bars represent standard deviation. | ||
In contrast with this combination of 6-carbon and 2-carbon spacer arm copolymer design, “separate center” random copolymers with cationic groups and hydrophobic alkyl groups present on separate repeat units show higher antibacterial activity as compared with “same center” homopolymers with both cationic centers and hydrophobic groups attached on the same repeat unit.21,44,46 Higher antibacterial activities in “separate center” random copolymers can be obtained through increasing the mol% of hydrophobic comonomers in random copolymers.
We postulated that an amphiphilic terpolymer architecture consisting of hydrophobic repeat unit copolymerized with 6-carbon and 2-carbon spacer arm counits – a combination of “separate center” and “same center” spacer arm design – can show unique biological activities resulting from judicious variation of spacer arm lengths and amphiphilicity conducive to permeability through bacterial cell membrane. In order to explore antibacterial activities of this terpolymer design, we synthesized a series of copolymers in this study to assess various combinations of spacer arm repeat units with hydrophobic side groups. PolyM6–M2 with 60 mol% of 6-carbon spacer arm (M6) repeat unit and 40 mol% of 2-carbon spacer arm repeat unit (M2) displayed moderately high antibacterial activity against E. coli (MIC = 125 μg mL−1). Incorporation of 12 mol% of repeat unit with methyl side group led to a significant increase in antibacterial activity, and the copolymer with approximately 24 mol% of methyl acrylate comonomer unit demonstrated high activity against E. coli (MIC = 32 μg mL−1). It should be noted that the mol% of M6 was kept constant at approximately 60% in all these copolymers and mol% of M2 was reduced to compensate for the inclusion of increasing mol% of hydrophobic comonomer.
Similar to the effects of adding methyl side groups, copolymerization of ethyl and butyl side groups led to increase in activity against E. coli, albeit to a much higher extent than incorporation of methyl side groups. Incorporation of just 12 mol% of comonomer units with butyl side group led to a dramatic increase in antibacterial activity towards E. coli (MIC = 15 μg mL−1). Longer alkyl side groups substantially increase the lipophilicity of copolymers leading to enhanced permeability of copolymers through the hydrophobic core of lipid bilayer.
Antibacterial activities of these polymers were also determined against Gram positive S. aureus. Interestingly, activities of these polymers demonstrate a subdued effect of hydrophobic comonomer inclusion on their activities against S. aureus. Copolymerization of up to 24 mol% of methyl side groups did not lead to any increment in the activity against S. aureus, whereas a dramatic increase in activity against E. coli was observed, as discussed above. Similarly, increasing the mol% of ethyl acrylate till 24% did not lead to substantial impact on the activity of polymer against S. aureus. Incorporation of butyl side groups resulted in higher activity against S. aureus, but the increase in activity against S. aureus was significantly lower as compared with increase in activity against E. coli at same mol% of butyl side groups. These observations indicate that a much higher level of lipophilicity is required to enhance activity of these polymers against S. aureus. Substantial differences in the cell surface morphology of S. aureus and E. coli may be a reason behind this observation. The cell wall of the S. aureus is made of a thick negatively charged peptidoglycan layer (15–80 nm thick), whereas the E. coli cell wall consists of a thin peptidoglycan layer sandwiched between the outer and inner cell membrane.47 Strong electrostatic interactions between these highly charged polymers and negatively charged peptidoglycan layer can hinder the permeabilization of polymers through thick cell wall of S. aureus. Thus, an increase in activity against S. aureus may require a substantially higher content of longer alkyl side groups, as compared with E. coli.
It was recently shown that sedimentation of bacterial and nanoparticle suspension can result due to adhesion of highly charged nanoparticles on the surfaces of negatively charged bacterial cells.48 This phenomena was shown to result in large errors and led to disqualification of MIC measurements. In contrast to this nanoparticle system, terpolymers in our study described here are readily soluble in aqueous broth/nutrient media used in MIC determinations and formed clear solutions rather than suspensions, as reported in earlier study. We did not observe any aggregations or formation of suspensions, except for the turbidity observed at below MIC concentrations (due to bacterial cell growth) and thus sedimentation of bacterial and polymer complexes is not expected. Moreover, as shown below, time-dependent killing efficiency studies confirmed bactericidal activity of the polymer PolyE-12% at 1× MIC concentration.
Hemolytic activity
Hemolytic activity has been widely used as a benchmark to assess the toxicity of synthetic amphiphilic polymers against mammalian cells. Hemolytic activities of polymers were determined against mice RBCs in terms of HC50 and are as shown in Fig. 2 and Table 1. HC50 is the minimum polymer concentration required to lyse 50% of RBCs within an incubation period of 1 h. The cell surface of erythrocytes is primarily comprised of zwitterionic lipid head groups and cholesterol, and lacks net negative charge in contrast with the net negatively charged surface of bacterial cells.10,11 Thus, the hemolytic activity of synthetic amphiphilic polymers mainly arises from the lipophilic interactions between the RBCs' lipid bilayer and hydrophobic groups in polymers.11 PolyM6–M2 with cationic charge on each repeat unit demonstrated non-hemolytic activity. High cationic charge density can be expected to reduce lysing ability of polymers towards RBCs, as high charge density can thwart the permeabilization of the polymers through lipophilic core of lipid bilayer.41,44 Higher degree of quaternization was shown to result in lower hemolytic activities of 1,3-thiazol and 1,2,3-triazole functionalized polymethacrylates.31 However, it has been reported earlier that homopolymer with 6-carbon spacer arm repeat unit and cationic charge on each repeat unit demonstrated very high hemolytic activity, and copolymerization of 2-carbon spacer arm units with 6-carbon spacer arm units led to drastic decline in hemolytic activity.41,45 These observations underline the crucial interplay in charge density and lengths of spacer arms in determining the hemolytic activity of synthetic amphiphilic polymers.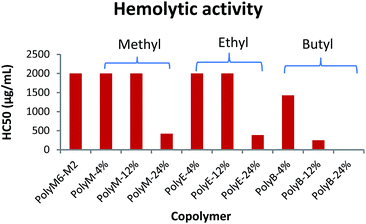 | ||
| Fig. 2 Hemolytic activity (HC50) of polymers towards mice RBCs. Results shown are averages of three experiments (size of error bars is negligible). | ||
In the terpolymer system described here, a third comonomer with alkyl side group i.e. methyl, ethyl, or butyl side group was copolymerized with M6 and M2 in various molar percentages. Incorporation of a hydrophobic comonomer should increase the hemolytic activity of these copolymers. However, optimization of mol% of hydrophobic comonomer and presence of higher cationic charge density originating from M6 and M2 can mitigate the RBC cell rupture ability of these polymers. Indeed, up to 12 mol% of methyl acrylate we did not observe an increase in hemolytic activity (HC50 > 2000 μg mL−1). At 24 mol% of methyl acrylate, the copolymer displayed higher hemolytic activity (HC50 = 423 μg mL−1). Similarly, no increase in hemolytic activity was observed with up to 12 mol% of ethyl acrylate repeat units but a higher mol% of 24% led to increased cell surface rupture ability against erythrocytes. This precipitous increase in hemolytic activity indicates that a certain mol% of M2 repeat units is required to prevent the penetration of these polymers through RBC's cell membrane. As mol% of M2 is reduced, there is probably not sufficient number of M2 comonomer units to prevent the “snorkel effect” and leads to copolymer's ability to penetrate through RBCs' lipid bilayer. Relative to incorporation of methyl and ethyl acrylate comonomers, addition of just 12 mol% of butyl acrylate resulted in substantial increase in hemolytic activity (HC50 = 250 μg mL−1). At approximately 24 mol% of butyl acrylate, PolyB-24% demonstrated high hemolytic activity (HC50 = 12.6 μg mL−1).
Polymethacrylate copolymers of 6-carbon spacer arm repeat units, M6, and comonomer units with ethyl side groups have been previously shown to possess very high hemolytic activity.45 High hemolytic activity of 6-carbon spacer arm homopolymer severely hinders the therapeutic potential of this highly antibacterial synthetic macromolecule. One of the reasons behind the high hemolytic activity of 6-carbon spacer arm homopolymer can be its high hydrophobicity resulting from hexyl spacer arms. Copolymerization of 6-carbon spacer arm monomer, M6, with a comonomer with low hydrophobicity can be expected to reduce its hemolytic effect. However, copolymerization of M6 comonomer units with ethyl side groups was shown to result in no reduction of hemolytic activity.45 In copolymer system reported here, it is apparent that inclusion of 2-carbon spacer arm comonomer units, M2, can substantially subdue the hemolytic activity arising from 6-carbon spacer arm counits. Furthermore, controlled incorporation of ethyl or methyl side groups did not lead to significant increase in hemolytic activity. As shown above, this terpolymerization of 6-carbon spacer arm, 2-carbon spacer arm, and alkyl acrylate repeat units have further improved the antibacterial activities with no detrimental effect on hemolytic activity at optimum levels of alkyl acrylate copolymerization.
Selectivity of copolymers
Selective activity of synthetic amphiphilic polymers against bacteria over human cells is highly desired for biomedical applications. The selectivity is defined as the ratio (HC50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MIC) of hemolytic activity to MIC and has been widely used in literature to assess the selective activity of synthetic amphiphilic polymers. As shown in Table 1, majority of polymers synthesized in this study displayed high selectivity towards both E. coli and S. aureus over RBCs. Incorporation of methyl or ethyl side groups led to higher selectivity against E. coli over RBCs. PolyB-12% and PolyB-24% showed low selectivity values due to their high hemolytic activities arising from higher hydrophobicity.
MIC) of hemolytic activity to MIC and has been widely used in literature to assess the selective activity of synthetic amphiphilic polymers. As shown in Table 1, majority of polymers synthesized in this study displayed high selectivity towards both E. coli and S. aureus over RBCs. Incorporation of methyl or ethyl side groups led to higher selectivity against E. coli over RBCs. PolyB-12% and PolyB-24% showed low selectivity values due to their high hemolytic activities arising from higher hydrophobicity.
Scanning electron microscopy analysis
To ascertain the bacterial cell surface rupture ability of these polymers, field emission scanning electron microscopy (FE-SEM) analysis was performed on E. coli and S. aureus cells after treatment with PolyE-12%. As shown in Fig. 3, control (without polymer treatment) E. coli and S. aureus cells show intact cell surface morphologies. Treatment of E. coli cells with PolyE-12% for 2 h led to drastic rupture of bacterial cell surface. Similarly, cell surface disruption ability of PolyE-12% was confirmed against S. aureus. FE-SEM analysis confirmed that these copolymers have high bacterial cell membrane rupture ability in addition to possible intracellular mechanisms11 of bacterial cell killing activity. Such severe bacterial cell surface damage caused by non-specific electrostatic and hydrophobic activity of these polymers indicates that the development of bacterial resistance would be highly hindered.Time dependent killing efficiency
Time-kill studies were performed for PolyE-12% against E. coli and S. aureus at 1× MIC and 2× MIC polymer concentrations. As shown in Fig. 4, more than 99.9% (ESI, Fig. S4†) of both E. coli and S. aureus CFUs were killed within 1 h of PolyE-12% treatment (1× MIC) and no viable CFUs mL−1 were observed of both E. coli and S. aureus after 2 h of PolyE-12% treatment (1× MIC), corresponding to a 5 log reduction of bacterial CFUs. During the same time period, substantial proliferation of bacterial CFU mL−1 in control experiment (without polymer treatment) was observed.These time kill studies demonstrate rapid bactericidal activity for our terpolymer system and substantiate very high bacterial killing potency. To the best of our knowledge, there are only few other types of cationic amphiphilic polymers that have shown rapid bacterial killing activity.31,49–51 Antibacterial polycarbonates synthesized with same centered approach achieved 100% killing efficiency after 4 h of polymer treatment.50 In another example, biodegradable polycarbonates (at 2× MIC concentration) could eliminate S. aureus after 2 h of polymer treatment.49 1,2,3-Triazole and 1,2-thiazole functionalized polymethacrylates have also shown rapid bactericidal activity.31
Rapid bactericidal activity of these terpolymers is in contrast to much slower bacterial killing kinetics of PEGylated polyacrylates with 6-carbon spacer arm repeat unit.40 After 1 h of challenging E. coli with PEGylated polyacrylates, only 20% to 50% of E. coli CFUs were killed and 5 log reduction in E. coli CFUs was obtained only after prolonged treatment for 6 h.40 Moreover, substantial number of S. aureus CFUs survived even after 8 h of bacterial incubation with PEGylated polyacrylates as shown in our earlier finding.40 These observations underline significantly different interaction of bacterial cells with PEGylated versus non-PEGylated synthetic amphiphilic polyacrylates reported in this study. Even though amphiphilic polyacrylates with non-ionic PEG side groups demonstrated high bacteriostatic ability (low MIC values); their significantly slower bacterial killing kinetics indicates the thwarted bacterial membrane rupture ability. Hydrogen bonding associations between PEG side groups and peptidoglycans in the cell wall of bacteria can hinder permeabilization of polymer chains through bacterial cell surface leading to reduced bacteria killing potency.
Conclusions
We have previously reported that amphiphilic polyacrylate copolymers with a combination of 6-carbon and 2-carbon spacer arms counits demonstrated high antibacterial activities with concomitant low hemolytic activities. In this study, we explored a novel amphiphilic terpolymer system with hydrophobic monomer copolymerized with 6-carbon and 2-carbon spacer arm comonomers – a combination of “separate center” and “spacer arm” design. Controlled incorporation of hydrophobic alkyl comonomer with corresponding replacement of 2-carbon spacer arm counit led to substantial increase in antibacterial activities without detrimental effects on hemolytic activities. This strategy led to polymers with highly selective antibacterial activities towards bacteria over RBCs. Furthermore, time-dependent bacterial killing efficiency studies demonstrated rapid bacterial killing action by polymers with 100% S. aureus killing efficiency achieved within one hour of treatment, corresponding to a 5-log reduction of bacterial CFU mL−1. These results demonstrate the potential of this new amphiphilic polymer architecture in the development of urgently required antibacterial agents to deter the severe threat of antimicrobial resistance.Acknowledgements
We acknowledge support from the Center of Engineered Polymeric Materials, the Graduate Center of the City University of New York, Department of Chemistry at the College of Staten Island, CUNY Research Foundation RF 68464-0046 and 66617-0044. We acknowledge Advanced Imaging Facility at CSI, CUNY, for providing facility support for FE-SEM analysis.References
- D. Jabes, Curr. Opin. Microbiol., 2011, 14, 564 CrossRef PubMed.
- H. Boucher, Clin. Infect. Dis., 2010, 50, S4 CrossRef PubMed.
- A. Y. Peleg and D. C. Hooper, N. Engl. J. Med., 2010, 362, 1804 CrossRef CAS PubMed.
- R. M. Klevens, J. R. Edwards, C. J. Richards Jr, T. C. Horan, R. P. Gaynes, D. A. Pollock and D. M. Cardo, Publ. Health Rep., 2007, 122, 160 CrossRef.
- P. W. Stone, E. C. Hedblom, D. M. Murphy and S. B. Miller, Am. J. Infect. Control, 2005, 33, 542 CrossRef PubMed.
- Review on Antimicrobial Resistance. Availabel online: http://www.amr-review.org/, accessed on 21 Oct 2016.
- A. Muñoz-Bonilla and M. Fernández-García, Prog. Polym. Sci., 2012, 37, 281 CrossRef.
- A. C. Engler, N. Wiradharma, Z. Y. Ong, D. J. Coady, J. L. Hedrick and Y. Yang, Nano Today, 2012, 7, 201 CrossRef CAS.
- M. Ganewatta and C. Tang, Polymer, 2015, 63, A1 CrossRef CAS.
- M. Zasloff, Nature, 2002, 415, 389 CrossRef CAS PubMed.
- K. Kuroda and G. A. Caputo, WIREs Nanomedicine and Nanobiotechnology, 2012, 5(1), 49 CrossRef PubMed.
- I. Sovadinova, E. F. Palermo, M. Urban, P. Mpiga, G. A. Caputo and K. Kuroda, Polymers, 2011, 3, 1512 CrossRef CAS.
- Y. Oda, S. Kanaoka, T. Sato, S. Aoshima and K. Kuroda, Biomacromolecules, 2011, 12, 3581 CrossRef CAS PubMed.
- K. Lienkamp and G. N. Tew, Chem.–Eur. J., 2009, 15, 11784 CrossRef CAS PubMed.
- E. F. Palermo and K. Kuroda, Biomacromolecules, 2009, 10, 1416 CrossRef CAS PubMed.
- J. Zhang, M. J. Markiewicz, B. P. Mowery, B. Weisblum, S. S. Stahl and S. H. Gellman, Biomacromolecules, 2012, 13, 323 CrossRef CAS PubMed.
- K. Kuroda and W. F. DeGrado, J. Am. Chem. Soc., 2005, 127, 4128 CrossRef CAS PubMed.
- P. H. Sellenet, B. Allison, B. M. Applegate and J. P. Youngblood, Biomacromolecules, 2007, 8(1), 19 CrossRef CAS PubMed.
- T. R. Stratton, J. A. Howarter, B. C. Allison, B. M. Applegate and J. P. Youngblood, Biomacromolecules, 2010, 11(5), 1286 CrossRef CAS PubMed.
- V. W. L. Ng, J. P. K. Tan, J. Leong, Z. X. Voo, J. L. Hedrick and Y. Y. Yang, Macromolecules, 2014, 47, 1285 CrossRef CAS.
- V. Sambhy, B. R. Peterson and A. Sen, Angew. Chem., 2008, 120, 1270 CrossRef.
- A. Song, S. G. Walker, K. A. Parker and N. S. Sampson, ACS Chem. Biol., 2011, 6, 590 CrossRef CAS PubMed.
- U. Baul, K. Kuroda and S. Vemparala, J. Chem. Phys., 2014, 141, 084902 CrossRef PubMed.
- J. Hoque, P. Akkapeddi, V. Yadav, G. Manjunath, D. Uppu, M. Konai, V. Yarlagadda, K. Sanyal and J. Haldar, ACS Appl. Mater. Interfaces, 2015, 7, 1804 CAS.
- D. Uppu, P. Akkapeddi, G. Manjunath, V. Yarlagadda, J. Hoque and J. Haldar, Chem. Commun., 2013, 49, 9389 RSC.
- D. Uppu, S. Samaddar, C. Ghosh, K. Paramanandham, B. Shome and J. Haldar, Biomaterials, 2016, 74, 131 CrossRef CAS PubMed.
- S. Chakraborty, R. Liu, Z. Hayouka, X. Chen, J. Ehrhardt, Q. Lu, E. Burke, T. Yang, B. Weisblum, G. C. L. Wong, K. Masters and S. H. Gellman, J. Am. Chem. Soc., 2014, 136, 14530 CrossRef CAS PubMed.
- A. Muñoz-Bonilla, O. León, M. Cerrada, J. Rodríguez-Hernández, M. Sánchez-Chaves and M. Fernández-García, Eur. Polym. J., 2015, 62, 167 CrossRef.
- M. Álvarez-Paino, A. Muñoz-Bonilla, F. López-Fabal, J. Gómez-Garcés, J. Heuts and M. Fernández-García, Biomacromolecules, 2015, 16, 295 CrossRef PubMed.
- R. Tejero, D. López, F. López-Fabal, J. Gómez-Garcés and M. Fernández-García, Polym. Chem., 2015, 6, 3449 RSC.
- R. Tejero, D. López, F. López-Fabal, J. Gómez-Garcés and M. Fernández-García, Biomacromolecules, 2015, 16, 1844 CrossRef CAS PubMed.
- A. Abd-El-Aziz, C. Agatemor, N. Etkin, D. Overy, M. Lanteigne, K. McQuillan and R. Kerr, Biomacromolecules, 2015, 16, 3694 CrossRef CAS PubMed.
- H. Wang, X. Shi, D. Yu, J. Zhang, G. Yang, Y. Cui, K. Sun, J. Wang and H. Yan, Langmuir, 2015, 31, 13469 CrossRef CAS PubMed.
- C. Yang, X. Ding, R. J. Ono, H. Lee, L. Y. Hsu, Y. W. Tong, J. Hedrick and Y. Y. Yang, Adv. Mater., 2014, 26, 7346 CrossRef CAS PubMed.
- Z. X. Voo, M. Khan, K. Narayanan, D. Seah, J. L. Hedrick and Y. Y. Yang, Macromolecules, 2015, 48, 1055 CrossRef CAS.
- K. D. Roberts, M. A. K. Azad, J. Wang, A. S. Horne, P. E. Thompson, R. L. Nation, T. Velkov and J. Li, ACS Infect. Dis., 2015, 1, 568 CrossRef CAS PubMed.
- Z. Z. Deris, J. D. Swarbrick, K. D. Roberts, M. A. K. Azad, J. Akter, A. S. Horne, R. L. Nation, K. L. Rogers, P. E. Thompson, T. Velkov and J. Li, Bioconjugate Chem., 2014, 25, 750 CrossRef CAS PubMed.
- P. Ferruti, N. Mauro, L. Falciola, V. Pifferi, C. Bartoli, M. Gazzarri, F. Chiellini and E. Ranucci, Macromol. Biosci., 2013, 14, 390 CrossRef PubMed.
- E. Martinelli, E. Guazzelli, C. Bartoli, M. Gazzarri, F. Chiellini, G. Galli, M. E. Callow, J. A. Callow, J. A. Finlay and S. Hill, J. Polym. Sci., Part A: Polym. Chem., 2015, 53, 1213 CrossRef CAS.
- A. Punia, A. Mancuso, P. Banerjee and N.-L. Yang, ACS Macro Lett., 2015, 4, 426 CrossRef CAS.
- A. Punia, E. He, K. Lee, P. Banerjee and N.-L. Yang, Chem. Commun., 2014, 50, 7071 RSC.
- A. Punia and N.-L. Yang, RSC Adv., 2015, 5, 80318 RSC.
- A. Punia, K. Lee, E. He, S. Mukherjee, A. Mancuso, P. Banerjee and N.-L. Yang, Int. J. Mol. Sci., 2015, 16(10), 23867 CrossRef CAS PubMed.
- A. Punia, P. R. Debata, P. Banerjee and N.-L. Yang, RSC Adv., 2015, 5, 95300 RSC.
- E. F. Palermo, S. Vemparala and K. Kuroda, Biomacromolecules, 2012, 13, 1632 CrossRef CAS PubMed.
- G. J. Gabriel, J. A. Maegerlein, C. F. Nelson, J. M. Dabkowski, T. Eren, K. Nusslein and G. N. Tew, Chem.–Eur. J., 2009, 15, 433 CrossRef CAS PubMed.
- K. Lienkamp, K. Kumar, A. Som, K. Nüsslein and G. N. Tew, Chem.–Eur. J., 2009, 15, 11710 CrossRef CAS PubMed.
- B. Fang, Y. Jiang, K. Nusslein, V. M. Rotello and M. M. Santore, Colloids Surf., B, 2015, 125, 255 CrossRef CAS PubMed.
- L. M. Thoma, B. R. Boles and K. Kuroda, Biomacromolecules, 2014, 15(8), 2933 CrossRef CAS PubMed.
- W. Chin, C. Yang, V. W. L. Ng, Y. Huang, J. Cheng, Y. W. Tong, D. J. Coady, W. Fan, J. L. Hedrick and Y. Y. Yang, Macromolecules, 2013, 46(22), 8797 CrossRef CAS.
- A. C. Engler, J. P. K. Tan, Z. Y. Ong, D. J. Coady, V. W. L. Ng, Y. Y. Yang and J. L. Hedrick, Biomacromolecules, 2013, 14, 4331 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: 1H NMR of monomers and polymers, calculation of mol% of comonomer repeat units, gel permeation chromatography results, and time-kill results are provided. See DOI: 10.1039/c7ra00047b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2017 |