Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecular and chemotypic variability of forskolin in Coleus forskohlii Briq., a high value industrial crop collected from Western Himalayas (India)†
Sharad Srivastava *a,
Ankita Misraa,
Priyanka Mishra
*a,
Ankita Misraa,
Priyanka Mishra b,
Pushpendra Shuklaa,
Manish Kumara,
Velusamay Sundaresanb,
Kuldeep Singh Negic,
Pawan Kumar Agrawald and
Ajay Kumar Singh Rawata
b,
Pushpendra Shuklaa,
Manish Kumara,
Velusamay Sundaresanb,
Kuldeep Singh Negic,
Pawan Kumar Agrawald and
Ajay Kumar Singh Rawata
aPharmacognosy and Ethnopharmacology Division, CSIR-National Botanical Research Institute Lucknow, U.P.226001, India. E-mail: sharad_ks2003@yahoo.com; Fax: +91-522-2205836; Tel: +91-522-2297818
bDepartment of Plant Biology and Systematics, CSIR-Central Institute of Medicinal and Aromatic Plants, Research Centre Bangalore, Karnataka, India
cNBPGR-R/S, Bhowali, Nainital, Uttarakhand, India
dNASF, ICAR, KAB-II, New Delhi-110011, India
First published on 27th January 2017
Abstract
C. forskohlii (willd.) Briq. is an industrially viable medicinal crop and is widely exploited for the therapeutic potential of its bioactive metabolite, forskolin. The present investigation aimed to explore the chemotypic variability of forskolin content and existing molecular diversity in the wild population of C. forskohlii from the Western Himalayan region of India. Twelve germplasm(s) from different populations were assessed for molecular fingerprinting (ISSR marker) and densitometeric quantification of forskolin. Two elite germplasms viz. NBC-24 (0.728%) and NBC-16 (0.641%) were obtained as the highest accumulator of forskolin with high genetic variability (92%). The UPGMA hierarchical clustering patterns revealed strong genetic grouping between the individuals corresponding to their geographical ranges. Mantel tests showed positive correlation (r = 0.354, p = 0.003) between molecular and chemical fingerprints that reflects the feasibility of the ISSR markers in analyzing genome information related to forskolin biosynthesis from varied phytogeography. Pearson correlation coefficient (0.102) between forskolin content with altitude gradient also denoted a positive correlation. However, the association of both genetic and chemical fingerprinting data with the geographic distance matrix was apparently negative (r = −0.234, p = 0.054; r = −0.067, p = 0.584) which meant that distance might be a predictor of population differentiation. Our study signifies the utility of metabolic and molecular fingerprints for identification of elite accessions and provides a lead to industry for commercial exploitability of Coleus species including its location specific commercial cultivation.
1 Introduction
Coleus forskohlii (willd.) Briq. (family Lamiaceae) is an industrially valuable medicinal plant that grows perennially in the tropical, subtropical and temperate parts of India, Nepal, Burma, Sri Lanka, Thailand, Pakistan and Sri Lanka, countries of the Arabic region, Africa and South America. It comes up well on the sun exposed dry hill slopes from 600 m to 1800 m altitude. In India, the well drained medium fertile soil of Tamil Nadu, Karnataka, Gujarat, Maharashtra, Rajasthan states are the major cultivation sites of C. forskohlii.1 The leaves, roots and tubers of this species are a rich source of biologically active labdane diterpene compounds named forskolin (7-beta-acetoxy-8,13-epoxy-1a,6β,9a-trihydroxy-labd-14-en-11-one). It has a unique property to resensitize the cell receptors by activating the enzyme, adenylyl cyclase which increases the levels of cyclic adenosine monophosphate (cAMP) and results in a broad range of positive physiological and biochemical effects. The Coleus crop has great economic potential as a source of forskolin used for treating wide range of clinical disorders viz. glaucoma, cardiovascular disorders, certain types of cancers, asthma, eczema, psoriasis, angina, obesity, hypertension etc.2–6The world market for botanical pharmaceuticals, fragrances, flavors and color ingredients exceed several billion dollars per year and the classic examples include taxol, vincristine, vinblastine, colchicines, artemisinin, podophylotoxin etc. The demand for forskolin has also been increased in recent time due to its high medicinal value and huge demand from pharmaceutical industries. However, a number of threats like habitat loss, climate change, unsustainable use and exhaustive collection of wild sources along with inadequate attempts for its replenishment and/or cultivation have enlisted this species as endangered.1 However, in recent years its cultivation had scaled up due to commercial importance of forskolin. To overcome this, large scale agro-cultivation practices of C. forskohlii have resulted in an annual production of about 100 tons from 700 ha in India.7
Diversity analysis is a prerequisite factor for exploiting the available genetic resources for improvement of plants species producing bioactive secondary metabolites having commercial applications. Molecular markers in association with biochemical traits have demonstrated their potential and wide range of applications in identifying genetic purity of germplasm stocks, understanding genome organization, genetic relationships, chromosome mapping, trait tagging, inheritance pattern and molecular breeding etc.8,9 To assess the genetic diversity in plants, PCR-based markers such as RAPD, ISSR, DAMD, and AFLP are the most common, as their application does not need any prior sequence information.9,10 ISSR is a powerful tool for identifying genetic information and has been widely used in genetic diversity studies with relatively lower analysis cost.11,12 A number(s) of chromatographic methods such as TLC, HPTLC, HPLC, GLC etc. are common methods employed for the quantification of forskolin in C. forskohlii.3 HPTLC has become a simple, rapid, reproducible, efficient and economic technique for forskolin quantification.13,14
Despite being a high value commercial crop, information pertaining to interrelationship between morphology, chemistry and molecular fingerprinting available on C. forskohlii are meagre. Few available reports suggest the variation in forskolin content is due to the climatic and genetic factors.15–17 However majority of these studies are restricted to the southern states of India such as Tamil Nadu and Karnataka where the main focus was given on crop cultivation. Therefore, data on the morphological, chemical and molecular diversity of C. forskohlii is sparsely available. In our recent report, we also demonstrated the phytogeographical variation and chemotaxonomic profile in natural population of C. forskohlii.15 In this present investigation, we aimed towards documenting the level of molecular diversity and variability in the content of potential metabolite forskolin in the wild population of C. forskohlii from the Western Himalayan region of India. We studied the molecular fingerprinting using ISSR marker and chemical profiling of forskolin by HPTLC. This study represents the first exploration of the genetic and chemical diversity of natural populations to identify elite chemotype(s) from Western Himalayas, India.
2 Material and methods
2.1 Ethics statement
All necessary permits were obtained from the concerned authorities for the described field studies and plant collection. The permit was issued by PCCF, Uttarakhand and J & K, India.2.2 Plant materials
Fresh leaves and root of C. forskohlii were collected from twelve different natural locations of Western Himalayas, India. A random sampling of 5–10 individuals was collected from each population (collection site) and harvested tubers were subjected for extraction process. The number of sample(s) collected from individual site was restricted (5–10) due to holistic and judicial approach of collection keeping in mind that the study will not affect the biodiversity of targeted species and also to restore the ecological balance of collection sites. The passport data sheet was prepared for each sample after proper authentication by Dr Sharad Srivastava, Principal Scientist, Pharmacognosy and Ethnopharmacology Division, CSIR-NBRI and the voucher specimens were submitted at the herbarium of CSIR-NBRI, Lucknow, India. Leaf samples were dried directly on silica gel and held at −80 °C for DNA extraction and also preserved (80% ethanol) for the study on trichome. Root samples were air-dried under a shade and stored at 20 °C for chemical extraction.2.3 Anatomical studies and trichome micro-morphology
Samples (tubers) of C. forskohlii were prepared by standard freehand sectioning.18 The sections were cut with smooth stokes and transferred from the blade into a drop of water on a microscope slide. The water from the drop was then partially and neatly absorbed by tissue paper and processed the section further for staining. Double staining procedure was adopted to stain the section before the photomicrography which follows; finely sectioned piece of tuber was treated with 30% ethyl alcohol, followed by 70% ethyl alcohol for 2 min each. The section was than stained with safranin (50% in ethyl alcohol and water) for 2–3 min, followed to treatment with 50, 70 and 90% ethyl alcohol. Now, the safranin stained section was subjected to staining with fast green (90% in ethyl alcohol and water) for 2 min, followed by washing with 90% and 100% ethyl alcohol. The doubled stained section was than treated with four gradient dilutions (50, 70, 90 and 100% in ethyl alcohol) of xylene. Finally, the section was sealed with Canada balsam (synthetic, microscopic adhesive grade) on glass slide. The morphology, structure and number of trichome(s) were analysed by light microscopy (Nikon Advance Research Microscope, Eclipse 80i, Nikon Instruments Inc., Americas).192.4 Standards and solvents for chemical analysis
The source for the forskolin standards was Sigma-Aldrich, USA and the other solvents (HPLC grade) viz. toluene, ethyl acetate and methanol were procured from Merck, Mumbai, India.2.5 Extraction of plant material and preparation of stock solution(s)
The shade dried roots of each C. forskohlii genotype were pulverised and screened through 40 mesh (USP standard) sieve. About 2 g fine powder sample(s) was macerated separately in 25 ml of methanol for 24 h. The methanol extract was then filtered through Whatmann no. 41 filter paper to remove any insoluble components. The procedure was repeated thrice for each sample. The pooled extracts were then concentrated under vacuum and yield (%) was calculated.Stock solution (1 mg ml−1) of standard forskolin and samples (10 mg ml−1) were prepared in methanol and diluted accordingly for further analytical work.
2.6 HPTLC condition(s) and quantification of marker
Working solution of samples (1 mg ml−1) and standard forskolin (0.1 mg ml−1) were prepared freshly from stock on same day for analytical work. Spot were applied as 6 mm-wide bands positioned 15 mm from the bottom and 15 mm from side of the plate, using Camag Linomat V automated TLC applicator (CAMAG Chemie-Erzeugnisse & Adsorptionstechnik AG, Muttenz, Switzerland) with nitrogen flow providing a delivery speed of 150 nl s−1 from application syringe.In the chromatographic profile, band (10 μl, methanolic extract) of different genotypes (1 mg ml−1) were applied in triplicate along with the reference standard of forskolin (5 μl). The chromatogram was scanned at 545 nm after development.20 These conditions were kept constant throughout the analysis of samples. Quantification was carried out on the basis of regression equation of area vs. concentration of standard curve.
2.7 DNA isolation and PCR amplification
Total genomic DNAs from individual genotypes were isolated from the fresh leaf tissue using the modified cetyl trimethyl ammonium bromide (CTAB) protocol with minor modifications.21 Isolated DNA was checked for its quality and quantity by running on 0.8% agarose gel electrophoresis and spectrophotometric analysis (Nano Drop, ND-1000, USA), respectively. A set of 25 ISSR primers procured from the University of British Columbia (Vancouver, Canada) for DNA profiling and repeatability of amplification for the respective primers. Out of 25 primer pairs, a set of 10 ISSRs were selected for the present study based on their reproducibility, clarity and highly polymorphic nature of amplified products (Table 3). The reaction mixture (20 ml) was consisted of 25–30 ng of template DNA, 1× PCR Buffer, 200 μm each dNTPs, 10 pmol of each primer and 1 Unit of Taq DNA polymerase. Amplification was performed in a thermal cycler (Applied Biosystems, USA) programmed for initial denaturation at 94 °C for 5 min followed by 45 cycles of 94 °C for 1 min, annealing for 1 min at respective annealing temperature (Tm), an extension of 72 °C for 2 min and a final extension at 72 °C for 7 min. PCR amplified products were separated on 1.2% agarose gel stained with ethidium bromide. The gels were digitally photographed by Bio-Imaging Systems (Syngene, USA). Lambda DNA EcoR1-HindIII double digest was used as marker (Bangalore Genei, Bangalore, India) to know the size of the fragments.| Genotypes | Collection site | Voucher no. | Collection stage | GPS information | Season of collection | Soil type | ||
|---|---|---|---|---|---|---|---|---|
| Altitude (m) | Latitude (N) | Longitude (E) | ||||||
| NBC-07 | Chamba | 305407 | Flowering | 1480 | 30°20′52.93′′ | 78°23′49.88′′ | August | Loamy |
| NBC-08 | Dhanaulti road | 305408 | Flowering | 2089 | 30°25′31.17′′ | 78°14′13.58′′ | August | Sandy–loamy |
| NBC-11 | Pauri | 305411 | Flowering | 1693 | 30°08′51.52′′ | 78°46′28.75′′ | October | Stony |
| NBC-16 | Dwarikhal | 305416 | Flowering | 448 | 29°54′8.00′′ | 78°40′25.6′′ | January | Stony |
| NBC-17 | Bachhetikhal | 305417 | Flowering | 246 | 30°05′42.20′′ | 78°37′54.30′′ | January | Stony |
| NBC-18 | Kandikhal | 305418 | Flowering | 436 | 30°20′45.70′′ | 78°37′54.30′′ | January | Stony |
| NBC-19 | Budha kedar | 305419 | Flowering | 420 | 30°33′39.50′′ | 78°38′39.00′′ | January | Stony |
| NBC-23 | Diwakhal/Pauri | 305423 | Flowering | 457 | 29°48′41.30′′ | 79°02′26.70′′ | January | Stony |
| NBC-24 | Pabo | 305424 | Flowering | 336 | 30°06′14.10′′ | 78°52′7.90′′ | January | Stony |
| NBC-25 | Bhowali | 305425 | Flowering | 506 | 22°14′17.21′′ | 79°31′12.00′′ | January | Loamy |
| NBC-01 | Jammu-01 | 305401 | Flowering | 92 | 32.7°50′01′′ | 75°8′12.00′′ | August | Loamy |
| NBC-14 | Jammu-02 | 305414 | Flowering | 94 | 33°30′12′′ | 75°18′30.00′′ | December | Sandy–loamy |
| S. No. | Genotypes | Forskolin contenta |
|---|---|---|
| a Values are mean ± S.D (n = 3). | ||
| 1 | NBC-07 | 0.116 ± 0.05 |
| 2 | NBC-08 | 0.189 ± 0.01 |
| 3 | NBC-11 | 0.563 ± 0.01 |
| 4 | NBC-16 | 0.641 ± 0.01 |
| 5 | NBC-17 | 0.098 ± 0.01 |
| 6 | NBC-18 | 0.159 ± 0.01 |
| 7 | NBC-19 | 0.061 ± 0.01 |
| 8 | NBC-23 | 0.235 ± 0.05 |
| 9 | NBC-24 | 0.728 ± 0.05 |
| 10 | NBC-25 | 0.474 ± 0.01 |
| 11 | NBC-01 | 0.013 ± 0.01 |
| 12 | NBC-14 | 0.039 ± 0.025 |
| Marker | Sequence (5′-3′) | Tm °C | TNB | NPB (PPB%) | PIC | EMR | MI | RP |
|---|---|---|---|---|---|---|---|---|
| a R = (A, G), Y = (C, T), Tm, annealing temperature; TNB, total number of bands; NPB, number of polymorphic bands; PPB%, percentage of polymorphism; PIC, polymorphic information content; EMR, effective multiplex ratio; MI, marker index; RP, resolving power. | ||||||||
| UBC 826 | (AC)8 C | 52.5 | 5 | 5 (100.00%) | 0.43 | 5.40 | 2.32 | 3.50 |
| UBC 828 | (TG)8 A | 52 | 5 | 3 (60.00%) | 0.26 | 5.52 | 1.44 | 1.99 |
| UBC 841 | (GA)8 YC | 52 | 4 | 3 (75.00%) | 0.31 | 5.06 | 1.59 | 2.16 |
| UBC 844 | (CT)8 RC | 54 | 7 | 5 (71.42%) | 0.32 | 6.32 | 2.08 | 3.66 |
| UBC 845 | (CT)8 RG | 53 | 5 | 4 (80.00%) | 0.36 | 6.24 | 2.09 | 2.50 |
| UBC 848 | (CA)8 RG | 52.5 | 6 | 4 (66.66%) | 0.28 | 4.55 | 1.29 | 2.83 |
| UBC 855 | (AC)8 YT | 53 | 4 | 3 (75.00%) | 0.31 | 5.25 | 1.64 | 2.00 |
| UBC 856 | (AC)8 YA | 53 | 5 | 4 (80.00%) | 0.36 | 6.56 | 2.38 | 2.83 |
| UBC 866 | (CTC)6 | 57.2 | 6 | 5 (83.33%) | 0.40 | 5.69 | 2.28 | 4.16 |
| UBC 876 | (GATA)2(GACA)2 | 48 | 6 | 3 (50.00%) | 0.21 | 4.08 | 0.86 | 2.16 |
| Total | 53 | 39 | ||||||
| Average/primer | 5.3 | 3.9 (74.14%) | 0.32 | 5.46 | 1.80 | 2.78 | ||
2.8 Data and statistical analysis
ISSR-amplified DNA fragments (bands) were scored for the presence (1) or absence (0) irrespective of band intensity since each product of identical molecular weight was supposed to represent a single locus. A pair-wise matrix of similarity between genotypes was determined for the banding pattern using Jaccard's similarity coefficient22 in SPSS Statistics 17.0 software.23 Treecon software24 was used to assess the genetic relatedness within and among the species population using un weighted pair-group method with arithmetic average (UPGMA) based on Nei's (1973) pair-wise genetic distance.24 The robustness of each UPGMA node was evaluated by a bootstrapping analysis of each data set over loci for 1000 replicates.25 The polymorphic information content (PIC) was calculated26 for each primer. In order to determine the utility of each of the marker system, resolving power (RP), effective multiplex ratio (EMR) and marker index (MI) were also calculated.10 The software program POPGENE v1.31 was used to estimate genetic diversity parameters viz. Shannon's information index, percentage of polymorphic loci (PPL) and genetic distance.27 PCA was performed to highlight the resolving power of the ordination based on similarity coefficient of data realized from ISSR average similarity indices using SPSS statistics 17.0 software (SPSS Corporation, Chicago, IL, USA).2.9 Statistical analysis
Hierarchical cluster analysis of the samples was performed based on the variation pattern of forskolin content using the Pearson linear correlation coefficient in PAST software, version 2.15.28 Further principal component analysis (PCA) was performed to understand the grouping of genotypes based on correlation coefficient of data/variables. Karl Pearson correlation coefficient was determined to relate the forskolin content (%) with altitude (m) of each genotype.For correlation analysis, Mantel test29 was performed using 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 permutations carried out in XLSTAT©-Pro version 7.5 (2004, Addinsoft Inc., Brooklyn, NY, USA). The significance level was set at α = 0.05 to compute the matrix correlation (r) between the similarity matrices generated from different assays to test the goodness of fit between the molecular and chemical marker system. Finally, isolation by distance based geographic matrix was compared with genetic distance matrix using a Mantel test. Geographic distance matrix was realized based on latitude and longitude coordinates using Geographic Distance Matrix Generator (version 1.2.3).30
000 permutations carried out in XLSTAT©-Pro version 7.5 (2004, Addinsoft Inc., Brooklyn, NY, USA). The significance level was set at α = 0.05 to compute the matrix correlation (r) between the similarity matrices generated from different assays to test the goodness of fit between the molecular and chemical marker system. Finally, isolation by distance based geographic matrix was compared with genetic distance matrix using a Mantel test. Geographic distance matrix was realized based on latitude and longitude coordinates using Geographic Distance Matrix Generator (version 1.2.3).30
3 Results
The samples of C. forskohlii were collected from twelve different locations of Western Himalayas (Table 1) with variable altitude of 92.98–2089.9 meter(s). No distinct morphological features were observed among all the samples based on visual and metric measurement of foliage and tuber(s) (ESI Fig. 1a and b†). Roots were fibrous in nature with primary, secondary and tertiary rootlets, which on microscopic evaluation (T.S) revealed insignificant variation in anatomical features (ESI Fig. 2†).Surface imaging of leaves (upper and lower surface) showed the presence of both glandular (peltate and capitates) and non glandular type of trichomes (ESI Fig. 3†). Number of trichomes (per mm2 of leaf) on lower surface varied from 17 to 23 whereas in upper surface trichome frequency was slightly higher (25 to 35) (ESI Table 1†). However, the variation in trichome frequency was statistically insignificant (p > 0.05) among the germplasms.
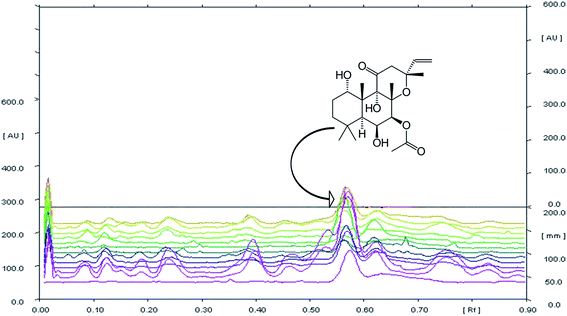
The chemical profiling of C. forskohlii was carried out by HPTLC to check the existing variability among the metabolites of collected germplasms. This aids in quality check and authentication of standard sample of Coleus in terms of forskolin, the major metabolite of industrial demand. The development of chromatogram was carried out by using different solvents (during method development) and the system consisting of toluene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate
ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (9
methanol (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05) was selected as for optimum separation of bioactive marker forskolin with high resolution and compact spots at Rf 0.56 ± 0.01.
0.05) was selected as for optimum separation of bioactive marker forskolin with high resolution and compact spots at Rf 0.56 ± 0.01.
Calibration of the methods used was performed on the basis of peak area versus reference standard concentration, when subjected to regression analysis. Method was calibrated at five different dilutions of standard forskolin (20–100 ng per spot). The linear regression equation was obtained with coefficient greater than 0.9998. Limit of detection (LOD) (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and limit of quantification (LOQ) (10
1) and limit of quantification (LOQ) (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) values were within the limit of acceptance and other statistical parameters were in accordance with the International Council for Harmonisation (ICH) guidelines. Stability of method was evaluated by repeated (n = 5) analysis of standard at single level (0.1 mg ml−1), standard deviation (6.083), RSD (0.357%) and variance (37.013) revealed that the method was stable under chromatographic conditions.15
1) values were within the limit of acceptance and other statistical parameters were in accordance with the International Council for Harmonisation (ICH) guidelines. Stability of method was evaluated by repeated (n = 5) analysis of standard at single level (0.1 mg ml−1), standard deviation (6.083), RSD (0.357%) and variance (37.013) revealed that the method was stable under chromatographic conditions.15
HPTLC chromatogram revealed the variable fingerprint profile of collected germplasm with respect to presence of known (forskolin) and unknown markers. It is noteworthy to mention that, only forskolin was taken into consideration for analysis of chemotypic variability among collected germplasm, firstly due to high relative abundance of forskolin in each germplasm(s) as compared to other unknown chemical marker(s). Secondary, forskolin is potentially bioactive and is majorly responsible for therapeutic activity of the species. In addition to this, the species was also targeted for commercial/industrial exploitation due to its forskolin content only. Overlay spectra of chromatogram (Fig. 1) showed the presence of forskolin in each sample and densitometeric scanning revealed the variation in the content of metabolite (Table 2). The existing variation among the germplasms was found to be significant (p > 0.01). Forskolin content varied from 0.013–0.728% (dry weight of root). The maximum forskolin content was found in NBC-24 (Pebo) and minimum in NBC-01 (Jammu).
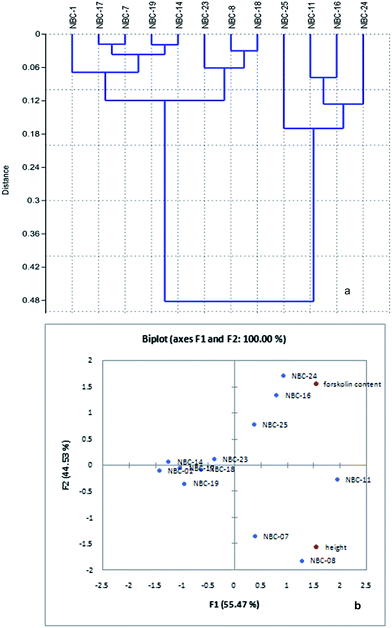
Karl Pearson correlation coefficient (0.102) between forskolin content with altitude revealed that there is a positive but week correlation exists among the two parameters. Cluster analysis of C. forskohlii genotypes based on forskolin content bifurcate the genotype group into two major clusters, cluster I and cluster II (Fig. 2a). NBC-25, NBC-11, NBC-16 and NBC-24 were grouped into single node in respect to relative concentration of forskolin (%) with mean value of 0.474, 0.563, 0.641 and 0.728% respectively. The remaining eight germplasms (NBC-01, NBC-17, NBC-07, NBC-19, NBC-14, NBC-23, NBC-08 and NBC-18) were clustered together in other branch. NBC-01 from Jammu and NBC-24 from Himachal Pradesh region showed maximum diversion in terms of forskolin as depicted in the cluster analysis. PCA analysis based on chemical distances has provided a biplot representation of the 12 germplasms accordingly. PCA derived on the basis of mean variables illustrated that the first two components, F1 and F2 were accounted for 55.47 and 44.53% variation respectively. The segregation of the population based on forskolin content with respect to altitude gradient has been shown in Fig. 2b.
C. forskohlii is an important medicinal plant with high commercial value due to its active phytochemical forskolin. In the present investigation, quantification of forskolin by validated HPTLC method was carried out for identification of the elite chemotype(s) to provide basic lead to industry for commercial exploitability including its location specific commercial cultivation. ISSR technique was employed for measuring genetic diversity within the collected genotypes. Besides, correlation between altitude and forskolin content was also established. The HPTLC analysis of twelve collected germplasms showed significant variation in forskolin content. Four elite germplasms viz. NBC-24 (0.728%), NBC-16 (0.641%), NBC-11 (0.563%) and NBC-25 (0.474%) of C. forskohlii were observed from our study were located in Himachal Pradesh. The genotypes from Jammu (NBC-01 and NBC-14) exhibited least amount (0.013 and 0.039%) of forskolin content. Previously, a number of investigators used different chromatographic techniques to determine forskolin content in C. forskohlii genotypes collected from the central and southern parts of India and they also found large variation of forskolin content.31–35 The variation in metabolite content may be due to several eco-geographical factors viz. climate, soil profile and microbial flora.36 Since there is a linear relationship exists between secondary metabolite content and maturity of the plants, all the genotypes were collected at the flowering stage. This was further confirmed by the findings from the anatomical and trichome studies. It is invariably evident in several reports that trichome morphology has significant effect on essential oil and secondary metabolite content of plant species at different stage of growth.37–39 Khatun et al. (2011) reported the pharmacognostic value of trichome morphology in C. forskohlii.40 We therefore investigated trichome characters and morpho-anatomical investigation to justify our study in correlation with existing literature. The presence of glandular trichome is a characteristic feature of the family Lamiaceae, to which the targeted species belongs. The insignificant (p > 0.05) variation in trichome frequency among the germplasms suggest that the collection was done at similar stage of maturity. This was further supported by morpho-anatomical observation, where no distinct variation was found among the samples. The germplasms were collected from the same phyto-geographical zone of Western Himalayas, where altitude and edaphic factors would be the key factors responsible for chemical variation15 and other environmental factors such as precipitation, humidity, temperature and light had no effect.
Molecular fingerprinting using ISSR marker showed high degree of polymorphism in C. forskohlii germplasm(s) from different regions of Western Himalayas. Ten ISSR primers generated a total of 53 products with an average of 5.3 products per primer. Among them, 39 (73.58%) products with an average of 3.9 products per primer were polymorphic and 14 (26.41%) products were monomorphic. The oligonucleotide sequences of these primers and the resultant multiple band patterns are summarized in Table 3. The representative ISSR profile of primer UBC 848 amplification has been shown in Fig. 3 and full-length blots/gels are presented in ESI Fig. 4.† The analysis showed the percentage of polymorphic band (PPB) ranging from 50% (UBC 876) to 100% (UBC 826) with an average value of 74.14% polymorphism per primer at the species level. PIC value was found in the range of 0.21 (UBC 876) to 0.43 (UBC 826) with an average of 0.32 per primer. Other parameters like effective multiplex ratio (EMR), marker index (MI) and resolving power (RP) were also found to be significant with an average value of 5.46, 1.08 and 2.78 per primer respectively. The observed number of alleles (na) ranged from 1.50 to 2.0 with mean value of 1.74 (SD = 0.44), while the effective number of allele (ne) ranged from 1.46 to 1.78 with mean value of 1.59 (SD = 0.41). The Nei's genetic diversity (h) and Shannon index (I) were estimated as 0.32 (SD = 0.20) and 0.46 (SD = 0.29), respectively (Table 4).
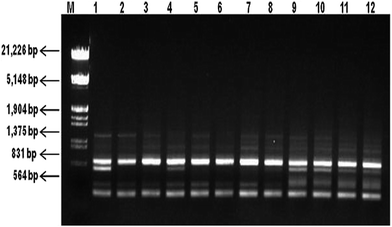 | ||
| Fig. 3 ISSR patterns (cropped) of Coleus forskohlii generated by primer UBC 848. Lane M is double-digested λ DNA (EcoRI and HindIII) DNA ladder and lanes 1–12 represent different Coleus forskohlii genotypes as listed in Table 1 (full-length gel is presented in ESI Fig. 4†). | ||
| S. No. | Diversity indices | Mean value (std. deviation) |
|---|---|---|
| 1 | na (observed number of alleles) | 1.74 (0.44) |
| 2 | ne (expected number of alleles) | 1.59 (0.41) |
| 3 | h (Nei's gene diversity) | 0.32 (0.20) |
| 4 | I (Shannon's information index of genetic diversity) | 0.46 (0.29) |
| 5 | NPL (no. of polymorphic loci) | 3.9 |
| 6 | PPL (percentage of polymorphism) | 74.14% |
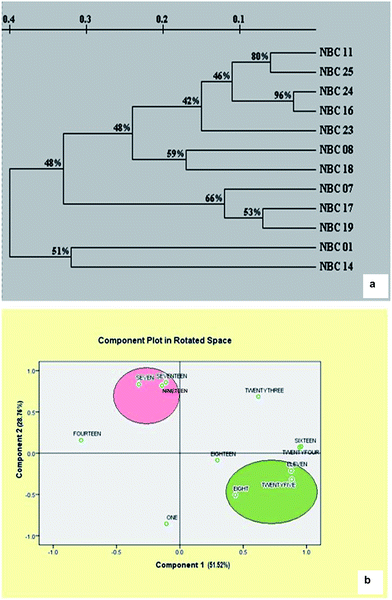
Cluster analysis revealed that the similarity index ranged from 0.02 (NBC-14 and NBC-11) to 0.92 (NBC-16 and NBC-24) with the mean value of 0.47 suggesting high level of genetic variability within the species. Dendrograms representing most probable genetic relationship between germplasms corresponding to molecular assay were presented in Fig. 4a. The dendrogram obtained from ISSR profiles showed two major clusters placing twelve germplasms into two major groups. It was interesting to observe that both the cluster grouped the genotypes similar to their geographical locations. Cluster I framed all the accessions from different places of Himachal Pradesh region whereas cluster II grouped both of the two genotypes from Jammu region (NBC-01 and NBC-14). Cluster I again sub-clustered into three major clusters with few outliers. Genotypes NBC-24 and NBC-16 from Himachal Pradesh regions shared the closest genetic relationships with 92% genetic similarity index which was also supported by 95% reliability node resulting through the bootstrap analysis. The PCA analysis based on genetic distances provided a spatial representation of the 12 genotypes following ISSR DNA fingerprinting (Fig. 4b). The two-dimensional PCA plot clearly differentiated all genotypes from Himachal Pradesh and Jammu region, thereby illustrating that the first three principal coordinate components accounted for 51.52, 28.76 and 9.06% variation, respectively and accounted for 89.34% of the genetic similarity variance.
A week but positive correlation (r = 0.354, p = 0.003) was achieved from the analysis of genetic and chemical fingerprint similarity matrices. Association of both genetic and chemical fingerprinting data with geographic distance matrix was apparently negative (r = −0.234, p = 0.054; r = −0.067, p = 0.584 respectively).
Genetic differences are considered as a standard and discrete means of identification as they get linked to genotypes for their genetic constitution.11 Molecular markers can more widely sample the genome in comparison to allozymes which reflect only a subset of genes. Highly polymorphic markers are essential to confidently identify genetic diversity. During the present study, a total of ten ISSR primers were screened on twelve C. forskohlii genotypes and the level of polymorphism (71.14%) suggested that these ISSR primers are viable markers to detect the genetic diversity in C. forskohlii at molecular level. The earlier work carried out on similar aspects lend support to our study, which also recorded high polymorphism (RAPD = 61.39%, ISSR = 68.75% and AFLP = 70.81%) in C. forskohlii genotypes collected from different places of central India.41 Recent report by Ahmad et al. (2013) also indicates high genetic diversity in C. forskohlii from central region of India but a lesser chemical diversity among the same samples.42
The result of the present study using ISSR markers revealed high level of genetic diversity with an average Nei's genetic diversity estimated as 0.32 (SD = 0.20). This high genetic diversity of C. forskohlii can be accounted predominantly to the out-crossing trait existing in this perennial herb.23 Along with this, life history traits i.e. seed dispersal mechanism, reproduction and geographical factors have great influence on the levels and distribution of genetic diversity. Geographically widespread species tend to maintain more genetic diversity than species with small geographical ranges.43 C. forskohlii germplasms collected from Western Himalayas, have wide topographical heterogeneity in its habitat which causes substantial changes in the environment, thus resulting in a strong isolation of populations because of drastic differences in phenology due to several factors like elevation gradients and mountain barriers which may have restricted the gene flow between the populations resulting in complex and varied genetic signatures.44
It is well known that the genetic structure and chemical fingerprints of plants are influenced by heredity and environmental factors. A correlation analysis was performed to gain a better understanding of their inter-relationships. Chemical profile and molecular signature were weekly correlated (r = 0.354, p = 0.004) with each other. Earlier work carried out on similar aspects lends support to our study.36 Therefore, the studied markers could play a vital role in dissecting the genome information related to forskolin biosynthesis and its influence by environmental factors. In agreement with previous reports, the present study also revealed negative correlation between genetic and chemical fingerprinting data with geographic distance matrix which implies that distance could be a predictor of population differentiation due to an isolation-by-distance effect, whereby geographically closer populations exchange more genes than more distant populations.36,45,46
The natural distribution of C. forskohlii extends from the sun-exposed arid and semi-arid hill slopes of the Himalayas to the eastward in Sikkim and Bhutan, Deccan Plateau, Eastern Ghats, Eastern Plateau and Western Ghats, India. The commercial cultivation of Coleus is mostly concentrated in Tamil Nadu and Karnataka states of India with some promising high yielding varieties.47 From our results, the positive correlation of forskolin content with altitude gradient was relevant and supported by fact plant species adapted to higher altitude contain increased concentration of active principles.48,49 This will also prompt future efforts towards good agronomic practices for large scale commercial cultivation of this plant adapted to higher elevations.
4 Conclusion
C. forskohlii is a commercially important medicinal plant and its chemotaxonomic investigation along with molecular fingerprinting would be helpful towards proper identification of elite chemotype(s) of forskolin for its pharmaceutical and therapeutic properties. The positive correlation between genetic and chemical fingerprints reflects the feasibility of the ISSR markers in analyzing genome information related to forskolin biosynthesis in C. forskohlii germplasms from different phytogeographical locations. In addition, C. forskohlii needs more attention toward the conservation and, it would be wise to save its population by developing better agronomy package and cultivation practices for different regions to facilitate large scale cultivation. Therefore, this report will serve as a platform for chemotaxonomic study of Coleus species, identification of elite accessions with known molecular makeup and to explore the best possible reason for chemical variation within and between the populations. This will also serve as good authentication tool for Coleus and also aid to upsurge the commercial cultivation using elite accessions and best agronomic practices for meeting industrial demand.Author contribution
Sharad Srivastava and Ankita Misra has done collection, chemotypic analysis, manuscript writing; Priyanka Mishra and V. Sundaresan contributed with genotypic studies; Pushpendra Shukla and Manish Kumar has done botanical and phytochemical studies; KS Negi helped in collection from Uttarakhand; Pawan K. Agarwal and AKS Rawat has done extensive editing and manuscript framing along with help in collection from Himalayan regions.Competing financial interests
There is no competing interest.Acknowledgements
The authors are thankful to Directors, CSIR-NBRI and CSIR-CIMAP, Lucknow, India for providing the facilities to carry out this research. Authors are also thankful to NASF, Indian Council of Agriculture Research, New Delhi, India for financial support (No. 7938).References
- R. Singh, S. P. Gangwar, D. Singh, R. Singh, R. Pandey and A. Kalra, Medicinal Plants, 2011, 3, 1–7 CAS.
- V. D. Wagh, P. N. Patil, S. J. Surana and K. V. Wagh, J. Postgrad. Med., 2012, 58, 199–202 CrossRef CAS PubMed.
- M. B. Patel, East Cent. Afr. J. Pharm. Sci., 2010, 13, 25–32 CAS.
- R. H. Rupp, N. J. De Souza, and A. N. Dohadwalla, Proceedings of the international symposium on forskolin: its chemical, biological and medical potential. Hoechst India Limited, Bombay, 1986 Search PubMed.
- K. Kamini, S. Ashashri and G. P. N. Lalit, Int. J. Of Ayur. Herbal Med., 2013, 3, 1106–1113 Search PubMed.
- G. M. A. Lakshmanan and S. Manikandan, Int. Lett. of Nat. Sci., 2015, 28, 1–9 Search PubMed.
- B. S. Shivkumar, R. Manjunath, A. N. S. Chandrashekhar and C. K. Suresh, J. Med. Aromat. Plant Sci., 2006, 28, 589–592 Search PubMed.
- P. Pandotra, A. P. Gupta, M. K. Husain, Gandhiram and S. Gupta, Biochem. Sys. Ecol., 2013, 48, 281–287 CrossRef CAS.
- M. Sarwat, G. Nabi, S. Das and P. S. Crit, Crit. Rev. Biotechnol., 2012, 32, 74–92 CrossRef CAS PubMed.
- A. Kumar, P. Mishra, S. C. Singh and V. Sundaresan, Plant Syst. Evol., 2014, 300, 1409–1420 CrossRef.
- B. Bornet and M. Branchard, Plant Mol. Biol. Rep., 2001, 19, 209–215 CrossRef CAS.
- W. Yang, A. C. de-Oliveira, I. Godwin, K. Schertz and J. L. Bennetzen, Crop Sci., 1996, 36, 1669–1676 CrossRef CAS.
- S. Malathy and J. S. Pap, Journal of Spices and Aromatic Crops, 2011, 8, 153–157 Search PubMed.
- N. Soni, P. Patidar, D. Dubey and K. Dashora, Asian J. Res. Chem., 2012, 5, 1029–1032 Search PubMed.
- A. Misra, S. Srivastavaa, P. Srivastava, P. Shukla, P. K. Agrawal and A. K. S. Rawat, Ind. Crops Prod., 2016, 84, 50–58 CrossRef CAS.
- M. Govarthanan, A. Guruchandar, S. Arunapriya, T. Selvankumar and K. Selvam, Int. J. Biotechnol. Mol. Biol. Res., 2011, 2, 202–208 CAS.
- L. Hegde, T. V. Kumar and K. Himabindu, in Bioprospecting & Ethnopharmacology, ed. J. Bernáth, É. Németh, L. E. Craker and Z. E. Gardner, ISHS, Acta Hort 675, 2005 Search PubMed.
- S. E. Ruzin, Plant microtechnique and microscopy, Oxford University Press, New York, 1999 Search PubMed.
- W. C. Evans, Trease and Evans Pharmacognosy, Saunders Company Ltd., London, 14th edn, 1996 Search PubMed.
- S. Srivastava, S. Verma, A. Gupta, P. Kushwaha and A. K. S. Rawat, Chemotaxonomic studies of Forskolin in Coleus forskohlii Briq. from Different Phyto-Geographical Zones of India, in Traditional Medicine and Globalization: The Future of Ancient Systems of Medicine, ed. P. K. Mukherjee, Maven Publishers, 2014, pp. 322–330 Search PubMed.
- S. P. S. Khanuja, A. K. Shasany, M. P. Darokar and S. Kumar, Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils, Plant Mol. Biol. Rep., 1999, 17, 1–7 CrossRef.
- P. Jaccard, Bull. Soc. Vaudoise Sci. Nat., 1908, 44, 223–270 Search PubMed.
- Y. Van-De-Peer and R. Wachter, Comput. Appl. Biosci., 1994, 10, 569–570 CrossRef CAS.
- M. Nei, Proc. Natl. Acad. Sci. U. S. A., 1973, 70, 3321–3323 CrossRef CAS.
- J. Felsenstein, Evolution, 1985, 39, 783–791 CrossRef.
- R. I. Ruiz, J. Dendauw, E. Vanbockstaele, A. Depicker and M. De-Loose, Mol. Breed., 2000, 6, 125–134 CrossRef.
- F. Yeh, R. Yang, and T. Boyle, POPGENE version 1.31–Microsoft Windows-based freeware for population genetic analysis, Quick users' guide, University of Alberta, Edmonton Search PubMed.
- Q. Hammer, D. A. T. Harper and P. D. Ryan, Palaeontologia Electronica, 2001, 4, 1–9 Search PubMed.
- N. Mantel, Cancer Res., 1967, 27, 209–220 CAS.
- P. J. Ersts, Geographic Distance Matrix Generator (version 1.2.3). American Museum of Natural History, Centre for Biodiversity and Conservation, http://biodiversityinformatics.amnh.org/open_source/gdmg, accessed 08 June 2015 Search PubMed.
- S. Ahmad, E. T. Tamboli, M. Garg, M. Singh, Y. T. Kamal and M. Mujeeb, Planta Med., 2011 DOI:10.1055/s-0031-1273620.
- P. Narayanan, K. S. Laddha and K. G. Akamanch, Curr. Sci., 2002, 83, 945–946 Search PubMed.
- A. M. Saleem, P. B. Dhasan and M. R. M. Rafiullah, Pharmacogn. Mag., 2005, 1, 89–92 Search PubMed.
- B. T. Schaneberg and I. A. Khan, J. AOAC Int., 2003, 86, 467–470 CAS.
- E. T. Tamboli, M. Singh, Y. T. Kamal, M. Garg, R. Parveen, M. Mujeeb and S. Ahmad, Nat. Prod. Res., 2013, 27, 1737–1742 CrossRef CAS PubMed.
- W. Zhao, X. Shi, J. Li, W. Guo and C. Liu, PLoS One, 2014, 9 DOI:10.1371/journal.pone.0112869.
- R. Leandra, Ind. Crops Prod., 2013, 43, 692–700 CrossRef.
- E. Valkama, J. P. Salminen, J. Koricheva and K. Pihlaja, Ann. Bot., 2003, 91, 643–655 CrossRef CAS PubMed.
- S. Kokkini, R. Karousou and D. Vokou, Biochem. Syst. Ecol., 1994, 22, 517–528 CrossRef.
- S. Khatun, U. Cakilcioglu and N. C. Chatterjee, Biol. Diversity Conserv., 2011, 4, 165–171 Search PubMed.
- N. Tripathi, N. Saini and S. Tiwari, J. Crop Sci. Biotech., 2013, 16, 253–261 CrossRef.
- S. Ahmad, E. T. Tamboli and R. Parveen, Planta Med., 2013, 79, 103 Search PubMed.
- J. L. Hamrick and M. J. W. Godt, Philos. Trans. R. Soc. London, Ser. B, 1996, 351, 1291–1298 CrossRef.
- S. Yashaswini and M. Vasundhara, Int. Res. J. Pharm., 2011, 2, 47–58 Search PubMed.
- M. Slatkin, Evolution, 1993, 47, 264–279 CrossRef.
- Z. F. Wang, J. L. Hamrick and M. J. Godt, J. Hered., 2004, 95, 234–243 CrossRef CAS PubMed.
- http://agritech.tnau.ac.in/crop_improvement/variety_released/variety_release_2013.pdf.
- G. Korekar, P. Dolkar, H. Singh, R. B. Srivastava and T. Stobdan, LWT--Food Sci. Technol., 2014, 55, 157–162 CrossRef CAS.
- P. K. Bajpai, A. R. Warghat, P. Dhar, A. Kant, R. B. Srivastava and T. Stobdan, LWT--Food Sci. Technol., 2014, 59, 981–988 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra26190f |
| This journal is © The Royal Society of Chemistry 2017 |