Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Identification of DNA single-base mismatches by resistivity of poly(N-isopropylacrylamide)-block-ssDNA copolymer brush films at dual temperatures†
Yi-Zu Liua,
Karthikeyan Manivannana,
Ai-Wei Leeb,
Yan-Jiun Huangc,
Po-Li Weid and
Jem-Kun Chen *a
*a
aDepartment of Materials Science and Engineering, National Taiwan University of Science and Technology, 43, Sec 4, Keelung Rd, Taipei, 106, Taiwan, Republic of China. E-mail: jkchen@mail.ntust.edu.tw; Fax: +886-2-27376544; Tel: +886-2-27376523
bDepartment of Anatomy and Cell Biology, School of Medicine, College of Medicine, Taipei Medical University, Taipei 110, Taiwan
cDepartment of Surgery, College of Medicine, Division of Colorectal Surgery, Taipei Medical University Hospital, Taipei Medical University, The PhD Program for Translational Medicine, College of Science and Technology, Academia Sinica, Taipei, Taiwan
dCancer Center, Division of General Surgery, Department of Surgery, Taipei Medical University Hospital, College of Medicine, Graduate Institute of Cancer Biology and Drug Discovery, Taipei Medical University, Taipei, Taiwan
First published on 26th April 2017
Abstract
We grafted azido-terminated poly(N-isopropylacrylamide) (PNIPAAm) brushes onto thin gold films on silicon as a bottom electrode. A probe of single-stranded DNA (ssDNA) presenting a 4-pentynoic acid succinimidyl ester unit was grafted onto the azido-terminated PNIPAAm brushes through a click reaction, resulting in block copolymer brushes. The PNIPAAm-b-ssDNA copolymer brushes formed homogeneous complexes stabilized via bio-multiple hydrogen bonds (BMHBs), which enhanced proton transfer and thereby decreased the resistivity of the structures. The homogeneous complex state of the PNIPAAm-b-ssDNA copolymer brushes transformed into a phase-separated state after hybridization with 0.5 pg nL−1 of its target DNA, which resulted from competition between the BMHBs and complementary HBs; this phase transformation of the PNIPAAm and probe segments inhibited proton transfer and significantly increased the resistivity. Furthermore, hybridization with mismatched DNA sequences generated sufficient “proton leakage” to decrease the resistivity at temperatures below the lower critical solution temperature (LCST), especially for adenine and guanine units. Sequences with thymine and cytosine mismatches could be distinguished from the target at temperatures above the LCST due to disruption of the BMHBs at increased temperatures (80 °C). The ability to detect label-free DNA and recognize sequence mismatches suggests the potential use of these novel materials in bioelectronics.
1. Introduction
In the past decade, sequence-specific detection of DNA has garnered considerable interest due to its application in clinical diagnosis, gene expression analysis, and biomedical studies.1–3 Identification and quantification of specific DNA sequences related to disease is the purpose of DNA detection. Mostly, detection of DNA has been from DNA sequencing4 and DNA microarrays.5 Also, multiple new methods, such as fluorescent,6 electrochemical,7 and chemiluminescent8 methods, have been developed to minimise the equipment cost and enable easier operation. Along with biomedical studies, DNA can be used in scaffolds, “nanomachines” (i.e., DNA nanotechnology), and drug carriers, which has increased interest in its accurate detection as well as the modeling and control of hybridization processes.9In recent years, DNA microarrays have been used on surface-immobilized single-stranded DNA (ssDNA) probes, which has aided their flexibility and enabled considerable data generation.10 However, the fluorescent emission of labelled target DNA cannot be detected by conventional methods. Hence, label-free methods are useful for supporting quantitative detection of DNA binding, scalability, and multiplexing, as well as the expense of the operation.11 Herein, for the first time, we report a label-free technique for DNA detection. DNA block copolymers are a new emerging class of functional hybrid polymers composed of an oligonucleotide strand covalently attached to another polymer. Based on their amphiphilicity and molecular recognition properties,12 DNA block copolymers can be assembled into various types of nanostructures, which are useful for several applications, ranging from DNA-templated syntheses13 to gene therapy14 and drug delivery.15 For example, Mirkin and coworkers showed that spherical micelles of DNA-block-polystyrene (DNA-b-PS) can be assembled into macroscopic polymer networks showing cooperative sharp melting transitions.16 The simultaneous self-assembly of DNA-b-PS and magnetic nanoparticles is used to fabricate nanoparticle-loaded DNA block copolymer assemblies. This process can dramatically enhance DNA-binding properties.17 Researchers have also shown that DNA block copolymers can be assembled into various morphologies by utilizing the molecular recognition property of DNA.18,19 Gianneschi and co-workers synthesized a DNA brush copolymer that undergoes a reversible sphere-to-cylinder morphology change by DNA cleavage and hybridization.20 Herrmann et al. reported the formation of rodlike assemblies of DNA-block-poly(propylene oxide) (DNA-b-PPO) using a long repetitive complementary DNA strand as a template.21 The rod-shaped aggregates showed significantly higher cellular uptake than spheres,22 demonstrating that the ability to control the morphology of DNA assemblies is important for their biological and medical applications.23–25
Alemdaroglu et al. observed the strength of the bio-multiple hydrogen bonds (BMHBs) in poly(N-isopropylacrylamide) (PNIPAAm) and nucleobases to follow the sequence guanine (G) > adenine (A) > thymine (T) > cytosine (C) > uracil (U). The presence of BMHBs can enhance the degree of H+ transport in a PNIPAAm film, which is caused by a notable change in electronic conductivity. The insulator PNIPAAm can be altered into a semiconducting form via a simple method.13 In the present study, we formed novel DNA recognition layers, with PNIPAAm-b-ssDNA copolymer brushes acting as aptamers, that changed conductivity upon homogeneous complexation and phase separation of the PNIPAAm and ssDNA segments. Copolymer brushes can change structure in situ on the surface during hybridization. PNIPAAm and ssDNA, which are regarded as an insulator and conductor, respectively, are miscible due to BMHBs. Our target was the DNA strand complementary to the ssDNA segment of the copolymer brushes.26 Upon grafting polymerization with ssDNA, homogeneous complexes were generated to facilitate proton transport in the films, leading to a decline in resistance. Because the HB between the target and probe (ssDNA) was much stronger than the BMHBs between the PNIPAAm and ssDNA segments, hybridization of the target with the PNIPAAm-b-ssDNA copolymer brushes changed the morphology significantly. The probe segments preferred to release from the complex and bond to the target. The isolated PNIPAAm segments blocked proton transport from the bottom to the top electrodes, resulting in an increase in resistance (Scheme 1). We examined the selectivity and sensitivity of the complexation/decomplexation of PNIPAM-b-ssDNA copolymer brushes by subjecting them to a target featuring single-nucleic-acid mismatches at various temperatures. The developed aptamers are potentially applicable for DNA recognition sensors.
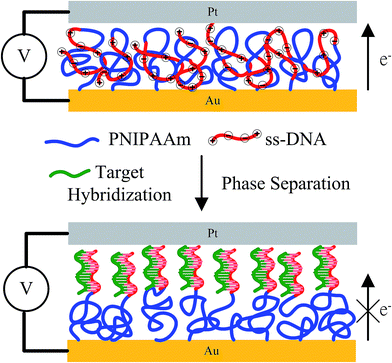 | ||
| Scheme 1 Homogeneous complexation, stabilized through BMHBs, and phase separation of PNIPAAm and ssDNA segments after hybridization with the target. | ||
2. Experimental section
2.1. Materials
One-side polished single-crystal silicon wafers (Si(100), with a diameter of 6 inches) were procured from Hitachi (Japan). N-Isopropylacrylamide (NIPAAm; Acros Organics, Belgium) was purified by hexane/toluene (50%, v/v) for use. 3-Aminopropanethiol (AT), 2-bromo-2-methylpropionyl bromide (BIBB), triethylamine (TA), CuBr, CuBr2, 1,1,4,7,7-pentamethyldiethylenetriamin (PMDETA), and sodium azide (NaN3) were obtained from Acros Organics. PMDETA, AT, and BIBB was purified by vacuum distillation. All solvents and chemicals were purchased from Sigma-Aldrich Chemicals (USA) and were of reagent grade. The target sequence was 5′-GACT TGCC ATCG TAGA ACTG-3′; its complementary probe sequence was NH2-(CH2)6-5′-CAGT TCTA CGAT GGCA AGTC-3′. The mismatched sequences were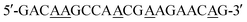 (AM) featuring A units,
(AM) featuring A units, 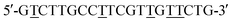 (TM) featuring T units,
(TM) featuring T units, 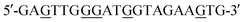 (GM) featuring G units, and 5′-
(GM) featuring G units, and 5′-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ACTT
ACTT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) CCATC
CCATC![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) TA
TA![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) AACT
AACT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) -3′ (CM) featuring C units. These DNA oligomers (the target, AM, TM, GM, and CM) were purchased from TriLink Biotechnologies (HPLC-purified for highest purity; USA).27–29 4-Pentynoic acid succinimidyl ester was prepared as described previously.30
-3′ (CM) featuring C units. These DNA oligomers (the target, AM, TM, GM, and CM) were purchased from TriLink Biotechnologies (HPLC-purified for highest purity; USA).27–29 4-Pentynoic acid succinimidyl ester was prepared as described previously.30
2.2. PNIPAAm-b-ssDNA copolymer brushes
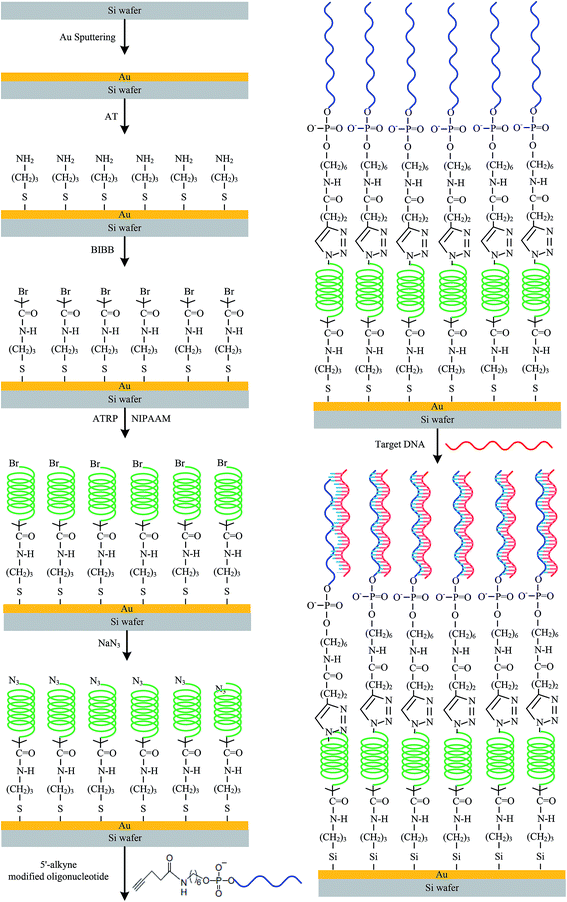
The length of the polymer was varied to “tune” the BMHBs between the PNIPAAm and ssDNA segments in PNIPAAm-b-ssDNA copolymer brushes. Fig. 1 outlines the synthetic pathway for anchoring initiators on the sputtered gold (Au) thin film of a silicon substrate, which acted as the bottom electrode. An NH2-terminated self-assembled monolayer (SAM) was first generated and then reacted with BIBB to obtain initiators for atom-transfer radical polymerization (ATRP). To immobilize the ATRP initiator, the as-prepared substrate was dipped in AT solution (0.5% wt%) and refluxed with toluene for 2 h at 50 °C. The AT units gathered on the Au surface through their thiol groups. The sample was mixed in a 2% (v/v) solution of BIBB and TA in toluene at room temperature for 8 h. Then, the as-prepared samples were placed in a Soxhlet apparatus to remove non-grafted material. Surface-initiated ATRP grafting of NIPAAm was performed in N2. NIPAAm, PMDETA, CuBr, and CuBr2, were dissolved in dimethylformamide (DMF).31,32 The solution was sonicated for 2 min, and added to AT/BIBB-functionalized wafers. Polymerization was carried out at 25 °C under N2 for 4, 8, 12, 16, or 20 h. Then, the bromo-PNIPAAm-grafted samples were removed from the solution and rinsed with copious amounts of deionized water to remove any unreacted monomer, followed by drying under a flow of N2 at 25 °C. Azido-PNIPAAm brushes were obtained on the surfaces after treatment of bromo-PNIPAAm brushes with NaN3.The substitution reaction was carried out overnight by exposing the bromo-terminated substrates to a saturated solution of NaN3 in DMF in a covered container. Then, the sample was rinsed with DMF followed by methanol and deionized water before drying in a N2 stream. After these reactions, the wafers were placed in a Soxhlet apparatus to remove non-grafted materials and then dried under N2 prior to use. A solution of 4-pentynoic acid succinimidyl ester-conjugated probe (100 nmol) in 2.5 mL phosphate buffer (PB) (pH 7.2, 20 mM) and 1 × 1 cm2 grafted azido-PNIPAAm samples were added to a 10 mL plastic culture tube. A stock solution (0.1 mL) of catalyst/ligand (1.0 mmol of CuSO4, 1.1 mmol of TBTA) in dimethyl sulfoxide (DMSO)/water (v/v, 1/1) and 4.0 mmol (0.79 mg) of sodium ascorbate were then added. The reaction mixture was agitated for 20 h at 15 °C. The as-prepared samples were incubated overnight at room temperature in the solution and then washed thrice with washing buffer [10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.05% Tween 20] and thrice with water. The samples were blow-dried under N2 to ensure that their surfaces were dry and free of dust particles. In addition, ssDNA was grafted to an azido-modified Au substrate without PNIPAAm in a blank experiment to analyze the thickness.
Herein, the surfaces presenting PNIPAAm that had been grafted for 4, 8, 12, 16, and 20 h were denoted PN4, PN8, PN12, PN16, and PN20, respectively, and sequentially grafted with ssDNA to form aptamers of PNIPAAm-b-ssDNA copolymer brushes, which were denoted PN4D, PN8D, PN12D, PN16D, and PN20D, respectively. The label-free target, AM, TM, GM, and CM were diluted with a hybridization buffer [comprising SSC (3 M NaCl, 0.3 M sodium citrate·2H2O, pH 7), Denhardt's solution (1% bovine serum albumin), 2% Ficoll400, 2% polyvinylpyrollidone, and 0.5% sodium dodecyl sulfate] to a final concentration of 50 μM. Two small strips of adhesive tape were affixed along the rim of the substrate and then covered with a clean microscope slide cover glass. The cavity formed between the chip and cover glass was filled by slowly loading the hybridization solution (≈30 μL) under capillary force. After removal of the cover glass, the chip was washed thrice with washing buffer and then with copious amounts of water and blow-dried under N2. Control experiments were performed in the same manner using AM, TM, GM, and CM, which had lengths similar to that of the target but only partial complementarity. Water contact angles of PNIPAAm-b-ssDNA copolymer brushes were measured after they had been dried under a flow of N2. The chemical compositions of the modified silicon surfaces were determined through X-ray photoelectron spectrometry (XPS; Scientific Theta Probe, UK).33 The thicknesses of the copolymer grafts on the silicon substrates were measured using ellipsometry (SE-5; Sopra, France). The tethered copolymer brushes were stripped from the surface through immersion in HF solution (5 wt%) for 5 min at room temperature. The stripped copolymers were analyzed using gel permeation chromatography after purification through extensive dialysis against deionized water, which was performed using a VISCOTEK-DM400 instrument equipped with a LR 40 refractive index detector. Monodisperse polymer standards (Polymer Lab; Agilent Technologies, USA) were used to generate a calibration curve.
2.3. Surface properties of PNIPAAm-b-ssDNA copolymer brushes
Static water contact angles (SWCAs) were estimated using a contact angle meter (Sindatek Instruments, Taiwan) and, during measurements, the temperature of the glass slide was controlled by a water bath at 25 °C (below the lower critical solution temperature (LCST)) or 45 °C (above the LCST).34 The morphologies were studied by scanning electron microscopy (SEM) using a JSM 6500F instrument (JEOL, Japan) operated at 15 kV and atomic force microscopy (AFM; Veeco Dimension 5000 scanning probe microscope).The electroactivities of the PNIPAAm-b-ssDNA copolymer brushes were studied by I–V curve measurements. That is, platinum (Pt) electrodes (thickness: 200 nm) were coated onto the sample surfaces via stainless-steel shadow masks as the top electrode. Then, the samples were lyophilized 24 h prior to use of I–V curves from the top to the bottom electrode of the Au substrate (Scheme 2). All the measurements were performed at room temperature with a relative humidity of 35–45%. Repeated measurements were within 5% of the average value for each sample. To prevent metal contamination, the interval between the Pt electrodes was ≈6.5 mm. The resistivity of each copolymer film was measured at 25 and 80 °C; voltage sweeps were recorded in the range from −0.2 to +0.2 V.
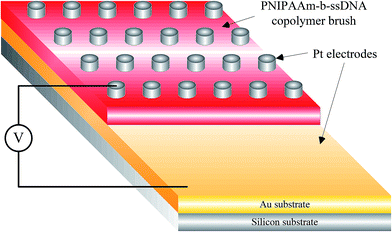 | ||
| Scheme 2 Schematic representation of the structured device used to measure the resistivities of PNIPAAm-b-ssDNA copolymer brushes. | ||
3. Results and discussion
3.1. Surface properties of PNIPAAm-b-ssDNA copolymer brushes
We used XPS to determine the chemical compositions of the Au surfaces at different stages throughout the surface modification process as well as in the presence of grafted PNIPAAm-b-ssDNA copolymer brushes.35 The oxygen (O)/carbon (C), nitrogen (N)/C and phosphorus (P)/C ratios of PNIPAAm and ssDNA segments were different, which could be exploited to monitor the surface compositions according to Fig. S1.† From the high-resolution C 1s spectra shown in Fig. 2a, we determined the O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N
N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C molar ratio for PNIPAAm-N3 and ssDNA to be 12.1
C molar ratio for PNIPAAm-N3 and ssDNA to be 12.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12.9
12.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75.5 and 7.4
75.5 and 7.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6.2
6.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14.1, respectively, consistent with the anticipated ratio of 12.5
14.1, respectively, consistent with the anticipated ratio of 12.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12.5
12.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75.0 and 8
75.0 and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 for PNIPAAm and the probe, respectively. Fig. 2 summarizes the O/C, N/C and P/C ratios of PN4D, PN8D, PN12D, PN16D and PN20D at 25, 45 and 80 °C, respectively. Compared with PNIPAAm segments, ssDNA segments possessed higher O/C, N/C and P/C ratios: 0.62, 0.4 and 0.1, respectively. The O/C, N/C and P/C ratios were close to the average of the molar ratios for PNIPAAm and ssDNA segments at 25 °C, respectively, suggesting that the ssDNA segments were “buried” within the PNIPAAm segments to generate homogeneous complexes stabilized through BMHBs. The O/C, N/C and P/C ratios increased abruptly upon a temperature increase from 25 to 45 °C, indicating the ssDNA segments were distributed predominately over the surface. These results suggest that the hydrogen bonding among PNIPAAm segments strengthened at 45 °C to repel the ssDNA segments from the matrix of PNIPAAm segments up to the surface, thereby resulting in the increase of N/C and O/C. Upon increasing the temperature from 45 to 80 °C, the O/C, N/C and P/C ratios decreased significantly, indicating that the thermal effect enhanced the motion of polymer chains, resulting in the rearrangement of PNIPAAm and ssDNA segments. The O/C, N/C and P/C ratios did not change significantly with the length of PNIPAAm segments at 25 and 45 °C, whereas these ratios decreased with the length of PNIPAAm segments at 80 °C.36 These results indicate that the length of the PNIPAAm segment facilitates the rearrangement of the copolymer, verifying the specific thermal effect for PNIPAAm-b-ssDNA brushes.
13 for PNIPAAm and the probe, respectively. Fig. 2 summarizes the O/C, N/C and P/C ratios of PN4D, PN8D, PN12D, PN16D and PN20D at 25, 45 and 80 °C, respectively. Compared with PNIPAAm segments, ssDNA segments possessed higher O/C, N/C and P/C ratios: 0.62, 0.4 and 0.1, respectively. The O/C, N/C and P/C ratios were close to the average of the molar ratios for PNIPAAm and ssDNA segments at 25 °C, respectively, suggesting that the ssDNA segments were “buried” within the PNIPAAm segments to generate homogeneous complexes stabilized through BMHBs. The O/C, N/C and P/C ratios increased abruptly upon a temperature increase from 25 to 45 °C, indicating the ssDNA segments were distributed predominately over the surface. These results suggest that the hydrogen bonding among PNIPAAm segments strengthened at 45 °C to repel the ssDNA segments from the matrix of PNIPAAm segments up to the surface, thereby resulting in the increase of N/C and O/C. Upon increasing the temperature from 45 to 80 °C, the O/C, N/C and P/C ratios decreased significantly, indicating that the thermal effect enhanced the motion of polymer chains, resulting in the rearrangement of PNIPAAm and ssDNA segments. The O/C, N/C and P/C ratios did not change significantly with the length of PNIPAAm segments at 25 and 45 °C, whereas these ratios decreased with the length of PNIPAAm segments at 80 °C.36 These results indicate that the length of the PNIPAAm segment facilitates the rearrangement of the copolymer, verifying the specific thermal effect for PNIPAAm-b-ssDNA brushes.
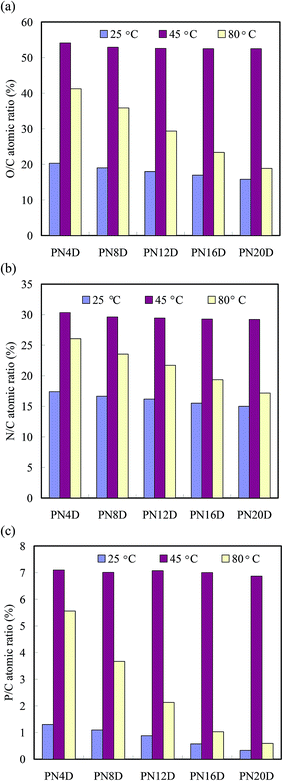 | ||
| Fig. 2 (a) O/C, (b) N/C and (c) P/C atomic ratios of the PNIPAAm-b-DNA copolymer brushes at 25, 45 and 80 °C. | ||
The brush parameters of the dry PNIPAAm-b-ssDNA brushes were estimated by the ellipsometric dry layer thickness (h),37 PNIPAAm and ssDNA bulk density of 1.1 and 1.7 g cm−3, respectively. These calculations were made with their roughnesses (Ra), grafting densities (σ), distance between grafting sites (dg), molecular weight, and surface coverage (Sc). Furthermore, we examined the temperature-sensitive activities of these PNIPAAm-b-ssDNA films in contact with aqueous solutions using the SWCA measurements. Table 1 presents the calculated data of layer parameters for ssDNA, PNIPAAm, and PNIPSSm-b-ssDNA films. The layer parameters (molecular weights (Mn and Mw), Sc, dg, and σ) increased gradually with low polydispersity index (PDI) values upon increasing the polymerization times for tethered PNIPAAm films. Slight increases in these layer parameter were obtained after ssDNA immobilization at the chain end. The thicknesses of the PNIPAAm-b-ssDNA layers decreased slightly with increasing temperature from 25 to 45 °C because the intramolecular HBs among PNIPAAm groups was stronger than the BMHBs between the PNIPAAm and ssDNA segments. In addition, the SWCAs of PN4D, PN8D, PN12D, PN16D, and PN20D brushes exhibited thermally responsive switching between hydrophilic and hydrophobic states at temperatures below and above the LCST, respectively. The SWCAs of the PN4D brushes were 78.5 and 46.9° at 25 and 45 °C, respectively. This thermal responsivity of PN4D was completely opposite to that of pure PNIPAAm (Table 1), which can be explained by considering the miscibility of the PNIPAAm and ssDNA segments at 25 °C. The HB interactions between the PNIPAAm and ssDNA segments resulted in the hydrophilic groups residing within these molecules, thereby exposing the hydrophobic groups on the surface at 25 °C. The HB interactions between the PNIPAAm and ssDNA segments weakened significantly at 45 °C. This was due to the predominance of intramolecular HBs of the PNIPAAm segments,38 which drove the ssDNA segments to the surface, leading to a hydrophilic surface state. Upon increasing the thickness of the PNIPAAm segment, the SWCA at 25 °C decreased from 78.5° for PN4D to 64.5° for PN8D, whereas the SWCA at 45 °C increased from 46.9 to 51.7°. The SWCAs of PN12D at 25 and 45 °C were 49.8 and 56.3°, respectively. These observations showed that PNIPAAm-b-ssDNA exhibited minimal thermoresponsive behavior. Notably, the thermoresponsive behavior of the copolymer brushes became similar to that of tethered PNIPAAm films when grafting of the PNIPAAm segment had been performed for >12 h (Table 1). The difference in the SWCAs between the hydrophilic (34°) and hydrophobic (62.1°) states reached 28.1° for PN16D. For PN20D, the thermal responsive behavior was close to that of pure PNIPAAm brushes. Our results indicate that when its thickness was >300 nm, the PNIPAAm segment predominated the surface properties.
| Samples | Mn/1000 (g mol−1) | Mw/1000 (g mol−1) | PDI (Mw/Mn) | Tga (°C) | Scb (mg m−2) | dgc (nm) | σd (nm−2) | he (nm) | Raf (nm) | SWCA (degree) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 45 °C | 25 °C | 45 °C | 25 °C | 45 °C | ||||||||
a Tg was obtained from DSC data (Fig. S2).b Sc = ρh.c 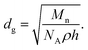 d σ = dg−2.e Thickness (h) was obtained from ellipsometry.f Roughness (Ra) was obtained from AFM. d σ = dg−2.e Thickness (h) was obtained from ellipsometry.f Roughness (Ra) was obtained from AFM. |
|||||||||||||
| ssDNA | 6.2 | 6.7 | 1.08 | — | 8.3 | 0.35 | 8.03 | 75.4 | 75.4 | 2.6 | 2.5 | 23.5 ± 3 | 22.1 ± 4 |
| PN4 | 11.1 | 14.9 | 1.34 | 133.6 | 14.5 | 0.36 | 7.84 | 131.9 | 86 | 9.3 | 7.5 | 34.6 ± 4 | 79.7 ± 4 |
| PN8 | 32.3 | 40.1 | 1.24 | 134.2 | 22.4 | 0.49 | 4.16 | 203.6 | 135 | 8.4 | 6.7 | 35.9 ± 4 | 84.3 ± 4 |
| PN12 | 57.4 | 67.3 | 1.17 | 134.3 | 32.9 | 0.54 | 3.44 | 299.1 | 202 | 7.8 | 6.1 | 32.5 ± 3 | 85.8 ± 4 |
| PN16 | 75.6 | 87.7 | 1.16 | 134.8 | 38.8 | 0.57 | 3.08 | 352.4 | 241 | 7.2 | 5.8 | 33.6 ± 3 | 86.4 ± 3 |
| PN20 | 100.7 | 116.2 | 1.15 | 134.9 | 46.9 | 0.60 | 2.80 | 426.6 | 298 | 6.9 | 5.3 | 31.1 ± 3 | 86.5 ± 3 |
| PN4D | 17.3 | 21.1 | 1.22 | 103.3 | 14.5 | 0.45 | 5.04 | 132.1 | 97.3 | 5.2 | 3.6 | 78.5 ± 4 | 46.9 ± 4 |
| PN8D | 38.5 | 46.3 | 1.20 | 110.1 | 22.4 | 0.54 | 3.49 | 203.8 | 145.8 | 4.8 | 3.5 | 64.5 ± 4 | 51.7 ± 4 |
| PN12D | 63.6 | 73.5 | 1.16 | 116.5 | 32.9 | 0.57 | 3.11 | 299.3 | 212.5 | 4.2 | 3.4 | 49.8 ± 3 | 64.3 ± 4 |
| PN16D | 81.8 | 93.9 | 1.15 | 121.4 | 38.8 | 0.59 | 2.84 | 352.6 | 251.4 | 3.9 | 3.2 | 34.2 ± 3 | 77.1 ± 3 |
| PN20D | 106.9 | 122.4 | 1.14 | 124.8 | 46.9 | 0.62 | 2.64 | 426.8 | 308.3 | 3.6 | 3.2 | 32.2 ± 3 | 83.3 ± 3 |
3.2. PNIPAAm-b-ssDNA copolymer brushes for label-free target DNA sensing
Pure PNIPAAm showed uneven microsphere- and “wicker”-like “globule” structures, with sizes of 200 nm to 1 μm,19 consistent with tough intramolecular HBs in the dry state. When DNA units complexed with PNIPAAm, the wicker-like globule structures vanished, suggesting that the intramolecular HB of PNIPAAm was damaged slightly as a result of the BMHBs formation. Fig. 3 shows field emission-SEM images of the surface of PN8D before and after hybridization with the target DNA. Obviously stacked globules appeared for PN8D, without aggregated particles, prior to hybridization (Fig. 3a); a particle structure appeared on the surface after hybridization with the target. This observation suggests that strong HB between the ssDNA unit and the target led to phase separation on the surface. In other words, hybridizing the target with the PNIPAAm-b-ssDNA copolymer brushes significantly changed the morphology as a result of competitive HB. The ssDNA segments preferred to be released from the homogeneous complex and bond to the target, resulting in a globule structure on the surface. The surface morphology change from homogeneous complex to phase separation was more obvious with a decreasing PNIPAAm chain length. Except PN4D, PNIPAAm-b-ssDNA copolymer brushes exhibited a regular morphology change. These subtle changes in surface morphology that change the BMHBs nature suggest the potential for relating such systems to preparation of DNA recognition and biocompatible materials.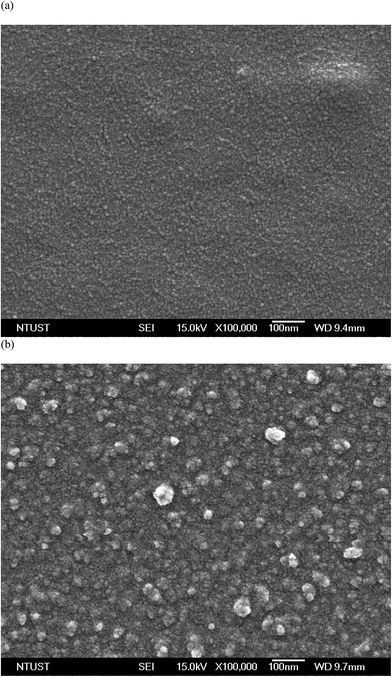 | ||
| Fig. 3 Surface SEM images of PN8D copolymer brushes (a) before and (b) after hybridization with the target. | ||
BMHBs can improve H+ transport in PNIPAAm-b-ssDNA copolymer brushes,18,19 so we focused mainly on determining the temperature-responsive resistance of our functionalized surfaces. The linear I–V curves exhibited ohmic behavior at 25 and 80 °C during device use (Scheme 2). The slopes of I–V curves corresponded to the resistivities of the surfaces at particular temperatures. Fig. 4 shows the logarithms of the average resistivities for PN4D, PN8D, PN12D, PN16D, and PN20D brushes at different temperatures from 20 to 90 °C. In particular, there were no I–V curves for the neat PNIPAAm film because proton transport did not occur. Therefore, we could not measure the resistivity of the neat PNIPAAm brushes, which could be regarded as an insulator. Upon grafting polymerization with ssDNA, proton transport in the films increased significantly, a result of the homogeneous complexes that were generated, leading to a decline in resistance. The resistivity of the PNIPAAm-b-ssDNA copolymer brushes increased upon increasing the thickness of the PNIPAAm segment, which is associated with the strength of BMHBs, confirming that the increases in conductivity of PNIPAAm arose from proton transport induced by BMHBs. The length ratio of ssDNA to PNIPAAm segments revealed that the improved conductivity at <30 °C occurred due to proton transport induced through BMHBs. The resistivities of the PNIPAAm-b-ssDNA copolymer brushes increased upon increasing the temperature at 70 °C, demonstrating that proton transport from the bottom to the top electrode was blocked as a result of phase separation of the PNIPAAm and ssDNA segments. The resistivities decreased abruptly from 70 to 90 °C, suggesting that increased vibrations of the PNIPAAm chains was facilitated by proton transport in the PNIPAAm-b-ssDNA copolymer brushes at >70 °C. For PN4D, PN8D and PN12D, the resistivity increased suddenly upon increasing the temperature from 30 to 40 °C due to phase separation of the PNIPAAm and ssDNA segments. The dependence of resistivity on temperature increased slowly for PN16D and PN20D, symptomatic of the fact that the effects of the increased vibration of the polymer chains at 30 to 40 °C were more evident as a result of weaker BMHBs. The resistivity transition temperature of PNIPAAm-b-ssDNA copolymer brushes were superior than the LCST because the BMHBs were improved slightly in the dry state exclusive of the association of water molecules.36 Grafting ssDNA on the PNIPAAm chains can offer increased proton transport and considerably enhanced conductivity. The ssDNA segment could be regarded as a “minority” carrier in the complexed layer, predominantly determining the resistivities of the films.39 Moreover, the variation in resistivity with temperature can be reversible for five cycles between 20 and 90 °C for all PNIPAAm-b-ssDNA copolymer brushes, indicating stable resistivity against temperature.
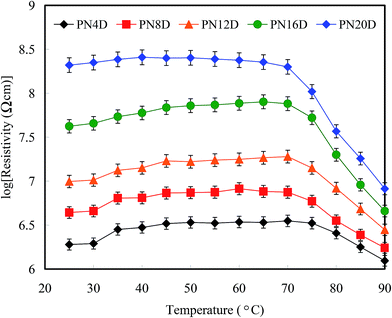 | ||
| Fig. 4 Resistivities of the PNIPAAm-b-DNA copolymer brushes (logarithm scale) recorded with respect to temperature at 25 to 90 °C. | ||
Fig. 5a and b display the logarithms of the average resistivities of the PNIPAAm-b-ssDNA copolymer brushes upon hybridization with the label-free target at various concentrations at 25 and 80 °C, respectively. Notably, the resistivity might be influenced readily by the humidity or roughness of the surface. The inaccuracy ranges were marked as indices for recognition of hybridization with the target. A change in resistivity without overlapping the inaccuracy range is defined herein as a “distinguishable” value. We observed approximately linear increases in the resistivity of the grafted copolymer layer upon increasing the concentration of the target to 8 pg nL−1 at 25 °C. The resistivity reached a plateau for all samples, indicating saturation of the hybridization between the probe segment (ssDNA) and the target; the strong complementary HB resulted in phase separation of the PNIPAAm and ssDNA segments, leading to an increase in resistivity. In other words, the target could be exploited to eliminate the proton transfer of complexation at 25 °C. Interestingly, the resistivity of the copolymer brushes after hybridization with the target at 80 °C decreased linearly upon increasing the concentration, indicating that the high temperature accelerated proton transfer to overcome the barrier of phase separation. A low concentration of the minor carriers (ssDNA segments) could generate a significant change in resistivity at 80 °C. The lowest concentrations for target recognition for PN4D, PN8D, PN12D, PN16D, and PN20D brushes at 25 °C were 0.5, 0.5, 1, 2, and 4 pg nL−1, respectively; these values at 80 °C were 4, 1, 1, 0.5, and 0.5 pg nL−1, respectively. We found that the sensitivities of PN4D and PN8D for hybridization with the target were greater than those of the other samples; thus, a higher ratio of probe![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNIPAAm segments enhanced the sensitivity significantly at 25 °C. In contrast, the sensitivities of PN16D and PN20D for target detection were higher than those of the other samples at 80 °C; thus, thermal effects facilitated proton transfer of the minor carriers (ssDNA segments) within longer polymer chains at 80 °C. Furthermore, we used AM, TM, GM, and CM to investigate the ability of our surfaces to distinguish mismatched hybridization from that of the target. Fig. 6a displays the logarithms of the average resistivities of PN4D before and upon hybridization at 25 °C with the target, AM, TM, GM, and CM at a concentration of 0.5 pg nL−1. The resistivity of blank PN4D ranged from 5.69 to 6.05 Ω cm and shifted to a range from 6.09 to 6.47 Ω cm upon hybridization at 25 °C with the target. A mismatched hybridization could be regarded as resulting in “proton leakage,” leading to a decrease in resistivity. Compared with blank PN4D, the resistivity of PN4D after hybridization with AM and GM at 0.5 pg nL−1 did not change significantly at 25 °C because of proton leakage, implying that it was distinguishable from the target (Fig. 6a).40 The proton leakage increased with respect to the strength of the BMHBs, but weakened with respect to the length of the PNIPAAm segment (Fig. S3†). The mismatches of AM and GM at 0.5 pg nL−1 could be distinguished from the target when using PN4D at 25 °C due to its high ratio of ssDNA
PNIPAAm segments enhanced the sensitivity significantly at 25 °C. In contrast, the sensitivities of PN16D and PN20D for target detection were higher than those of the other samples at 80 °C; thus, thermal effects facilitated proton transfer of the minor carriers (ssDNA segments) within longer polymer chains at 80 °C. Furthermore, we used AM, TM, GM, and CM to investigate the ability of our surfaces to distinguish mismatched hybridization from that of the target. Fig. 6a displays the logarithms of the average resistivities of PN4D before and upon hybridization at 25 °C with the target, AM, TM, GM, and CM at a concentration of 0.5 pg nL−1. The resistivity of blank PN4D ranged from 5.69 to 6.05 Ω cm and shifted to a range from 6.09 to 6.47 Ω cm upon hybridization at 25 °C with the target. A mismatched hybridization could be regarded as resulting in “proton leakage,” leading to a decrease in resistivity. Compared with blank PN4D, the resistivity of PN4D after hybridization with AM and GM at 0.5 pg nL−1 did not change significantly at 25 °C because of proton leakage, implying that it was distinguishable from the target (Fig. 6a).40 The proton leakage increased with respect to the strength of the BMHBs, but weakened with respect to the length of the PNIPAAm segment (Fig. S3†). The mismatches of AM and GM at 0.5 pg nL−1 could be distinguished from the target when using PN4D at 25 °C due to its high ratio of ssDNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNIPAAm segments. However, the resistivities of PN4D after hybridization with TM and CM at 0.5 pg nL−1 shifted to overlap the range of that with the target at 25 °C; therefore, TM and CM cannot be distinguished from that of the target due to their weaker BMHBs. These results indicate that the copolymer can distinguish the target from G and A mismatch, but not T and C mismatch at 25 °C. To identify the T and C mismatch, the temperature was increased to 80 °C to observe the changes in resistivities. The resistivity of blank PN20D ranged from 7.87 to 8.36 Ω cm, and decreased to the range from 7.34 to 7.79 Ω cm upon hybridization with the target at 80 °C. Because of proton leakage at 80 °C, the resistivities of PN20D after hybridization with AM and GM at 0.5 pg nL−1 decreased to the similar range as well, implying them to be undistinguishable from the target. In contrast, the resistivities of PN20D after hybridization with CM and TM at 0.5 pg nL−1 did not change significantly at 80 °C because the weak BMHBs were disrupted completely at 80 °C, implying them to be distinguishable from the target (Fig. 6b). These results suggest that selectivity for target detection was achieved by resistivity measurement at 25 and 80 °C to eliminate a false-positive mismatch. We increased the concentrations of AM, TM, GM, and CM for hybridization with the copolymer brushes until the resistivity could be distinguished from that of the target. Fig. 7 displays the lowest concentrations of distinguishable mismatch (LCDMs) from the target for all of the copolymer brushes at 25 and 80 °C. The LCDMs of AM, TM, GM, and CM from the target increased gradually upon increasing the length of PNIPAAm segment at 25 °C, verifying that the degree of proton leakage had a predominant effect on the LCDM. In contrast, the LCDMs of AM, TM, GM, and CM from the target decreased upon an increasing length of the PNIPAAm segments, indicating that thermal effects on the minor carriers determined the LCDM at 80 °C. These findings suggest that the mismatches of AM and GM could be distinguished efficiently from the target at temperatures below the LCST of PNIPAAm, whereas the mismatches of CM and TM could be distinguished predominantly at 80 °C, especially if the copolymer had a longer PNIPAAm segment. Notably, the thermal effect of the minor carriers was obvious at >80 °C. Furthermore, the variation in resistivity with respect to temperature was reversible for five cycles between 25 and 80 °C for all copolymer brushes, suggesting the stability of their resistivities to temperature. The electrical properties of the as-prepared copolymers may could be used to detect specific DNA sequences without the need to label them “smart materials”.
PNIPAAm segments. However, the resistivities of PN4D after hybridization with TM and CM at 0.5 pg nL−1 shifted to overlap the range of that with the target at 25 °C; therefore, TM and CM cannot be distinguished from that of the target due to their weaker BMHBs. These results indicate that the copolymer can distinguish the target from G and A mismatch, but not T and C mismatch at 25 °C. To identify the T and C mismatch, the temperature was increased to 80 °C to observe the changes in resistivities. The resistivity of blank PN20D ranged from 7.87 to 8.36 Ω cm, and decreased to the range from 7.34 to 7.79 Ω cm upon hybridization with the target at 80 °C. Because of proton leakage at 80 °C, the resistivities of PN20D after hybridization with AM and GM at 0.5 pg nL−1 decreased to the similar range as well, implying them to be undistinguishable from the target. In contrast, the resistivities of PN20D after hybridization with CM and TM at 0.5 pg nL−1 did not change significantly at 80 °C because the weak BMHBs were disrupted completely at 80 °C, implying them to be distinguishable from the target (Fig. 6b). These results suggest that selectivity for target detection was achieved by resistivity measurement at 25 and 80 °C to eliminate a false-positive mismatch. We increased the concentrations of AM, TM, GM, and CM for hybridization with the copolymer brushes until the resistivity could be distinguished from that of the target. Fig. 7 displays the lowest concentrations of distinguishable mismatch (LCDMs) from the target for all of the copolymer brushes at 25 and 80 °C. The LCDMs of AM, TM, GM, and CM from the target increased gradually upon increasing the length of PNIPAAm segment at 25 °C, verifying that the degree of proton leakage had a predominant effect on the LCDM. In contrast, the LCDMs of AM, TM, GM, and CM from the target decreased upon an increasing length of the PNIPAAm segments, indicating that thermal effects on the minor carriers determined the LCDM at 80 °C. These findings suggest that the mismatches of AM and GM could be distinguished efficiently from the target at temperatures below the LCST of PNIPAAm, whereas the mismatches of CM and TM could be distinguished predominantly at 80 °C, especially if the copolymer had a longer PNIPAAm segment. Notably, the thermal effect of the minor carriers was obvious at >80 °C. Furthermore, the variation in resistivity with respect to temperature was reversible for five cycles between 25 and 80 °C for all copolymer brushes, suggesting the stability of their resistivities to temperature. The electrical properties of the as-prepared copolymers may could be used to detect specific DNA sequences without the need to label them “smart materials”.
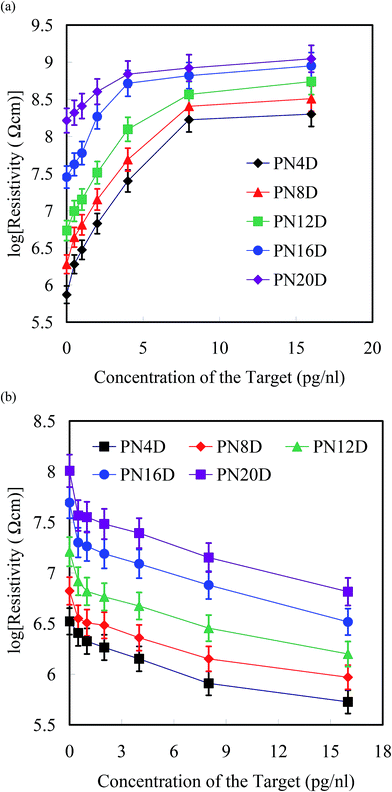 | ||
| Fig. 5 Resistivities of PNIPAAm-b-ssDNA copolymer brushes (logarithm scale) plotted with respect to the hybridization concentration of the target (0.5–16 pg nL−1) at (a) 25 and (b) 80 °C. | ||
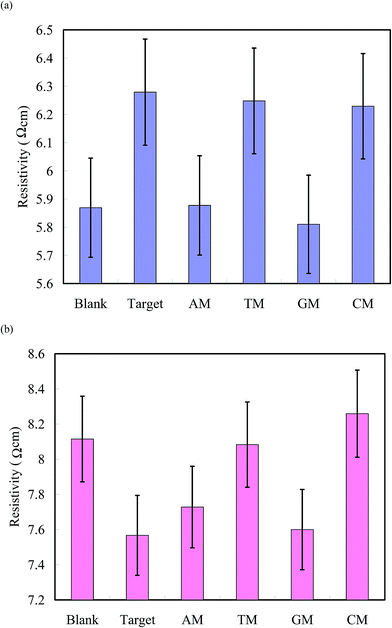 | ||
| Fig. 6 Resistivities (logarithm scale) of (a) PN4D at 25 °C and (b) PN20D at 80 °C before and after hybridization with the target, AM, TM, GM, and CM at 0.5 pg nL−1. | ||
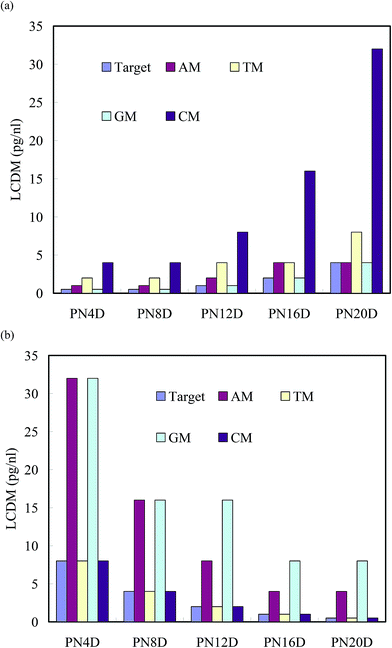 | ||
| Fig. 7 LCDMs of AM, TM, GM, and CM from the target for hybridization with PNIPAAm-b-ssDNA copolymer brushes at (a) 25 and (b) 80 °C. | ||
4. Conclusions
Specific BMHBs between PNIPAAm and nucleobases can generate non-covalently interacted colloidal supramolecular systems within a single chain of a copolymer brush. These interactions between the PNIPAAm and nucleobase segments result in differences in the thermoresponsive hydrophobicity and resistivity of the surface. The mode of HB of PNIPAAm-b-ssDNA copolymer brushes can be “switched” by hybridization with a target DNA, with significant changes in electrical resistivity occurring for the complexed and phase-separated states. The presence of A and G mismatches from the target could be determined efficiently through measurements of resistivity at temperatures below the LCST due to proton leakage; in contrast, T and C mismatches could be differentiated efficiently from the target at 80 °C. Accordingly, PNIPAAm/nucleobase copolymer brushes could be used in the label-free detection of DNA.Conflict of interest
The authors declare no competing financial interest.Acknowledgements
Authors are thankful to the Ministry of Science and Technology of the Republic of China and Taipei Medical University – National Taiwan University of Science and Technology Joint Research Program (TMU-NTUST-106-01) for supporting this research financially.References
- S. Bi, J. Zhang and S. Zhang, Ultrasensitive and Selective DNA Detection Based on nicking Endonuclease Assisted Signal Amplification and its Application in Cancer Cell Detection, Chem. Commun., 2010, 46, 5509–5511 Search PubMed.
- J.-K. Chen and J. Y. Li, Synthesis of Tethered poly-(N-isopropyl-acrylamide) for Detection of Breast Cancer Recurrence DNA, J. Colloid Interface Sci., 2011, 358, 454–461 Search PubMed.
- J. Zhu, Y. Lu, C. Deng, G. L. Huang, S. Y. Chen, S. K. Xu, Y. Lv, K. Mitchelson and J. Cheng, Assessment of Fluorescence Resonance Energy Transfer for Two-Color DNA Microarray Platforms, Anal. Chem., 2010, 82, 5304–5312 Search PubMed.
- Y. Hirano, M. Ikegami, K. Kowata and Y. Komatsu, Bienzyme reactions on cross-linked DNA scaffolds for electrochemical analysis, Bioelectrochemistry, 2017, 113, 15–19 Search PubMed.
- L. Fabris, M. Dante, G. Braun, S. J. Lee, N. O. Reich, M. Moskovits, T.-Q. Nguyen and G. C. Bazan, A Heterogeneous PNA-Based SERS Method for DNA Detection, J. Am. Chem. Soc., 2007, 129, 6086–6087 Search PubMed.
- X. W. He and N. Ma, A General Strategy for Label-Free Sensitive DNA Detection Based on Quantum Dot Doping, Anal. Chem., 2014, 86, 3676–3681 CrossRef CAS PubMed.
- F. Xuan, X. T. Luo and I.-M. Hsing, Ultrasensitive Solution-Phase Electrochemical Molecular Beacon-Based DNA Detection with Signal Amplification by Exonuclease III-Assisted Target Recycling, Anal. Chem., 2012, 84, 5216–5220 CrossRef CAS PubMed.
- J. Lee, K. Yun, G. Lim, S. Lee, S. Kim and J. Park, DNA biosensor based on the electrochemiluminescence of Ru(bpy)32+ with DNA-binding intercalators, Bioelectrochemistry, 2007, 70, 228–234 CrossRef CAS PubMed.
- T. Bellini, R. Cerbino and G. Zanchetta, DNA-Based Soft Phases, Top. Curr. Chem., 2012, 318, 225–279 CrossRef CAS PubMed.
- F. R. R. Teles and L. P. Fonseca, Trends in DNA Biosensors, Talanta, 2008, 77, 606–623 CrossRef CAS.
- E. Ozkumur, S. Ahn, A. Yalcin, C. A. Lopez, E. Cevik, R. J. Irani, C. DeLisi, M. Chiari and M. S. Unlu, Label-Free Microarray Imaging for Direct Detection of DNA Hybridization and Single-Nucleotide Mismatches, Biosens. Bioelectron., 2010, 25, 1789–1795 CrossRef PubMed.
- G. Fuks, R. M. Talom and F. Gauffre, Biohybrid Block Copolymers: Towards Functional Micelles and Vesicles, Chem. Soc. Rev., 2011, 40, 2475–2493 RSC.
- F. E. Alemdaroglu, K. Ding, R. Berger and A. Herrmann, DNA-Templated Synthesis in Three Dimensions: Introducing a Micellar Scaffold for Organic Reactions, Angew. Chem., Int. Ed., 2006, 45, 4206–4210 CrossRef CAS PubMed.
- D. Cukjati, D. Batiuskaite, F. André, D. Miklavčič and L. M. Mir, Real time electroporation control for accurate and safe in vivo non-viral gene therapy, Bioelectrochemistry, 2007, 70, 501–507 CrossRef CAS PubMed.
- F. E. Alemdaroglu, N. C. Alemdaroglu, P. Langguth and A. Herrmann, DNA Block Copolymer Micelles – A Combinatorial Tool for Cancer Nanotechnology, Adv. Mater., 2008, 20, 899–902 CrossRef CAS.
- Z. Li, Y. Zhang, P. Fullhart and C. A. Mirkin, Reversible and Chemically Programmable Micelle Assembly with DNA Block-Copolymer Amphiphiles, Nano Lett., 2004, 4, 1055–1058 CrossRef CAS.
- X.-J. Chen, B. L. Sanchez-Gaytan, S. E. N. Hayik, M. Fryd, B. B. Wayland and S.-J. Park, Self-Assembled Hybrid Structures of DNA Block-Copolymers and Nanoparticles with Enhanced DNA Binding Properties, Small, 2010, 6, 2256–2260 CrossRef CAS PubMed.
- H. W. Yang, A. Lee, C. Huang and J. K. Chen, Characterization of poly(N-isopropylacrylamide)–Nucleobase Supramolecular Complexes Featuring Bio-Multiple Hydrogen Bonds, Soft Matter, 2014, 10, 8330–8340 RSC.
- H.-W. Yang, J. K. Chen, C. C. Cheng and S. W. Kuo, Association of poly(N-isopropylacrylamide) Containing Nucleobase Multiple Hydrogen Bonding of Adenine for DNA Recognition, Appl. Surf. Sci., 2013, 271, 60–69 CrossRef CAS.
- M.-P. Chien, A. M. Rush, M. P. Thompson and N. C. Gianneschi, Programmable Shape-Shifting Micelles, Angew. Chem., Int. Ed., 2010, 49, 5076–5080 CrossRef CAS PubMed.
- K. Ding, F. E. Alemdaroglu, M. Boersch, R. Berger and A. Herrmann, Engineering the Structural Properties of DNA Block Copolymer Micelles by Molecular Recognition, Angew. Chem., Int. Ed., 2007, 46, 1172–1175 CrossRef CAS PubMed.
- F. E. Alemdaroglu, N. C. Alemdaroglu, P. Langguth and A. Herrmann, Cellular Uptake of DNA Block Copolymer Micelles with Different Shapes, Macromol. Rapid Commun., 2008, 29, 326–329 CrossRef CAS.
- J. K. Chen, G. Y. Zhou, C. J. Chang and C. C. Cheng, Label-Free Detection of DNA Hybridization Using Nanopillar Arrays Based Optical Biosensor, Sens. Actuators, B, 2014, 194, 10–18 CrossRef CAS.
- J.-K. Chen and J.-Y. Li, Fabrication of DNA Extraction Device with Tethered poly(N-isopropylacrylamide) Brushes on Silicon Surface for a Specific DNA Detection, Sens. Actuators, B, 2010, 150, 314–320 CrossRef CAS.
- J.-K. Chen and J.-Y. Li, Detection of Specific DNA Using a Microfluidic Device Featuring Tethered poly(N-isopropylacrylamide) on a Silicon Substrate, Appl. Phys. Lett., 2010, 97, 063701 CrossRef.
- J. K. Chen, J. H. Wang, S. Fan and J. Chang, Reversible Hydrophobic/Hydrophilic Adhesive of PS-b-PNIPAAm Copolymer Brush Nanopillar Arrays for Mimicking the Climbing Aptitude of Geckos, J. Phys. Chem. C, 2012, 116, 6980–6992 CAS.
- J. K. Chen, G. Y. Zhou, C. Chang, A. Lee and F. Chang, Label-free DNA Detection Using Two-Dimensional Periodic Relief Grating as a Visualized Platform for Diagnosis of Breast Cancer Recurrence After Surgery, Biosens. Bioelectron., 2014, 54, 35–41 CrossRef CAS PubMed.
- C. Y. Lee, H. E. Canavan, L. J. Gamble and D. G. Castner, Evidence of Impurities in Thiolated Single-Stranded DNA Oligomers and their Effect on DNA Self-Assembly on Gold, Langmuir, 2005, 21, 5134–5141 CrossRef CAS PubMed.
- J. K. Chen, B. J. Bai and F. C. Chang, Diagnosis of Breast Cancer Recurrence Using a Microfluidic Device Featuring Tethered Cationic Polymers, Appl. Phys. Lett., 2011, 99, 013701 CrossRef.
- M. Slater, M. Snauko, F. Svec and J. M. J. Frechet, “Click Chemistry” in the Preparation of Porous Polymer-Based Particulate Stationary Phases for μ-HPLC Separation of Peptides and Proteins, Anal. Chem., 2006, 78, 4969–4975 CrossRef CAS PubMed.
- T. Y. Chen and J. K. Chen, Ferritin Immobilization on Patterned poly(2-hydroxyethyl methacrylate) Brushes on Silicon Surfaces from Colloid System, Colloid Polym. Sci., 2011, 289, 433–445 CAS.
- J. K. Chen, J. H. Wang, C. C. Cheng and J. Y. Chang, Reversibly Thermoswitchable Two-Dimensional Periodic Gratings Prepared from Tethered poly(N-isopropylacrylamide) on Silicon Surfaces, ACS Appl. Mater. Interfaces, 2013, 5, 2959–2966 CAS.
- J. K. Chen, Z. Y. Chen, H. C. Lin, P. D. Hong and F. C. Chang, Patterned poly(2-hydroxyethyl methacrylate) Brushes on Silicon Surfaces Behave as “Tentacles” to Capture Ferritin from Aqueous Solution, ACS Appl. Mater. Interfaces, 2009, 1, 1525–1532 CAS.
- E. Burdukova, H. Li, N. Ishida, J. P. O'Shea and G. V. Franks, Temperature Controlled Surface Hydrophobicity and Interaction Forces Induced by poly(N-isopropylacrylamide), J. Colloid Interface Sci., 2010, 342, 586–592 CrossRef CAS PubMed.
- J. K. Chen, C. Y. Hsieh, C. F. Huang and P. M. Li, Characterization of Patterned poly(methyl methacrylate) Brushes Under Various Structures Upon Solvent Immersion, J. Colloid Interface Sci., 2009, 338, 428–434 CrossRef CAS PubMed.
- A.-W. Lee, C.-C. Hsu, Y.-Z. Liu, P.-L. Wei and J.-K. Chen, Supermolecules of poly(N-isopropylacrylamide) complexating Herring sperm DNA with bio-multiple hydrogen bonding, Colloids Surf., B, 2016, 148, 422–430 CrossRef CAS PubMed.
- J. K. Chen and T. Y. Chen, Fabrication of High-Aspect-Ratio poly(2-hydroxyethyl methacrylate) Brushes patterned on silica surfaces by very-large-scale integration process, J. Colloid Interface Sci., 2011, 355, 359–367 CrossRef CAS PubMed.
- L. Chen, M. Liu, H. Bai, P. Chen, F. Xia, D. Han and L. Jiang, Antiplatelet and Thermally Responsive poly(N-isopropylacrylamide) Surface with Nanoscale Topography, J. Am. Chem. Soc., 2009, 131, 10467–10472 CrossRef CAS PubMed.
- T. L. Chang, C. Y. Tsai, C. C. Sun, R. Uppala, C. C. Chen, C. H. Lin and P. H. Chen, Electrical Detection of DNA Using Gold and Magnetic Nanoparticles and Bio Bar-Code DNA Between Nanogap Electrodes, Microelectron. Eng., 2006, 83, 1630–1633 CrossRef CAS.
- J. T. Davis and G. P. Spada, Supramolecular Architectures Generated by Self-Assembly of Guanosine Derivatives, Chem. Soc. Rev., 2007, 36, 296–313 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra28270a |
| This journal is © The Royal Society of Chemistry 2017 |