Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Developmental, chemical and transcriptional characteristics of artificially pollinated and hormone-induced parthenocarpic fruits of Siraitia grosvenorii†
Dongping Tu‡
ab,
Zuliang Luo‡a,
Bin Wua,
Xiaojun Ma *a,
Hongwu Shia,
Changming Moc,
Jie Huangc and
Wenjuan Xied
*a,
Hongwu Shia,
Changming Moc,
Jie Huangc and
Wenjuan Xied
aInstitute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100193, China. E-mail: mayixuan10@163.com; Fax: +86 10 57833290; Tel: +86 10 57833281
bGuangxi University of Chinese Medicine, Nanning 530200, China
cGuangxi Botanical Garden of Medicinal Plants, Nanning 530023, China
dGuilin Medical University, Guilin 541004, China
First published on 21st February 2017
Abstract
Siraitia grosvenorii is a dioecious cucurbitaceae plant that is native to southern China and prevalent in Guangxi Province. Natural pollination of this species is difficult, and artificial pollination is therefore the main approach for its cultivation. The fruit set of the plants largely depends on the biosynthesis and crosstalk of phytohormones. Here, we show that parthenocarpic fruit can be induced by 1-(2-chloro-4-pyridyl)-3-phenylurea (CPPU, an active cytokinin) and gibberellin (GA3) in S. grosvenorii. In addition to pollination, similar changes were detected in the external quality and sweet mogrosides of parthenocarpic fruits. Furthermore, the transcriptome of S. grosvenorii fruits was assessed by RNA sequencing (RNA-Seq). Differentially expressed genes (DEGs) in the fruits set were compared with those in untreated ovaries. Excluding 2794 common DEGs, large numbers of genes expressed specifically in parthenocarpic (2281) or pollinated (6191) fruits were found. In conclusion, CPPU and GA3-induced parthenocarpic fruits offer novel insights for the large-scale cultivation of S. grosvenorii. This study yielded a number of candidate genes that can be applied in further studies to improve fruit quality and yield.
1. Introduction
Siraitia grosvenorii is a perennial vine of the cucurbitaceae family. It grows mainly in Guangxi Province in China and has been cultivated for more than 200 years. The fruits of S. grosvenorii, known as Luo Han Guo (LHG), have been used for thousands of years as a natural sweetener and as a folk medicine for the treatment of lung congestion, colds, and sore throats. In recent years, pharmacological investigations have shown that the extracts and individual compounds (especially mogroside V) isolated from LHG have specific biological effects, including anti-tumor,1,2 anti-diabetic,3–8 anti-inflammatory,9 and anti-oxidative10 properties. The primary medically active components of LHG are cucurbitane-type triterpenoids, known as mogrosides, which include mogroside III(MIII), mogroside IVa(MIVA), mogroside IV(MIV), mogroside V(MV), iso-mogroside V(IMV), 11-oxomogroside-V(OMV) and siamenoside I(SI), among others. Moreover, the majority of mogrosides in ripe fruits exhibit a high level of sweetness, are low in calories, and can serve as a substitute for sugar for obese and diabetic patients.11,12 Commercial extracts (mixtures of mogrosides) are approximately 300 times sweeter than sucrose.13 The purified sweet component MV has been approved as an intense sweetening agent in Japan,14 and extracts of S. grosvenorii fruits have achieved generally recognized as safe (GRAS) status in the USA as a non-nutritive sweetener and flavor enhancer.12Due to the high demand for non-caloric sweeteners from natural sources, LHG has become increasingly popular in international markets, and its use in industrial food and beverages has gradually increased.15 The Johnson & Johnson subsidiary McNeil Nutritionals LLC has introduced a zero-calorie tabletop sweetener, Nectresse, that is made with LHG extracts. In addition, leading manufacturers, such as Coke and Kashi, have introduced products containing LHG extracts. However, LHG accounts for a small share of the alternative sweetener market, remaining at 2.2% in natural supermarkets during the 2014–2015 period. The market share of LHG is small because of the limited supply of this extract.16 The development of the S. grosvenorii industry has been affected by problems related to its cultivation. First, S. grosvenorii is a dioecious cucurbitaceae plant, and a large number of male plants must therefore be planted to ensure contact between staminate and pistillate flowers. Second, natural pollination is difficult due to the specific flower structure and the sticky pollen of S. grosvenorii, which cannot be pollinated by wind or insects. Therefore, an artificial pollination technique is needed to increase the fruit set rate in this species. However, artificial pollination has certain drawbacks. For example, it is associated with a heavy workload and high labor costs. Moreover, blossoming is short and concentrated, and pollen viability is affected over time. In addition, the seeds account for nearly 40% of the fresh fruit weight following pollination of diploid S. grosvenorii. Notably, mogrosides are distributed mainly in the fleshy pulp of the fruits, which is almost devoid of seeds. Additionally, the seeds contain a large amount of oil and fat components, which increase the cost of extraction and purification.11 Fortunately, parthenocarpy provides an alternative approach to achieve high yields and seedless S. grosvenorii fruits.
Parthenocarpy is an important physiological event and largely depends on the coordinated action of inherent hormones produced in unpollinated ovaries or induced by the application of exogenous hormones.17,18 Fruit set, the decision of the ovary to become a fruit, is a complex developmental process that is of great relevance to both the yield and reproduction of crop plants.19,20 It is usually achieved through successful pollination and fertilization or via parthenocarpy.21 Although extensive studies have been conducted, the molecular mechanisms underlying fruit setting remain poorly defined.22 Studies examining several fruit crops have revealed that the endogenous hormone levels of parthenocarpic cultivars are much higher than those of non-parthenocarpic cultivars.21,23 Moreover, parthenocarpic fruit setting and growth can be induced by the application of diverse phytohormones to unpollinated ovaries, gibberellins (GAs), auxins, and cytokinins in particular. Exogenous hormones such as indole acetic acid (IAA),24 GAs,25 1-(2-chloro-4-pyridyl)-3-phenylurea (CPPU, an active cytokinin)26 and brassinosteroids (BRs)27 are widely used to study the induction of parthenocarpy. Additionally, experimental results have demonstrated that a mixture of two or more exogenous hormones can improve the quality of parthenocarpic fruits.28 At present, the available technology for chemically inducing parthenocarpy is based on seedless vegetables and fruits such as watermelon,29 tomato26,30 and grape.31
The aim of the present study was to investigate the effect of exogenous hormones on the induction of parthenocarpic fruit in S. grosvenorii and to perform a comparative characterization of the response of unpollinated S. grosvenorii ovaries to exogenous hormones (GA3 and CPPU) or pollination, involving morphological characteristics, cytological characteristics, and chemical components. In addition, we investigated the transcriptome of S. grosvenorii fruits and compared global transcriptional events in different S. grosvenorii fruits to distinguish the molecular mechanism of parthenocarpy from that leading to pollinated fruit setting. Furthermore, the global analysis of parthenocarpy helped us to identify valuable parthenocarpic genes to resolve the major challenges related to parthenocarpic fruit yield and quality.
2. Materials and methods
2.1 Plant materials and experimental treatments
A Siraitia grosvenorii (Swingle) C. Jeffery Cultivar (Yongqing No. 1) was cultivated in the Guangxi Botanical Garden of Medicinal Plants (Nanning, China). Fruits of different stages were used in the present analysis. In total, 500 pistillate flowers from twenty representative S. grosvenorii plants were selected. Each experiment was performed at 7:00 am on a sunny day and included three treatments: (i) untreated ovaries, (ii) artificial pollination, and (iii) hormone treatment of unpollinated ovaries. For artificial pollination, pollen was removed from staminate flowers using a bamboo stick and then added to the stigmas of pistillate flowers at anthesis. Hormone treatment was also performed at anthesis, during which the whole pistillate flower was immersed for 2 s in a solution containing 10 mg L−1 CPPU (Sigma-Aldrich, MO, USA) and 20 mg L−1 GA3 (Sigma-Aldrich), both of which were dissolved in a solution containing 5% ethanol and 0.1% Tween 80. The plants were then covered with plastic food bags for three days. As a control (CK), untreated ovaries were covered with plastic bags for three days. Fruits were collected after different growth periods (0, 1, 3, 5, 10, 20, 30, 40, 50, 60 and 70 days after anthesis (DAA)). All samples were stored on dry ice and transported to the laboratory as soon as possible. After determining fruit weight, length (vertical diameter) and width (transverse diameter), the sample was instantly frozen in liquid nitrogen and stored at −80 °C for further analysis.2.2 Morphological and cytological observations
Fruit samples collected at 0, 1, 3, 5, and 10 DAA were fixed in a mixture of 70% ethanol, formaldehyde, and acetic acid (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 by volume). A 5 mm thick slice was then removed from the samples and embedded in paraffin, and transverse 9 μm-thick sections were prepared from the slices using a microtome (Leica Microsystems SM2500, Wetzlar, Germany). The sections were finally mounted, stained with toluidine blue, and photographed under a high-resolution digital microscope (Leica DM2500; Leica Microsystems GmbH, Wetzlar, Germany). Fruit and seed characteristics were characterized in both parthenocarpic and pollinated fruits. The fruit length and maximum diameter were estimated using an Absolute Digital Caliper (Mitutoyo, Tokyo, Japan). The weights of the whole fruit, pulp and seeds were measured using a precision balance (Mettler-Toledo Inc., Greifensee, Switzerland).
5 by volume). A 5 mm thick slice was then removed from the samples and embedded in paraffin, and transverse 9 μm-thick sections were prepared from the slices using a microtome (Leica Microsystems SM2500, Wetzlar, Germany). The sections were finally mounted, stained with toluidine blue, and photographed under a high-resolution digital microscope (Leica DM2500; Leica Microsystems GmbH, Wetzlar, Germany). Fruit and seed characteristics were characterized in both parthenocarpic and pollinated fruits. The fruit length and maximum diameter were estimated using an Absolute Digital Caliper (Mitutoyo, Tokyo, Japan). The weights of the whole fruit, pulp and seeds were measured using a precision balance (Mettler-Toledo Inc., Greifensee, Switzerland).
2.3 Chemical analysis of mogrosides
Three biological replicates of dry fruit samples were powdered to a homogeneous size (ca. 50 mesh) using a disintegrator. Approximately 0.5 g of powder from each sample was accurately weighed and placed in a 50 mL capped conical flask with 25 mL of a methanol (Fisher, IA, USA)/water solution (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, v/v). The flask was then sealed and sonicated in an ultrasonic water bath for 30 min at room temperature. A duplicate extract was prepared, and the two extracts were mixed and transferred to a volumetric flask, after which they were diluted to 100 mL in methanol/water (80
20, v/v). The flask was then sealed and sonicated in an ultrasonic water bath for 30 min at room temperature. A duplicate extract was prepared, and the two extracts were mixed and transferred to a volumetric flask, after which they were diluted to 100 mL in methanol/water (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, v/v) and filtered through a 0.22 μm microporous membrane.
20, v/v) and filtered through a 0.22 μm microporous membrane.
Next, 2 μL samples of the extracts were analyzed via LC-MS/MS using an Agilent Technologies 1260 Series LC system (Agilent, CA, USA) equipped with an automatic degasser, a quaternary pump, and an autosampler. Chromatographic separations were performed in Agilent Poroshell 120 SB C18 columns (100 mm × 2.1 mm, 2.7 μm) via gradient elution using a mobile phase consisting of (A) water (containing 0.1% formic acid) and (B) acetonitrile (Fisher), with the following gradient procedure: 0 min, 26% B; 5 min, 30% B; 5.01 to 5.50 min, 80% B; and 5.51 to 10.0 min, 26% B, employing a flow rate of 0.25 mL min−1.
The column effluent was monitored using a 4500 QTRAP® LC-MS/MS (AB Sciex, Toronto, Canada). Ionization was achieved through electrospray ionization (ESI) in negative ion mode using nitrogen as the nebulizer. Multiple reaction monitoring (MRM) scanning was employed for quantification. The source settings and instrument parameters for each MRM transition were optimized to not only maximize the generated deprotonated analyte molecule ([M−H]−) of each targeted mogroside but also efficiently produce its characteristic fragment/product ions. The electrospray voltage was set at −4500 V, and the source temperature was 500 °C. The curtain gas (CUR), nebulizer gas (GS1), and heater gas (GS2) were set at 15, 50 and 40 psi, respectively. The compound-dependent instrumental parameters of two individual precursor-to-product ion transitions specific for each analyte, including the precursor ion, two product ions, the declustering potential (DP), the entrance potential (EP), the collision energy (CE) and collision cell exit potentials (CXPs), were optimized and are listed in ESI Table S1.† The dwell time was 400 ms for each MRM transition. OMV, MV, IMV, SI, MIVA, MIV, and MIII standards (purchased from Chengdu Must Bio-Technology Co., Ltd., Sichuan, China) were also used for quantification.
2.4 RNA isolation and transcriptome analysis
Two biological replicates of samples of 0 and 3 DAA pollinated and hormone-treated ovaries were collected for RNA-Seq analysis. For each sample, six individual ovaries were ground into powder and mixed in liquid nitrogen. Total RNA was isolated from each sample using the TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer's instructions, as described by Tang et al.32 The purity and concentration of each RNA sample was then measured using an Agilent 2100 Bioanalyzer. The isolated RNA samples were sent to the Beijing Genomics Institute (Shenzhen, China) for RNA-Seq analysis using an Illumina HiSeqTM 2000. Raw sequences were filtered by removing 3′ adaptor fragments as well as low-quality sequences (tags with unknown sequence “N”), several types of impurities and sequences that were too long or too short. Transcripts larger than 200 bp were compared with sequences in protein databases, including NCBI Nr, Swiss-Prot, KEGG, and COG, using blastX. We then employed the Blast2GO program33 for GO annotation of the transcripts. A strict algorithm was utilized to identify the differentially expressed genes between different transcriptomes.34 The P-value could be employed to identify differences in expression between two libraries, and false discovery rates (FDRs) were used to determine the threshold of the P value. We set an FDR ≤ 0.001 and an absolute value of the log2 ratio ≥ 1 as thresholds to determine the significance of the differences in gene expression according to a previously described method.35 Functional analysis of DEGs, comparing pollinated and hormone-treated samples, was performed using Venny (http://bioinfogp.cnb.csic.es/tools/venny/).2.5 Validation of differential gene expression via qRT-PCR
As described for the RNA-Seq analysis, total RNA was employed for qRT-PCR. cDNA was obtained using a reverse transcription kit (TaKaRa, Dalian, China). qRT-PCR was then performed with the SYBR Premix Ex TaqTM kit (Takara) using an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) and the following amplification conditions: 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 95 °C for 5 s and 60 °C for 34 s. The 18S rRNA gene served as an endogenous control. The primer sequences of the genes are shown in ESI Table S2.† The qRT-PCR results were obtained from three repeated reactions for each gene and sample. A close correlation was observed between the relative expression levels measured via RNA-Seq and qRT-PCR, validating the RNA-Seq methodology described above for the quantitative analysis of the S. grosvenorii transcriptome.2.6 Statistical analysis
Data (mean ± SD) from various samples were subjected to one-way nested analysis of variance. Two-sample analysis was performed by student's t-test using SPSS statistics 22.0 (IBM, Chicago, IL, USA). SIMCA-P+12.0 (Umetrics AB, Sweden) was employed to perform principal component analysis (PCA) of the results of chemical analysis.3. Results
3.1 Morphological and cytological observation of S. grosvenorii fruits
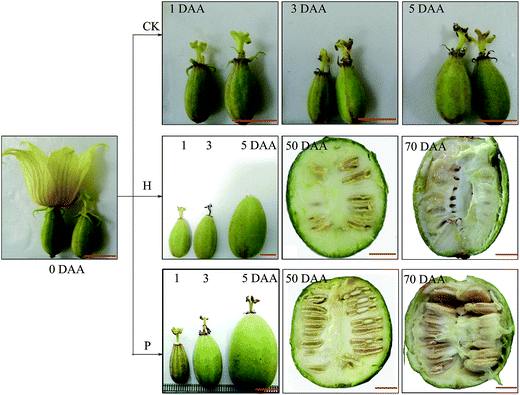
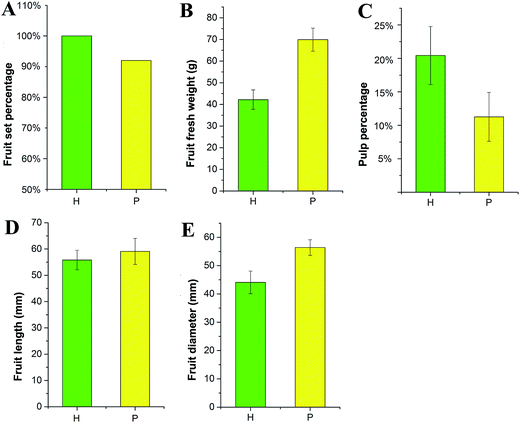
The morphological changes in S. grosvenorii ovaries that were left untreated (CK) or were pollinated (P) or hormone-treated (H) were observed from 0 to 70 DAA. Untreated ovaries did not grow after anthesis because all of the ovaries failed to set and eventually damped off (Fig. 1). Pollination resulted in gradual growth of the ovary, with a fruit set rate of 92% (n = 200) and fruit fresh weight (FW) of approximately 69 g (Fig. 2B). Interestingly, the application of hormones to unpollinated flowers induced parthenocarpic growth and the production of fruits that were similar to those of the pollinated flowers. The fruit set rate and fruit fresh weight were 100% (n = 200) and approximately 42 g, respectively. Furthermore, the lengths of the empty seeds in the parthenocarpic fruits were significantly shorter than in pollinated ovaries, whereas pollinated ovaries produced normal-sized and hard seeds that were able to germinate. The weights of the parthenocarpic fruits were lower than those of pollinated fruits, as observed for other plants. Remarkably, the pulp percentage of the parthenocarpic fruits was higher than that of pollinated fruits (Fig. 2C).Ovule development and pulp cells in S. grosvenorii fruits were observed via optical microscopy using a paraffin method (ESI Fig. S1†). Cytological observation revealed that the funiculus was connected to the placenta funicle; the nucellus was filled with parenchyma; and the micropyle was not closed in 0 DAA ovary samples (ESI Fig. S1A†). In fruitlets derived from induced parthenocarpy, embryo sac atrophy was apparent, with separation from the inner integument at 3 DAA, whereas there was a clear increase in pulp cells (ESI Fig. S1C, c†). In the ovules, center cell shrinkage and the formation of a hole in the center of the embryo sac were observed at 5 DAA, and the pulp cells continued to rapidly increase at 5–10 DAA. The integument also rapidly increased, and the embryo showed no further development at 10 DAA.
In contrast, the embryo sac began to grow after pollination, and the embryo and pulp cells gradually increased at 3 DAA. The outer shell composed of palisade cells was formed by the outer integument at 5 DAA, while the inner shell was formed by inner integuments; the germ cells were uniform and compact. The outer shell was thicker, while the inner shell was thinner at 10 DAA. The germinal center was dense and developing normally. Concomitantly, the pulp cells rapidly increased (ESI Fig. S1I, i†).
3.2 Quantitative analysis of mogrosides in S. grosvenorii fruits
Mogrosides, a group of triterpenoid glycosides isolated from S. grosvenorii fruits, are regarded as the main ingredients supplying the sweet taste and medicinal activity of these fruits. In this study, the developed LC-ESI-MS/MS method was applied to determine the content of seven mogrosides in different stages of S. grosvenorii fruits. Typical LC-MS/MS chromatograms of standard solutions and fruit samples obtained in MRM mode (ESI Fig. S2†). Seven mogrosides were identified through comparison of their retention times and their precursor and product ions. Quantitative analyses were performed using external standard methods, and the results are summarized in ESI Table S3† and via graphical representation (Fig. 3), showing the content of each targeted mogroside and the total content of all mogrosides.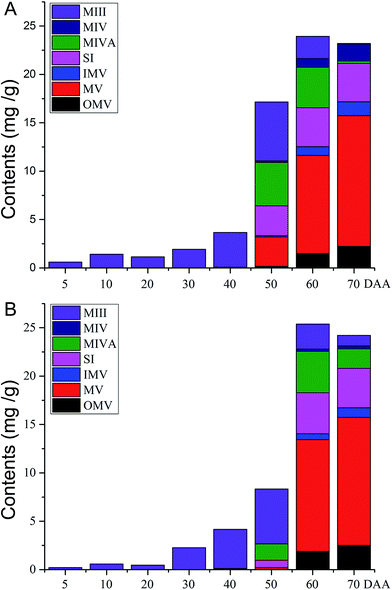 | ||
| Fig. 3 Graphical representation of the content of each mogroside and the total content in different fruit samples. (A) Samples of pollinated and (B) hormone-treated ovaries. | ||
The results showed that in all fruit samples, the mogroside contents exhibited marked differences in different growth stages. The PCA score plot (ESI Fig. S3†) revealed a clear separation of different groups of S. grosvenorii samples, indicating that the chemical compositions determined in various periods in samples subjected to different treatments were altered in the different growth stages. Unripe fruit (0–40 DAA) consisted of mainly MIII. With a longer growing time (50–70 DAA), other types of mogrosides were observed, such as MV, MIVA, MIV, SI, OMV and IMV. The major component of the ripe fruits was MV, and the highest accumulation of MV was observed at 70 DAA. This result demonstrated that the MV content significantly increased, while the level of MIII dramatically decreased with a longer growing time. The content of MV accounted for approximately 55% of the total content of seven mogrosides at 70 DAA.
Regarding the difference between pollinated and parthenocarpic fruits, we found that the contents of the seven targeted mogrosides in fruits in different growth stages were not significantly different (Fig. 3). However, the pollinated fruits exhibited high concentrations of MV, MIVA and SI at 50 DAA, compared with relatively low concentrations in parthenocarpic fruits. In addition, the contents of MIII, MIVA and MIV measured at 70 DAA differed between pollinated and parthenocarpic fruits. In general, the differences in contents between pollinated and parthenocarpic fruits were comparatively minor. Based on the current results, we suggest that pollinated and parthenocarpic fruits exhibit similar quality in terms of their main components.
3.3 Transcriptome analysis of S. grosvenorii fruits
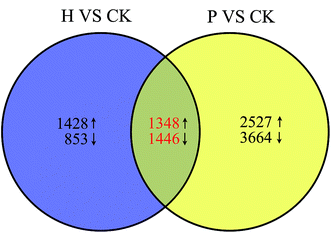
To characterize the developmental processes that were altered in parthenocarpic fruits and identify candidate genes responsible for parthenocarpy, transcriptomic comparisons were conducted between pollinated and parthenocarpic fruits. Based on the morphological and cytological observations, we found that S. grosvenorii ovaries exhibited critical developmental changes from 0 to 3 DAA. Thus, S. grosvenorii fruits (CK at 0 DAA and pollinated and parthenocarpic at 3 DAA) were harvested for transcriptome analyses.Genes showing differential expression in setting fruits were filtered according to the CK transcriptome (ESI Table S4†). In multiple tests, the false discovery rate (FDR) method was used to determine the P-value threshold, which corresponded to differential gene expression. Venn diagram analysis was used to screen parthenocarpy-specific gene expression (Fig. 4). An FDR ≤ 0.001 and an absolute value of |log2 ratio ≥ 1.0| were used as the thresholds for determining DEGs. To identify common characteristics and differences between the pollinated and parthenocarpic fruit sets, we compared DEGs based on CK. The results showed that 1348 up-regulated and 1446 down-regulated genes were common DEGs in the ovaries after pollination and hormone treatment, respectively (Fig. 4). These DEGs were considered to be key regulators of fruit setting and growth. Moreover, 2527 up-regulated and 3664 down-regulated genes displayed differential expression in the ovaries of pollinated plants, indicating a correlation between these genes and seed formation. There were 1428 up-regulated and 853 down-regulated genes that were only differentially expressed in the parthenocarpic fruits set, which were defined as parthenocarpy-specific genes. To validate the reliability of the RNA-Seq data, 12 phytohormone-related genes were selected for qRT-PCR analysis (ESI Table S2†). A good correlation (R = 0.9087) between the two methods was obtained (ESI Fig. S4†), supporting the high reliability of the RNA-Seq data obtained in this study.
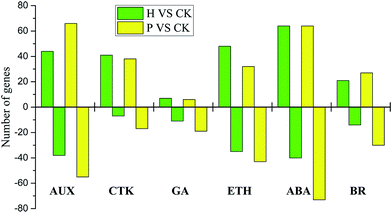
Auxin-related genes, which are well-established regulators of fruit setting, displayed dramatic changes in expression during pollinated and parthenocarpic fruit setting. Among these genes, the auxin influx carrier (AUX1/LAX), indole-3-acetaldehyde oxidase (AAO1), indole-3-pyruvate monooxygenase (YUCCA), auxin transporter-like protein (LOB) and ABC transporter B (ABCB) genes were up-regulated after pollination, whereas the MADS-box protein (MADS-box) and auxin response factor (ARF) genes were down-regulated, and the auxin-responsive protein (AUX/IAA) and small auxin-up RNA (SAUR) genes were both up- and down-regulated. The up-/down-regulated gene ratios were 8/6 and 6/4, respectively (ESI Fig. S5†). The expression analyses of genes assigned to GA regulation showed that the transcription of genes encoding gibberellin 3-beta-dioxygenase (GA3ox), DELLA protein (DELLA) and ent-kaurenoic acid hydroxylase (KAO1) was up-regulated following pollination. In contrast, the gibberellin receptor GID1 (GID1) and ent-kaurene oxidase (KO) genes were down-regulated; and 1 gibberellin 20-oxidase (GA20ox) gene was up-regulated, while 4 were down-regulated. Moreover, the expression of cytokinin genes, including adenylate isopentenyltransferase (IPT), cytokinin hydroxylase (CYP735A) and the two-component response regulator ARR-A (A-ARR), were increased, while the two-component response regulator ARR-B (B-ARR), cytokinin dehydrogenase (CKX) and histidine-containing phosphotransfer protein (AHP) were decreased. Five glucan endo-1,3-beta-glucosidase-like protein (CRE1) genes were up-regulated, and 3 were down-regulated. Ethylene plays a potential role in fruit setting. The RNA-Seq data revealed a decrease in the mRNA levels of the ethylene-related genes aminocyclopropanecarboxylate oxidase (ACO1), serine/threonine-protein kinase (CTR1), and ethylene-responsive transcription factor 1 (ERF1), whereas the expression of the ethylene-insensitive protein 2 (EIN2), ethylene receptor (ETR), and GDSL esterase (GDSL) genes were increased. Two ethylene-responsive transcription factor 1 (ERF1) genes were up-regulated, and 3 were down-regulated. In addition, the results for BR-related genes showed that the transcription of genes encoding brassinosteroid-6-oxidase (CYP85A1) was dramatically up-regulated after pollination, whereas the BR-signaling kinase (BSK) and brassinosteroid-resistant 1/2 (BZR1/2) genes were down-regulated. Four brassinosteroid-insensitive 1-associated receptor kinase 1 (BAK1) genes were up-regulated, and 2 were down-regulated; while 10 protein brassinosteroid-insensitive 1 (BRI1) genes were up-regulated, and 6 were down-regulated in the pollinated samples.
A similar expression pattern of genes including YUCCA, AUX/IAA, MADS-box, ARF, SAUR, GA3ox, DELLA, GID1, CYP735A, GDSL, CYP85A1, BAK1 and BRI1 was observed in the hormone-treated samples, potentially clarifying how endogenous hormones can stimulate fruit setting. However, some genes exhibited the opposite expression profile to that seen in the pollinated samples. Remarkably, the AUX1/LAX, ABCB, KAO1, and indole-3-acetic acid-amido synthetase (GH3) genes were markedly down-regulated during fruit setting, whereas CKX genes were largely up-regulated. Furthermore, analysis of the number of known genes related to phytohormones during fruit setting revealed that only approximately 1/3 of the genes were commonly expressed, while 2/3 were expressed in a pollination-specific or parthenocarpy-specific manner (ESI Table S5†).
4. Discussion
It is generally accepted that fruit setting and associated cell division occur as a result of the coordinated action of phytohormones produced in the ovary after pollination and/or fertilization.30 At present, researchers are increasingly focusing on the parthenocarpic fruit setting. It has been found that parthenocarpic growth can be induced by auxins, GAs or cytokinins.24,26,36,37 However, the regulatory mechanisms associated with parthenocarpic fruit setting remain unknown. The aims of the present study were to observe the changes that occur during ovary development after fertilization and hormone application (including CPPU and GA3) and to identify common characteristics and differences between pollinated and parthenocarpic fruit sets.As shown in Fig. 1, the unpollinated ovaries of S. grosvenorii grew parthenocarpically in response to the application of GA3 and CPPU. The external shape of the parthenocarpic fruits was similar to that of pollinated fruits. The empty seeds in the parthenocarpic fruits were smaller and less lignified compared with those in the pollinated fruits. Furthermore, the percentage of pulp was higher in parthenocarpic fruits than in pollinated fruits. Nevertheless, the weight and size of parthenocarpic fruits were lower than those of pollinated fruits, as observed in other plants.38,39 Optical microscopy revealed a difference in the seeds between pollinated and parthenocarpic fruits at 1 DAA (ESI Fig. S1B, F†). The exterior integument had developed as the testa, but the embryo showed almost no growth in the parthenocarpic fruits. As a result, the seeds were empty and underdeveloped. Fruit development is usually triggered by ovule fertilization, which requires coordination between seed development and the growth and differentiation of the ovary to host seeds.40 Crane41 noted that the seed is the center of auxin synthesis and that auxin moves outward and stimulates fruit growth. Dorcey42 showed that fertilization triggers an increase in the auxin response in ovules and induces subsequent activation of GA metabolism, specifically in young seeds. Furthermore, GAs synthesized in seeds are translocated to the pod to promote cell expansion and other differentiation processes. Therefore, the relationship between seed and fruit development merits a thorough analysis to improve the quality of S. grosvenorii fruits in the future.
In ripe fruits of S. grosvenorii, the most important compounds present in the fruit pulp are MV, MIV, SI and MVI.43–45 Using the LC-MS/MS method developed in this study, we simultaneously determined the contents of MIII, MIVA, MIV, MV, IMV, OMV, and SI during different growth stages of pollinated and parthenocarpic fruits. It is noteworthy that the content at 50 DAA was significantly different (P < 0.05) between pollinated and parthenocarpic fruits. Other types of mogrosides, such as MIVA, MV and SI, could also be detected, but the content in pollinated fruits was higher than in parthenocarpic fruits at this stage. This difference may be associated with the activity of related enzymes during this period. Fortuitously, the total content of the targeted mogrosides were not significantly different (P > 0.05) between pollinated and parthenocarpic fruits at 70 DAA.
According to the observed morphological and chemical characteristics, we suggest that the potential for parthenocarpic fruit induced by exogenous hormones to replace artificial pollination for the production of commercial S. grosvenorii should be evaluated. The possibility of the commercial application of exogenous hormones to induce parthenocarpic fruits has been discussed previously.46–48 However, few studies have explored the potential application of exogenous hormones as a replacement for artificial pollination. The present results clearly demonstrated that the application of hormones at anthesis resulted in greater fruit setting and a higher pulp percentage. In addition, the parthenocarpic fruits induced by exogenous hormones accumulated high concentrations of mogrosides, similar to the levels in artificially pollinated fruits. However, the small fruit size produced must be addressed to identify measures for increasing yields. Thus, the related genes during the process of fruit growth were observed to explore the cause of the small fruit size resulting from induced parthenocarpy.
Global transcriptome profiling showed that numerous genes were differentially expressed in developing ovaries in response to different treatments, among which a large number of genes involved in hormone biosynthesis and signaling were found to be altered during fruit setting. The capacity of auxins, cytokinins and GAs to induce parthenocarpic fruit setting in plants has been well documented.24,26,49,50 Our analyses showed that auxin plays a central role in promoting fruit growth. Genes that are associated with auxin, including YUCCA, LAX, ABAB, AUX/IAA, LOB, ARF, SAUR, GH3, MADS-box and AAO1 being identified in the transcriptome. Auxin is a key regulator of plant development. It influences cell division, cell elongation and programmed cell death, driving embryonic and post-embryonic development.51 To a certain extent, auxin functions by regulating a group of primary responsive genes: Aux/IAA, GH3 and SAUR.52 The auxin burst induced by pollination is the first positive signal in fruit setting, and the functional interaction between the Aux/IAA and ARF proteins is one of the best-characterized components of this mechanism.53 The presence of IAA promotes Aux/IAA protein ubiquitination and degradation; thus, ARFs are released from the repressive effect, and auxin response genes are ultimately activated.54 GH3 genes encode IAA-amido synthetases, which help to maintain auxin homeostasis by conjugating excess IAA to amino acids, thereby regulating plant growth, development and defense responses.55–57 Recent studies have proposed that SAUR proteins are able to modulate auxin transport and cell expansion via an unknown mechanism.58,59 MADS-box genes encode a family of transcription factors that control diverse developmental processes in flowering plants, in organs ranging from roots to flowers and fruits.60 In addition, the YUCCA, AAO1, ABCB and LAX genes have been reported to play important roles in IAA synthesis or transport.61–63 After the application of hormones, the expression of the major AUX/IAA genes was strongly down-regulated; ARFs were released; and transcription was activated to increase the effect of auxin. Additionally, the YUCCA, GH3, MADS-box, LAX and ABCB genes jointly maintained the dynamic balance of the IAA concentration in fruitlets (ESI Table S5†), ultimately resulting the formation of parthenocarpic fruit. In addition to the similar expression of the genes observed in parthenocarpic fruits, AAO1 and ABCB were strongly up-regulated after pollination.
GAs constitute a group of plant hormones that control developmental processes such as germination, shoot elongation, tuber formation, flowering, and fruit setting and growth in diverse species.64,65 The gene expression analyses conducted in the present study showed that the transcription of the genes encoding the GA20ox, GA2ox, GA3ox, GID1, DELLA, KAO1, and KS enzymes in the GA biosynthetic and signaling pathways was altered by pollination and hormone treatment (ESI Table S5†). GA20ox and GA3ox are crucial enzymes for the control of active GAs through two parallel pathways, whereas active GAs and their precursors can be irreversibly inactivated by GA2ox via the introduction of a hydroxyl at the 2β position.50,66 In Arabidopsis, the biosynthetic enzymes GA20ox and GA3ox are required for silique elongation, and increasing GA levels through genetic engineering led to the formation of parthenocarpic fruits.67 The biosynthesis of GAs in plants can be divided into seven steps, and KS and KAO are key enzymes in different steps of gibberellin biosynthesis.68,69 Endogenous GA is perceived by GA receptors in the GID1 family, which was first identified in rice.70 Three GID1-type GA receptors in Arabidopsis are known: AtGID1a, AtGID1b, and AtGID1c. After binding to its receptor, the GA-GID1 complex interacts with DELLA proteins, which are negative regulators of the GA signaling pathway.71,72 The increase in GA contents in parthenocarpic fruits likely resulted from the effect of this hormone on increasing the transcription of the genes encoding GA20ox, GA3ox and GA2ox. The increase in GA2ox expression may be a result of positive feedback regulation in response to increased GAs levels.73,74 The transcripts of KAO and GID1 were down-regulated, while DELLA was up-regulated in parthenocarpic fruits. In comparison with parthenocarpic fruits, pollinated fruits exhibited altered GA metabolism, characterized by up-regulation of KAO and GA3ox and a down-regulation of GA20ox and GA2ox gene transcription. In summary, we have shown that parthenocarpic fruit setting is mediated, at least in part, by GAs. Thus, the increase in active GA contents in the fruit via the up-regulation of genes encoding enzymes involved in GA biosynthesis (GA20ox and GA3ox) induces fruit setting.
In addition to GAs and auxins, other phytohormones, such as cytokinins and BRs, have also been shown to play roles in fruit setting and subsequent fruit growth.27,75 The expression of the cytokinin synthesis enzymes CKX, IPT and CYP735A were up-regulated (ESI Table S5†). Moreover, the expression level of A-ARR was increased, resulting in an increase in cytokinins and, consequently, constitutive cell division. Furthermore, the content of BR increased with the up-regulation of the expression of the BR synthesis genes CYP85A, BRI1 and BAK1, which induced fruit cell division and expansion. These processes affected the formation and development of the parthenocarpic fruit.
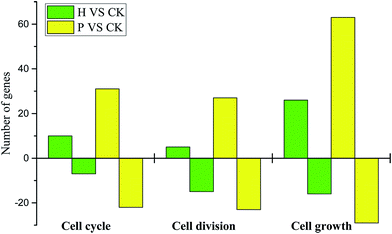
Briefly, a similar expression pattern of hormone-related genes was observed between pollinated and parthenocarpic fruits. The regulation of key genes in a similar pattern to that following pollination promoted the formation and development of parthenocarpic fruits. However, the process by which the ovary becomes a fruit indicates the involvement of complex gene regulation and expression. Thus, the application of exogenous hormones induced parthenocarpic fruits by regulating the expression of specific genes, but this process was unable to completely replace pollination by inducing all of the same changes. Furthermore, there were more cell cycle-, division- and growth-related DEGs in the pollinated fruits, which could explain why the growth rate and size of the pollinated fruits exceeded those of parthenocarpic fruits.
5. Conclusions
This study represents an integrative approach towards understanding the development of parthenocarpy in S. grosvenorii. Parthenocarpy provides an alternative and economical approach to achieve high yields and seedless S. grosvenorii fruits. The transcriptomic analyses revealed a similar expression pattern of key genes between pollinated and parthenocarpic fruits. Our study provides basic data on the biological regulation of fruit setting in S. grosvenorii. In the further work, genetic engineering would be employed to breed parthenocarpic cultivars of S. grosvenorii, which is coupled with our transcriptome data to elucidate the mechanism of hormone-induced parthenocarpic fruits.Abbreviations
| CPPU | 1-(2-Chloro-4-pyridyl)-3-phenylurea |
| cDNA | Complementary DNA |
| DEG | Differentially expressed gene |
| DAA | Days after anthesis |
| GA3 | Gibberellin A3 |
| GO | Gene ontology |
| GRAS | Generally recognized as safe |
| IMV | iso-Mogroside V |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LHG | Luo Han Guo |
| MRM | Multiple reaction monitoring |
| MIII | Mogroside III |
| MIVA | Mogroside IVa |
| MIV | Mogroside IV |
| MV | Mogroside V |
| NR | Non-redundant protein database |
| NT | Nucleotide database |
| OMV | 11-Oxomogroside-V |
| PCA | Principal component analysis |
| qRT-PCR | Real-time reverse transcription PCR |
| RNA-Seq | RNA sequencing |
| SI | Siamenoside I |
Acknowledgements
We acknowledge the Beijing Genomics Institute at Shenzhen for assistance with the original data processing and related bioinformatics analysis. We are grateful to all of the faculty involved at the Guangxi Botanical Garden of Medicinal Plants (Nanning, China) for their assistance with the sampling. This work was supported by the National Natural Science Foundation of China (Grant No. 81373914 & 81573521) and the Natural Science Foundation of Guangxi Province in China (Grant No. 2013GXNSFAA019084).References
- M. Takasaki, T. Konoshima, Y. Murata, M. Sugiura, H. Nishino, H. Tokuda, K. Matsumoto, R. Kasai and K. Yamasaki, Cancer Lett., 2003, 198, 37–42 CrossRef CAS PubMed.
- C. Liu, L. H. Dai, D. Q. Dou, L. Q. Ma and Y. X. Sun, Oncogenesis, 2016, 5, e217 CrossRef CAS PubMed.
- Y. A. Suzuki, Y. Murata, H. Inui, M. Sugiura and Y. Nakano, J. Agric. Food Chem., 2005, 53, 2941–2946 CrossRef CAS PubMed.
- Z. L. Luo, F. Qiu, K. L. Zhang, X. J. Qin, Y. H. Guo, H. W. Shi, L. X. Zhang, Z. L. Zhang and X. J. Ma, RSC Adv., 2016, 6, 7034–7041 RSC.
- X. Y. Qi, W. J. Chen, L. Q. Zhang and B. J. Xie, Nutr. Res., 2008, 28, 278–284 CrossRef CAS PubMed.
- Y. A. Suzuki, M. Tomoda, Y. Murata, H. Inui, M. Sugiura and Y. Nakano, Br. J. Nutr., 2007, 97, 770–775 CrossRef CAS PubMed.
- X. Y. Qi, W. J. Chen, L. G. Liu, P. Yao and B. J. Xie, Mol. Nutr. Food Res., 2006, 50, 732–738 CAS.
- F. F. Song, W. J. Chen, W. B. Jia, P. Yao, A. K. Nussler, X. F. Sun and L. G. Liu, Phytother. Res., 2006, 20, 552–560 CrossRef PubMed.
- M. H. Pan, J. R. Yang, M. L. Tsai, S. Sang and C. T. Ho, J. Funct. Foods, 2009, 1, 145–152 CrossRef.
- W. J. Chen, J. Wang, X. Y. Qi and B. J. Xie, Int. J. Food Sci. Nutr., 2007, 58, 548–556 CrossRef CAS PubMed.
- C. Li, L. M. Lin, F. Sui, Z. M. Wang, H. R. Huo, L. Dai and T. L. Jiang, Chin. J. Nat. Med., 2014, 12, 89–102 CAS.
- X. Yan, M. E. Rivero-Huguet, B. H. Hughes and W. D. Marshall, Food Chem., 2008, 107, 1022–1028 CrossRef.
- R. N. Philippe, M. D. Mey, J. Anderson and P. K. Ajikumar, Curr. Opin. Biotechnol., 2014, 26, 155–161 CrossRef CAS PubMed.
- W. Jakinovich Jr, C. Moon, Y. H. Choi and A. D. Kinghorn, J. Nat. Prod., 1990, 53, 190–195 CrossRef CAS.
- R. S. Pawar, A. J. Krynitsky and J. I. Rader, Anal. Bioanal. Chem., 2013, 405, 4397–4407 CrossRef CAS PubMed.
- J. Grebow, Stevia and monk fruit: what makes a natural sweetener natural? Data from: Nutritional Outlook, 2016, http://www.nutritionaloutlook.com/food-beverage/stevia-and-monk-fruit-what-makes-natural-sweetener-natural Search PubMed.
- A. Srivastava and A. K. Handa, J. Plant Growth Regul., 2005, 24, 67–82 CrossRef CAS.
- J. C. Serrani, E. Carrera, O. Ruiz-Rivero, L. Gallego-Giraldo, L. E. Peres and J. L. García-Martínez, Plant Physiol., 2010, 153, 851–862 CrossRef CAS PubMed.
- Y. L. Ruan, J. W. Patrick, M. Bouzayen, S. Osorio and A. R. Fernie, Trends Plant Sci., 2012, 17, 656–665 CrossRef CAS PubMed.
- T. Ariizumi, Y. Shinozaki and H. Ezura, Breed. Sci., 2013, 63, 3–13 CrossRef CAS PubMed.
- G. Gillaspy, H. Ben-David and W. Gruissem, Plant Cell, 1993, 5, 1439–1451 CrossRef PubMed.
- H. Wang, N. Schauer, B. Usadel, P. Frasse, M. Zouine, M. Hernould, A. Latché, J. C. Pech, A. R. Fernie and M. S. Bouzayen, Plant Cell, 2009, 21, 1428–1452 CrossRef CAS PubMed.
- J. Li, Z. Wu, L. Cui, T. L. Zhang, Q. W. Guo, J. Xu, L. Jia, Q. F. Lou, S. W. Huang, Z. G. Li and J. F. Chen, Plant Cell Physiol., 2014, 55, 1325–1342 CrossRef CAS PubMed.
- J. C. Serrani, O. Ruiz-Rivero, M. Fos and J. L. García-Martínez, Plant J., 2008, 56, 922–934 CrossRef CAS PubMed.
- C. Mesejo, C. Reig, A. Martínez-Fuentes and M. Agustí, Sci. Hortic., 2010, 126, 37–41 CrossRef CAS.
- J. G. Ding, B. W. Chen, X. J. Xia, W. H. Mao, K. Shi, Y. H. Zhou and J. Q. Yu, PLoS One, 2013, 8, e70080 CAS.
- F. Q. Fu, W. H. Mao, K. Shi, Y. H. Zhou, T. Asami and J. Q. Yu, J. Exp. Bot., 2008, 59, 2299–2308 CrossRef CAS PubMed.
- M. Koiwai, F. Okuyama, K. I. Tanaka, A. Yamazaki, H. Honma, K. Ikeda and S. Taira, Hortic. Res., 2012, 11, 87–95 CrossRef.
- J. V. Maroto, A. Miguel, S. Lopez-Galarza, A. S. Bautista, B. Pascual, J. Alagarda and J. L. Guardiola, Plant Growth Regul., 2005, 45, 209–213 CrossRef CAS.
- B. Gorguet, A. W. Heusden and P. Lindhout, Plant Biol., 2005, 7, 131–139 CrossRef CAS PubMed.
- L. J. Chai, Y. M. Li, S. W. Chen, A. Perl, F. X. Zhao and H. Q. Ma, Plant Sci., 2014, 229, 215–224 CrossRef CAS PubMed.
- Q. Tang, X. J. Ma, C. M. Mo, I. W. Wilson, C. Song, H. Zhao, Y. F. Yang, W. Fu and D. Y. Qiu, BMC genomics, 2011, 12, 343 CrossRef CAS PubMed.
- A. Conesa, S. Götz, J. M. García-Gómez, J. Terol, M. Talón and M. Robles, Bioinformatics, 2005, 21, 3674–3676 CrossRef CAS PubMed.
- S. Audic and J. M. Claverie, Genome Res., 1997, 7, 986–995 CAS.
- Y. Benjamini and D. Yekutieli, Ann. Stat., 2001, 29, 1165–1188 CrossRef.
- N. Tang, W. Deng, G. J. Hu, N. Hu and Z. G. Li, PLoS One, 2015, 10, e0125355 Search PubMed.
- C. Mesejo, R. Yuste, C. Reig, A. Martínez-Fuentes, D. J. Iglesias, N. Muñoz-Fambuena, A. Bermejo, M. A. Germanà, E. Primo-Millo and M. Agustí, Plant Sci., 2016, 247, 13–24 CrossRef CAS PubMed.
- C. Royo, P. Carbonell-bejerano, R. Torres-pérez, A. Nebish, Ó. Martínez, M. Rey, R. Aroutiounian, J. Ibáñez and J. M. Martínez-zapater, J. Exp. Bot., 2015, 67, 259–273 CrossRef PubMed.
- C. Martí, D. Orzáez, P. Ellul, V. Moreno, J. Carbonell and A. Granell, Plant J., 2007, 52, 865–876 CrossRef PubMed.
- M. Sotelo-Silveira, N. Marsch-Martínez and S. D. Folter, Planta, 2014, 239, 1147–1158 CrossRef CAS PubMed.
- J. C. Crane, Annu. Rev. Plant Physiol., 1964, 15, 303–326 CrossRef CAS.
- E. Dorcey, C. Urbez, M. A. Blázquez, J. Carbonell and M. A. Perez-Amador, Plant J., 2009, 58, 318–332 CrossRef CAS PubMed.
- H. Y. Zhang, H. H. Yang, M. Zhang, Y. R. Wang, J. R. Wang, L. F. Yau, Z. H. Jiang and P. Hu, J. Food Compos. Anal., 2012, 25, 142–148 CrossRef CAS.
- F. L. Lu, D. P. Li, C. M. Fu, J. L. Liu, Y. L. Huang, Y. Y. Chen, Y. X. Wen and T. Nohara, J. Nat. Med., 2012, 66, 70–76 CrossRef CAS PubMed.
- Y. Shen, S. J. Lin, C. Han, Z. O. Zhu, X. D. Hou, Z. Long and K. L. Xu, Microchem. J., 2014, 116, 142–150 CrossRef CAS.
- V. V. Yarushnykov and M. M. Blanke, Plant Growth Regul., 2005, 45, 21–27 CrossRef CAS.
- C. X. Zhang, U. Lee and K. Tanabe, Plant Growth Regul., 2008, 55, 231–240 CrossRef CAS.
- Q. F. Niu, T. Wang, J. Z. Li, Q. Q. Yang, M. J. Qian and Y. W. Teng, Plant Growth Regul., 2015, 76, 251–258 CrossRef CAS.
- L. Mariotti, P. Picciarelli, L. Lombardi and N. Ceccarelli, J. Plant Growth Regul., 2011, 30, 405–415 CrossRef CAS.
- J. C. Serrani, R. Sanjuán, O. Ruiz-rivero, M. Fos and J. L. García-martínez, Plant Physiol., 2007, 145, 246–257 CrossRef CAS PubMed.
- P. J. Davies, Biosynthesis, signal transduction, action!, Plant hormones, Springer, Netherlands, 2010, pp. 1–802, DOI:10.1007/978-1-4020-2686-7.
- G. Hagen and T. Guilfoyle, Plant Mol. Biol., 2002, 49, 373–385 CrossRef CAS PubMed.
- F. Ruiu, M. E. Picarella, S. Imanishi and A. Mazzucato, Plant Mol. Biol., 2015, 89, 263–278 CrossRef CAS PubMed.
- X. Tan, L. I. Calderon-Villalobos, M. Sharon, C. X. Zheng, C. V. Robinson, M. Estelle and N. Zheng, Nature, 2007, 446, 640–645 CrossRef CAS PubMed.
- J. E. Park, J. Y. Park, Y. S. Kim, P. E. Staswick, J. Jin, J. Yun, S. Y. Kim, J. Kim, Y. H. Lee and C. M. Park, J. Biol. Chem., 2007, 282, 10036–10046 CrossRef CAS PubMed.
- P. E. Staswick, B. Serban, M. Rowe, I. Tiryaki, M. T. Maldonado, M. C. Maldonado and W. Suza, Plant Cell, 2005, 17, 616–627 CrossRef CAS PubMed.
- J. H. Yang, S. J. Han, E. K. Yoon and W. S. Lee, Nucleic Acids Res., 2006, 34, 1892–1899 CrossRef CAS PubMed.
- T. Qiu, Y. Chen, M. M. Li, Y. Y. Kong, Y. B. Zhu, N. Han, H. W. Bian, M. Y. Zhu and J. H. Wang, Plant Signaling Behav., 2013, 8, e25283 CrossRef PubMed.
- N. Dharmasiri, Functions of SAUR genes in auxin response, 2007, https://digital.library.txstate.edu/handle/10877/2780 Search PubMed.
- Z. B. Zhang, H. Y. Li, D. F. Zhang, Y. H. Liu, J. Fu, Y. S. Shi, Y. C. Song, T. Y. Wang and Y. Li, J. Plant Physiol., 2012, 169, 797–806 CrossRef CAS PubMed.
- Y. Yamamoto, N. Kamiya, Y. Morinaka, M. Matsuoka and T. Sazuka, Plant Physiol., 2007, 143, 1362–1371 CrossRef CAS PubMed.
- S. Pollmann, A. Müller and E. W. Weiler, Plant Biol., 2006, 8, 326–333 CrossRef CAS PubMed.
- B. Péret, K. Swarup, A. Ferguson, M. Seth, Y. D. Yang, S. Dhondt, N. James, I. Casimiro, P. Perry, A. Syed, H. B. Yang, J. Reemmer, E. Venison, C. Howells, M. A. Perez-Amador, J. Yun, J. Alonso, G. T. Beemster, L. Laplaze, A. Murphy, M. J. Bennett, E. Nielsen and R. Swarup, Plant Cell, 2012, 24, 2874–2885 CrossRef PubMed.
- P. Hedden and Y. Kamiya, Annu. Rev. Plant Physiol. Plant Mol. Biol., 2003, 48, 431–460 CrossRef PubMed.
- N. Olszewski, T. P. Sun and F. Gubler, Plant Cell, 2002, 14, S61–S80 CAS.
- C. Dijkstra, E. Adams, A. Bhattacharya, A. F. Page, P. Anthony, S. Kourmpetli, J. B. Power, K. C. Lowe, S. G. Thomas and P. Hedden, Plant Cell Rep., 2008, 27, 463–470 CrossRef CAS PubMed.
- I. Rieu, O. Ruiz-Rivero, N. Fernandez-Garcia, J. Griffiths, S. J. Powers, F. Gong, T. Linhartova, S. Eriksson, O. Nilsson and S. G. Thomas, Plant J., 2008, 53, 488–504 CrossRef CAS PubMed.
- J. Song, B. J. Guo, F. W. Song, H. R. Peng, Y. Y. Yao, Y. R. Zhang, Q. X. Sun and Z. F. Ni, Gene, 2011, 482, 34–42 CrossRef CAS PubMed.
- H. Aach, H. Bode, D. G. Robinson and J. E. Graebe, Planta, 1997, 202, 211–219 CrossRef CAS.
- M. Ueguchi-Tanaka, M. Ashikari, M. Nakajima, H. Itoh, E. Katoh, M. Kobayashi, T. Chow, Y. C. Hsing, H. Kitano, I. Yamaguchi and M. Matsuoka, Nature, 2005, 437, 693–698 CrossRef CAS PubMed.
- D. E. Richards, K. E. King, T. Ait-Ali and N. P. Harberd, Annu. Rev. Plant Physiol. Plant Mol. Biol., 2001, 52, 67–88 CrossRef CAS PubMed.
- A. Voegele, A. Linkies, K. Müller and G. Leubner-metzger, J. Exp. Bot., 2011, 62, 5131–5147 CrossRef CAS PubMed.
- P. Hedden and A. L. Phillips, Trends Plant Sci., 2000, 5, 523–530 CrossRef CAS PubMed.
- M. Fos, K. Proaño, F. Nuez and J. L. García-Martínez, Physiol. Plant., 2001, 111, 545–550 CrossRef CAS PubMed.
- S. J. Trueman, Afr. J. Agric. Res., 2011, 5, 3402–3407 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra28341a |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |