Open Access Article
Open Access ArticleEnhanced proliferation and osteogenic differentiation of MC3T3-E1 pre-osteoblasts on graphene oxide-impregnated PLGA–gelatin nanocomposite fibrous membranes
Chuan Fu
a,
Haotian Bai†a,
Qi Hu†a,
Tianlin Gao†b and
Yunshen Bai† *a
*a
aDepartment of Orthopaedic Surgery, The Second Hospital of Jilin University, Changchun, TX: 130021, PR China. E-mail: BaiyunshenJilin@163.com
bSchool of Public Health, Jilin University, Changchun, 130021, P. R. China
First published on 27th January 2017
Abstract
Currently, combining biodegradable polymeric scaffolds with living cells for bone repair has received significant attention. Ideal bone tissue engineering scaffolds should be biocompatible, biodegradable, and mechanically robust and have the ability to regulate cell function. The aim of this study was to fabricate graphene oxide-impregnated poly(lactic-co-glycolic acid)–gelatin (GO–PLGA–Gel) nanofibrous matrices that stimulate osteoblast proliferation and differentiation for bone regeneration. The GO–PLGA–Gel nanofibrous matrices were comprised of interconnected continuous fibres with three-dimensional porous structure that were successfully fabricated via an electrospinning process. The morphology, surface properties, mechanical properties and chemical composition of nanofibrous matrices were characterized by SEM, ATR-FITR, XRD, a materials testing machine and water contact angle measurements. Subsequently, fluorescence staining and an MTT assay were utilized to observe the influence of the Gel and GO on the MC3T3-E1 cell proliferation and attachment in vitro. In addition, osteogenic differentiation was determined from the alkaline phosphatase activity (ALP), expression of osteogenic marker genes and alizarin red staining. The results demonstrated that the MC3T3-E1 cells attachment and proliferation on GO–PLGA–Gel nanofibrous matrices were much higher than on the pure PLGA nanofibrous matrices. More importantly, GO–PLGA–Gel nanofibrous matrices significantly increased the alkaline phosphatase activity (ALP), expression of osteogenesis-related genes and calcium deposition of MC3T3-E1 cells. Our data indicated that blending GO with Gel retained the osteogenesis nature of GO without negatively influencing the proliferation cell effect of the Gel. Therefore, it is concluded that the GO–PLGA–Gel nanofibrous matrices are versatile biocompatible scaffolds for bone tissue engineering.
Introduction
Tissue engineering is an interdisciplinary field for the restoration or regeneration of damaged tissues by the biomimetic fabrication of scaffolds with appropriate microenvironments to mimic the native extracellular matrix (ECM) and control cellular behaviours.1,2 In the last two decades, bone tissue engineering scaffolds with various architectural configurations and geometries have been developed for bone regeneration. To mimic the native extracellular matrix (ECM), various techniques have been employed, such as melt extrusion, emulsion templating, self-assembly, phase separation and electrospinning.3–6 Among these techniques, electrospinning is a simple and effective technique used to produce three dimensional fibrous meshes with diameters ranging from nanometres to micrometres, mimicking ECM structures such as collagen fibrils.7 Many studies have reported that various synthetic and natural polymers, including polycaprolactone, PLGA, collagen and chitosan, have been electrospun to fabricate potential bone scaffolds that are utilized as cell-supporting matrices for bone repair.8–10PLGA is one of the most promising degradable materials and attractive for tissue engineering applications due to its good mechanical properties, adjustable degradation rate, low immunogenicity, and acquisition of FDA approval for human therapy.11,12 PLGA-based electrospun nanofibrous scaffolds have been used for engineering a wide range of tissues such as skeletal muscle, cartilage and bone tissues.13–15 However, the poor bioactivity and hydrophobic properties of PLGA have limited its biological applications. One strategy to solve this problem is to blend PLGA with other biopolymers, such as silk fibroin, hyaluronic acid and gelatin to improve the bioactivity of PLGA.16–18 Gelatin is a natural biopolymer derived from high molecular weight collagen; it has been widely used in many aspects of tissue engineering because of its many advantages, including a biological origin, excellent biocompatibility and biodegradability.19 Furthermore, since gelatin is a denatured biopolymer, the selection of gelatin as a bone tissue engineering scaffold materials can avoid the immunogenicity and pathogen transmission problems related to collagen.20 Many studies have demonstrated that gelatin could efficiently improve the surface properties of electrospun nanofibrous scaffolds by enabling the cells to adhere on composite scaffolds more easily and proliferate to a sufficient number for tissue regeneration. Lee et al. fabricated poly(L-lactide-co-3-caprolactone) (PLCL)/gelatin composite electrospun nanofibrous scaffolds with different weight ratios and found that inclusion of 10% gelatin could significantly improve the cytocompatibility of PLCL/gelatin composite scaffolds.21 A systemic study reported that gelatin can enhance and improve the hydrophilicity, protein adsorption and bioactivity properties of electrospun scaffolds effectively; however, the gelatin alone possesses limited osteoinductive abilities that have restricted its regenerative stimulation of large bone defects. Therefore, to improve the osteogenic activity of gelatin-blended nanofibrous scaffolds, the addition of some other components to the composite with good osteoconductive properties might be needed.
Graphene oxide (GO) is an oxidized derivative of graphene and has many hydrophilic functional groups, such as hydroxyl and carboxyl groups, that present high dispersibility in aqueous solutions, better hydrophilicity and can server as anchor sites for binding with metal ions or nanoparticles.22 Previous studies have shown that GO can enhance both the mechanical properties of the substrates and the cellular behaviour.23–25 Moreover, it is worth mentioning that the GO can be efficiently functionalized with chitosan, dopamine, and other biomaterials through π–π stacking, electrostatic, and hydrophobic interactions.26,27 Furthermore, GO substrates can promote proliferation and differentiation of various cell lines, including induced pluripotent stem cells (iPSCs), C2C12 myoblasts and PC12 cells.28–31 Therefore, GO can act as an effective reinforcement filler in scaffold materials to improve their biological properties in areas such as cell adhesion, growth and differentiation. For example, Dai et al. found that incorporation of GO into PLGA nanofibers could facilitate the proliferation and osteogenic differentiation of mouse mesenchymal stem cells.32 We hypothesized that the GO can be used as a bio-building block for scaffold substrates to obtain an ideal scaffold with improved mechanical and biological properties.
In this study, hybrid nanofibers, from blending GO, PLGA and Gel, that are analogous to the natural ECM, were fabricated by an electrospinning technique. Our aim was to investigate whether these electrospun GO–PLGA–Gel nanofibrous matrices could be used as an ideal candidate for bone tissue engineering. For this purpose, we fully characterized the physicochemical and mechanical properties of GO–PLGA–Gel nanofibrous matrices using various techniques including scanning electron microscopy (SEM), attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR) spectroscopy, X-ray diffraction (XRD), contact angle measurements, and a materials testing machine. Furthermore, the GO–PLGA–Gel nanofibrous matrices were then subjected to cell culture (MC3T3-E1 cell) to evaluate the effect of GO and Gel on cell behaviours, including adhesion, proliferation, and differentiation.
Experiments
Materials
PLGA (molecular weight = 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, LA/GA = 75/25) was synthesized by the Changchun Institute of Applied Chemistry, Chinese Academy of Sciences (CIAC, China). GO was purchased from Chengdu Organic Chemicals Co. Ltd, China (thickness: 0.55–1.2 nm diameter: 0.5–3 μm). Bovine serum albumin (BSA) was obtained from Beijing Solarbio Science & Technology Co., Ltd. 1,1,1,3,3,3-Hexafluoroisopropanol (HFIP), gelatin, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT), and the BCA protein assay kit were purchased from Sigma-Aldrich (USA). The reagents for cell experiments were purchased from Gibco (USA).
000, LA/GA = 75/25) was synthesized by the Changchun Institute of Applied Chemistry, Chinese Academy of Sciences (CIAC, China). GO was purchased from Chengdu Organic Chemicals Co. Ltd, China (thickness: 0.55–1.2 nm diameter: 0.5–3 μm). Bovine serum albumin (BSA) was obtained from Beijing Solarbio Science & Technology Co., Ltd. 1,1,1,3,3,3-Hexafluoroisopropanol (HFIP), gelatin, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT), and the BCA protein assay kit were purchased from Sigma-Aldrich (USA). The reagents for cell experiments were purchased from Gibco (USA).
Fabrication of GO–PLGA–Gel fibrous matrices
GO–PLGA–Gel nanofibrous matrices were fabricated by electrospinning. Briefly, PLGA (20 wt%) was dissolved in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP). Gelatin was then dissolved in PLGA solution to prepare the composite solution of PLGA/Gel (PLGA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) gelatin = 9
gelatin = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The composite solution was then doped with 2 wt% graphene oxide, and homogenized for 12 h by ultrasonication to form a uniform suspension. After that, the mixture solution of PLGA, Gel and GO was directly electrospun onto the aluminium foil-covered collector (applied voltage: 20 kV, flow rate of the solution: 1 mL h−1, air gap distance: 20 cm; injection needle diameter: 0.61 mm). To remove the residual solvent, the GO–PLGA–Gel nanofibrous matrices were completely dried under vacuum at room temperature for 1 day. Meanwhile, the nanofibrous matrices were washed three times in absolute ethanol before vacuum drying in order to remove the HFIP completely.
1). The composite solution was then doped with 2 wt% graphene oxide, and homogenized for 12 h by ultrasonication to form a uniform suspension. After that, the mixture solution of PLGA, Gel and GO was directly electrospun onto the aluminium foil-covered collector (applied voltage: 20 kV, flow rate of the solution: 1 mL h−1, air gap distance: 20 cm; injection needle diameter: 0.61 mm). To remove the residual solvent, the GO–PLGA–Gel nanofibrous matrices were completely dried under vacuum at room temperature for 1 day. Meanwhile, the nanofibrous matrices were washed three times in absolute ethanol before vacuum drying in order to remove the HFIP completely.
Physicochemical and mechanical characterizations of GO–PLGA–Gel nanofibrous matrices
The surface morphology of the nanofibrous matrices was examined by a scanning electron microscope (SEM, XL 30 ESEM-FEG, FEI). The average fibre diameter was statistically analyzed with Image J software. The chemical properties and crystalline structure of the matrices were analyzed with attenuated total reflectance (ATR)-FTIR spectroscopy (ATR-FTIR, Perkin Elmer, ATR-FTIR-2000) and X-ray diffraction (XRD, D8 ADVANCE, Germany). To analyse the surface hydrophobicity, water contact angles of the matrices were measured using a contact angle measurement system (VAC2000, AST). The mechanical properties of the matrices were tested by a universal mechanical testing machine (Instron 1121, UK), using fibre matrices with a width of 10 mm and initial length 30 mm.Protein adsorption capacity of GO–PLGA–Gel nanofibrous matrices
Bovine serum albumin (BSA) is a common kind of model protein that is used widely to determine the protein adsorption capacity of tissue engineered materials.33,34 In this study, BSA was selected as a model protein to determine the adsorption efficiencies of synthesized nanofibrous matrices. The matrices were tailored into 15 mm discs, fitted into a tube, and incubated with 10 mL of BSA solution (10 mL, 2 mg mL−1) under stirring at 150 rpm for 24 h. The adsorbed BSA concentration was determined through the concentration reduction of BSA within the samples using a BCA protein assay (wave length = 562 nm). Furthermore, rhodamine B labelled BSA (Sigma) solution (1 mg mL−1) was incubated in the matrices for 2 h. Then the samples were rinsed in PBS three times and mounted for visualization with a fluorescence microscope (TE2000-U, Nikon).Cell spreading, attachment, and proliferation assays
MC-3T3-E1 cells (obtained from the Cell Culture Centre of the Institute of Basic Medical Sciences Chinese Academy of Medical Sciences, China) were cultured in high-glucose DMEM supplemented with 10% foetal bovine serum, 100 U mL−1 penicillin, and 100 mg mL−1 streptomycin. The culture conditions were maintained at 37 °C in a humidified atmosphere containing 5% CO2 and the medium was renewed every 2 days. For cell seeding, PLGA, GO–PLGA, PLGA–Gel, and GO–PLGA–Gel nanofibrous matrices were prepared to match the inside diameter of a 24-well cell culture plate. The matrices were sterilized by immersing in 70% alcohol for 30 min, and seeded with MC3T3-E1 cells at 3.5 × 104 cells per mLper well in a 24-well plate. The medium was changed every 2 days. For the cell spreading and attachment examination, after the stipulated time period (1, 3 and 7 days), cell/matrices samples were rinsed twice with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. After washing three times with PBS, the samples were incubated with 0.5% Triton-X 100 in PBS for 5 min. Then, actin filaments were stained by incubating the samples with rhodamine phalloidin (Invitrogen, Eugene, OR, USA) and the cell nucleus was stained using 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, USA). The cell/matrices matrix samples were observed under a fluorescence microscope (TE2000-U, Nikon).Cell proliferation on the nanofibrous matrices was determined using the MTT assay. After 1, 3 and 7 days post seeding, 100 μL of MTT solution (5 mg mL−1 in PBS) was added to each well and the cells were incubated for an additional 4 h. Then, the medium was removed and the formed formazan crystals were dissolved by 800 μL of acidified isopropanol (0.2 mL of 0.04 N hydrochloric acid (HCl) in 10 mL of isopropanol). After the incubation period, 200 mL of the solution in each well was pipetted out into another 96 well plate for absorbance measurement at 540 nm on the multifunctional microplate scanner (Infinite M200, TECAN). The mean value of the four parallel samples for each material was used as the final result.
Alkaline phosphatase (ALP) activity
The ALP activity of MC3T3-E1 cells incubated for 7 and 14 days was determined by quantitation of the enzyme activity. Briefly, the medium of each well was carefully removed and cells were washed three times with PBS. Then, the cells were lysed in RIPA buffer before freezing at −80 °C for 30 min and thawing at 37 °C. ALP activity was measured by mixing 50 μL of pNPP solution in each well at 37 °C for 30 min in the dark. Following the incubation period, the reaction was terminated with 3 M NaOH and the OD was measured at 405 nm using the multifunction microplate scanner. The relative ALP activity was represented as the average OD values.Gene expression by quantitative PCR (Q-PCR)
The MC3T3-E1 cells cultured on various nanofibrous matrices for 7 days were also collected for evaluation of the osteogenesis-related gene expression. The total RNA was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer's protocol. The total RNA concentration and purity were detected by a Nanodrop Assay (Tecan M200), and the first strand cDNA was synthesized by reverse transcriptase as described by the M-MLV manual (Promega). The expression of osteogenic markers was quantified by qPCR SYBRGreen Mix Kit (Takara). The specific primer sequences for the target gene used for qRT-PCR, including the anti-runt-related transcription factor 2 (RUNX2), osteopontin (OPN) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are listed in Table 1. The specificity of the listed oligonucleotides was checked by a BLASTN® (Basic Local Alignment Search Tool) against the mouse RefSeq RNA database at NCBI. The qPCR amplification was conducted as follows: initial denaturation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 30 s, 58 °C for 1 min, and 72 °C for 1 min. The comparative threshold cycle method was used to analyse the Q-PCR results using iCycleriQ Detection System software with GAPDH as the reference gene. All results were quantified using the ΔΔCt relative quantification method.| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| RUNX 2 | 5-GCCCTCATCCTTCACTCCAAG-3′ | 5-GGTCAGTCAGTGCCTTTCCTC-3′ |
| OPN | 5-TCAGGACAACAACGGAAAGGG-3′ | 5-GGAACTTGCTTGACTATCGATCAC-3′ |
| GAPDH | 5-CAACCTGGTCCTCAGTGTAGC-3′ | 5-CGTGCCGCCTGGAGAAACCTGCC-3′ |
Mineralization
Calcium deposition was determined by alizarin red S (ARS) staining of the MC3T3-E1 cells. All nanofibrous matrices were seeded with MC3T3-E1 cells at a density of 2 × 104 cells per cm2 in 24-well plates to evaluate cell mineralization at two time points (14 and 21 days). At every specific time point, the cells were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature and then washed with acidic PBS (pH 4.2) three times. The cell/matrix samples were stained with alizarin red s solution (50 mM) for 20 min at 37 °C. After removal of alizarin red s solution, the cell/matrix samples were washed three times with deionized water and observed under a light microscope. The calcium was quantified using a cetylpyridinium chloride (CPC) treatment. The ARS-stained samples were treated with 1 mL of 10% CPC solution for 1 h to desorb calcium ions, and the absorbance of the solution was read at 540 nm in a multifunction microplate scanner. Furthermore, after 21 days of culture, cell/matrix samples were washed away with PBS and fixed with 4% paraformaldehyde for 3 h at room temperature. Samples were then washed with PBS three times, dehydrated through a graded series of ethanol (50%, 70%, 80%, 90%, 95%, and 100%) and dried under vacuum. Finally, the samples were coated with gold and observed by SEM.Statistical analysis
All quantitative data were analysed with OriginPro 8.0 (Origin Lab Corporation, USA) and presented as the mean ± standard deviation. Statistical differences were performed by one-way analysis of variance (ANOVA). A value of p < 0.05 was considered to be significant.Results and discussion
Physicochemical characterizations of GO–PLGA–Gel nanofibrous matrices
GO–PLGA–Gel nanofibrous matrices were formed by electrospinning the admixture of PLGA and Gel blended with GO. ESEM, XRD and ATR-FTIR spectroscopy were used for detailed characterizations of the samples. As shown in Fig. 1, a three-dimensional network structure with a smooth surface was fabricated by electrospinning. And the PLGA nanofibers keeps a wide distribution range, with a mean diameter of 1269 ± 376 nm. However, the average diameter of the nanofibres dramatically decreased to 1051 ± 318, 882 ± 295 and 787 ± 242 nm when GO, Gel or both were blended with PLGA, respectively. The integration of a 3D framework composed of nanofibres that are similar to the topological structure of natural ECM could create a potential scaffold for promoting cell adhesion and growth. Furthermore, GO nanosheets on the surface of the smooth nanofibres could not be observed. This phenomenon can be explained partly by the fact that the GO nanosheets embedded in the nanofibres were overlapped and aligned along the axial direction of the shape anisotropy of in the nanofibres.32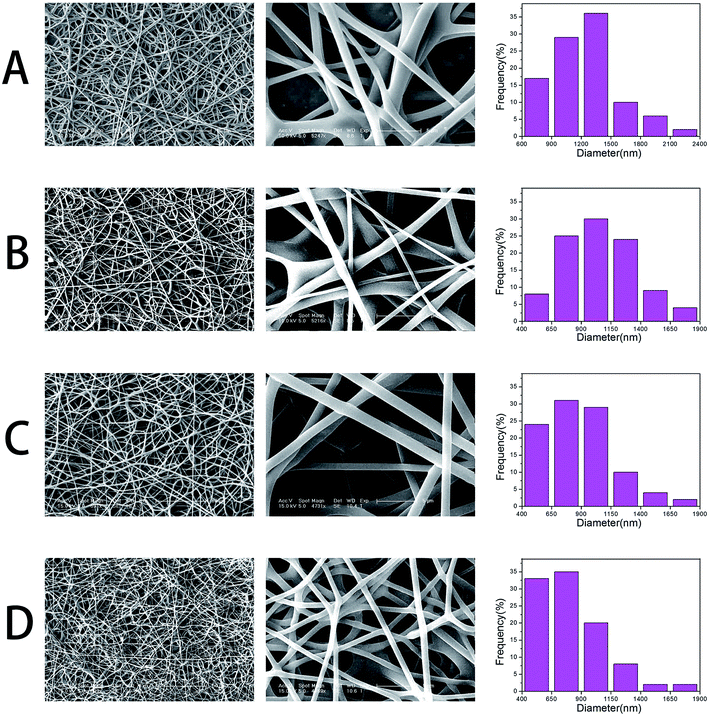 | ||
| Fig. 1 SEM micrographs and diameter distributions of nanofibrous matrices, (A) pure PLGA, (B) PLGA–Gel, (C) GO–PLGA and (D) GO–PLGA–Gel. | ||
The surface chemical properties of the PLGA, GO–PLGA, PLGA–Gel and GO–PLGA–Gel nanofibrous matrices were first characterized by ATR-FTIR spectra analysis. As shown in Fig. 2A, pure PLGA showed several characteristic peaks at 2990 and 2940 cm−1, 1753 cm−1, 1183 and 1083 cm−1 that were assigned to –CH3, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O, respectively. The FT-IR spectra of the PLGA–Gel and GO–PLGA–Gel nanofibrous matrices showed broad absorption bands at 3000–3600 cm−1 that corresponded to the symmetric stretching vibrations of –OH and –NH2. Furthermore, another band at 1550–1660 cm−1 was observed, which represented –NH2, which was derived from the Gel, and there were no –NH2 groups in the molecules of PLGA and GO. Compared with the PLGA nanofibrous matrices, the FTIR spectra of the GO–PLGA nanofibrous matrices showed no visible alteration, indicating that the GO was dispersed into the PLGA only by physical mixing instead of chemical reaction. XRD patterns of the GO and nanofibrous matrices were obtained to determine the mixture of GO in the PLGA nanofibrous matrices. As shown in Fig. 2B, the GO pattern shows a characteristic peak at 2θ ≈ 11°, corresponding to an interlayer spacing of 0.79 nm, which is the typical separation of layered GO.35 The XRD patterns of the GO–PLGA and GO–PLGA–Gel nanofibrous matrices exhibited a similar diffraction peak at 2θ ≈ 11°. In contrast, the PLGA and PLGA–Gel nanofibrous matrices did not show any diffraction peaks due to their amorphous structure. The results indicated the presence of GO and Gel in the GO-PLGA-Gel nanofibrous matrices.
O and C–O, respectively. The FT-IR spectra of the PLGA–Gel and GO–PLGA–Gel nanofibrous matrices showed broad absorption bands at 3000–3600 cm−1 that corresponded to the symmetric stretching vibrations of –OH and –NH2. Furthermore, another band at 1550–1660 cm−1 was observed, which represented –NH2, which was derived from the Gel, and there were no –NH2 groups in the molecules of PLGA and GO. Compared with the PLGA nanofibrous matrices, the FTIR spectra of the GO–PLGA nanofibrous matrices showed no visible alteration, indicating that the GO was dispersed into the PLGA only by physical mixing instead of chemical reaction. XRD patterns of the GO and nanofibrous matrices were obtained to determine the mixture of GO in the PLGA nanofibrous matrices. As shown in Fig. 2B, the GO pattern shows a characteristic peak at 2θ ≈ 11°, corresponding to an interlayer spacing of 0.79 nm, which is the typical separation of layered GO.35 The XRD patterns of the GO–PLGA and GO–PLGA–Gel nanofibrous matrices exhibited a similar diffraction peak at 2θ ≈ 11°. In contrast, the PLGA and PLGA–Gel nanofibrous matrices did not show any diffraction peaks due to their amorphous structure. The results indicated the presence of GO and Gel in the GO-PLGA-Gel nanofibrous matrices.
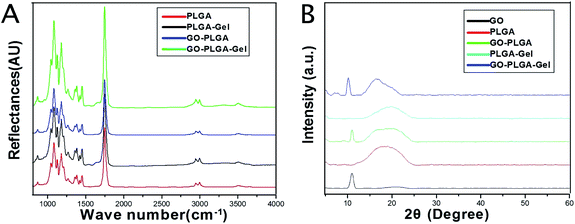 | ||
| Fig. 2 (A) FTIR spectra of PLGA, PLGA–Gel, GO–PLGA and GO–PLGA–Gel nanofibrous matrices. (B) XRD patterns of powdery GO, PLGA, PLGA–Gel, GO–PLGA and GO–PLGA–Gel nanofibrous matrices. | ||
The hydrophilicity of the materials played an important role in their interactions with the cells. We measured the water contact angles of these nanofibrous matrices surfaces to evaluate whether the hydrophilicity of the PLGA nanofibrous matrices was improved by the addition of GO and Gel. As shown in Fig. 3, the water contact angle was 99.8 ± 6.6° for the PLGA nanofibrous matrices, 89.5 ± 4.6° for the GO–PLGA nanofibrous matrices, 73.7 ± 3.5° for the PLGA–Gel nanofibrous matrices, and 59.4 ± 6.2° for the GO–PLGA–Gel nanofibrous matrices. The hydrophilicity of the PLGA nanofibrous matrices slightly increased by mixing with GO because of the presence of hydrophilic OH, C–O–C and COOH groups on the GO surface.23 The hydrophilic surface on the GO–PLGA nanofibrous matrices tends to be wetted easily by the water droplet. This is because the GO–PLGA nanofibrous matrices possesses expanded layers, consisting of hydrophilic GO sheets, bearing oxygen functional groups on their basal planes and edges. Furthermore, after blending with Gel, the contact angle of nanofibrous matrices was significantly decreased (p < 0.05) because gelatin is a more hydrophilic molecule than PLGA. Among the nanofibrous matrices, the lowest contact angle was obtained for the GO–PLGA–Gel nanofibrous matrices, indicating that the hydrophobic PLGA nanofibrous matrices are transformed into hydrophilic ones by blending with GO and Gel. The improvement in the hydrophilicity of the scaffold surface enhances the cellular behaviours, including the initial attachment, proliferation and differentiation.36 Therefore, GO–PLGA–Gel nanofibrous matrices can provide a suitable microenvironment for the attachment and proliferation of cells because they have a more hydrophilic surface than the other matrices.
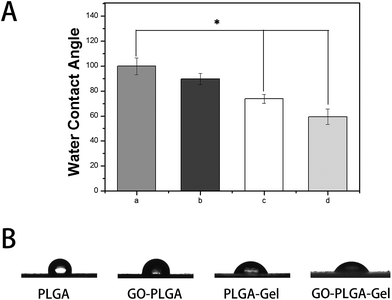 | ||
| Fig. 3 Water droplets (A) and contact angles (B) on the surface of nanofibrous matrices. (a) PLGA, (b) GO-PLGA, (c) PLGA-Gel, (d) GO-PLGA-Gel (p < 0.05, n = 5). | ||
Mechanical properties
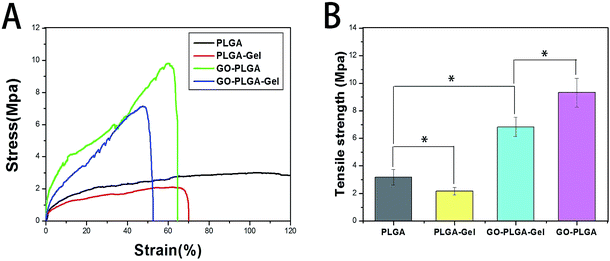
Scaffolds for tissue engineering should be mechanically robust. Fig. 4 shows the stress–strain behaviour and tensile properties of the PLGA, PLGA–Gel, GO–PLGA and GO–PLGA–Gel nanofibrous matrices. As shown in Fig. 4, the tensile strengths of the PLGA, PLGA–Gel, GO–PLGA and GO–PLGA–Gel nanofibrous matrices were approximately 3.18, 2.16, 6.82, and 9.32 MPa, respectively. Blending of the PLGA with gelatin caused a reduction in the mechanical strength. This decrease may be due to the weak physical properties of gelatin. In contrast, the tensile strength of the GO-impregnated nanofibrous matrices was significantly higher, by almost 2.9-fold and 3.1-fold, respectively, than those of the of the PLGA and PLGA–Gel nanofibrous matrices. Previous studies reported that the mechanical properties of the substrates were improved by the addition of GO because of the strong interfacial interactions between GO and the polymer matrix caused by enhanced chemical interactions due to hydrogen bonding between the GO nanosheets and PLGA polymer chains and by the high specific surface area of the nanosheets.37–39 Therefore, this result suggests that blending these nanofibrous matrices with GO improves the mechanical properties of the nanofibrous matrices, thereby making them more suitable for tissue engineering applications.Protein adsorptivity
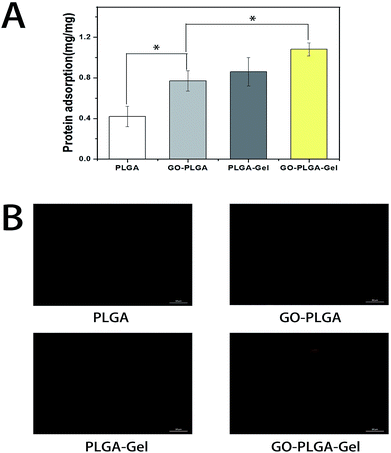
The adsorption of proteins onto the surface of materials is highly related to cell growth and colonization.40 Therefore, the adsorptions of bovine serum albumin (BSA) on the nanofibrous matrices was examined. As shown in Fig. 5A, it is apparent that the protein adsorption amounts on the PLGA nanofibrous were obviously lower than those of the GO or Gel incorporated ones. Conversely, the PLGA–Gel nanofibrous matrices exhibited higher protein adsorption capacities, nearly 2 times the adsorption rates than PLGA nanofibrous matrices (p < 0.05), consistent with the enhanced hydrophilicity of the surface. Moreover, it was found that GO–PLGA and PLGA–Gel nanofibrous matrices have similar protein adsorption capacities. Indeed, it has been reported that GO has a strong capability to adsorb various proteins, including cytochrome c, bovine serum albumin, ribonuclease A, and protein kinase A.41–43 In addition, the hydrophilic nature of the GO-incorporated nanofibrous matrices probably improved its affinity for proteins. Among the nanofibrous matrices, the highest protein adsorption was obtained for the GO–PLGA–Gel nanofibrous matrices, nearly 1.25 and 1.4 times of adsorption rates than those of PLGA–Gel and GO–PLGA nanofibrous matrices (p < 0.05), respectively. To directly observe the adsorbed proteins on different nanofibrous matrices, rhodamine B labelled BSA in PBS solution was incubated with the samples for 2 h. As shown in Fig. 5B, BSA adsorption on the nanofibrous matrices was clearly detected by fluorescence staining. However, there was weak fluorescence staining in the PLGA, indicating that there was a weak interaction between hydrophobic PLGA and protein molecules. On the other hand, the fluorescence intensity of the nanofibrous matrices was significantly enhanced by blending with Gel or GO (GO/Gel), indicating that the protein adsorption capacity of the PLGA nanofibrous matrices was significantly increased by blending with Gel and GO. The enrichment of proteins on the GO–PLGA–Gel nanofibrous matrices facilitated the cells adhesion and proliferation.Cell adhesion and proliferation
MC3T3-E1 cell adhesion and proliferation was investigated to evaluate whether the GO–PLGA–Gel nanofibrous matrices satisfied the basic requirements for bone tissue engineering. MC3T3-E1 cell proliferation on the nanofibrous matrices was investigated by an MTT assay. As shown in Fig. 6, the metabolic activity (OD value) of the MC3T3-E1 cells of all three groups increased gradually with the prolonged culture time. On the other hand, the proliferation of cells on the PLGA–Gel nanofibrous matrices with or without GO was significantly (p < 0.05) better than that on the other groups, indicating that the gelatin significantly stimulates cell proliferation. Moreover, it is noteworthy that the observed OD values of MC3T3-E1 cells on GO-incorporated nanofibrous matrices were higher than those of the pure PLGA (p < 0.05), which may be due to the improved hydrophilicity and protein adsorption capacity of nanofibrous matrices. The improvement in the hydrophilicity and protein adsorption capacity of the nanofibrous matrices surface can enhances the cellular behaviours, including the initial attachment, proliferation and differentiation.36,40 It was interesting that the GO–PLGA–Gel nanofibrous matrices presented a higher cellular activity than the GO–PLGA and PLGA–Gel nanofibrous matrices, indicating that the combination of GO and Gel had a synergistic effect in terms of cell proliferation.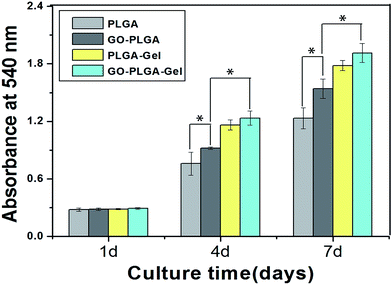 | ||
| Fig. 6 Proliferation of MC3T3-E1 cells cultured on the nanofibrous matrices for 1 to 7 days in vitro. (p < 0.05, n = 4). | ||
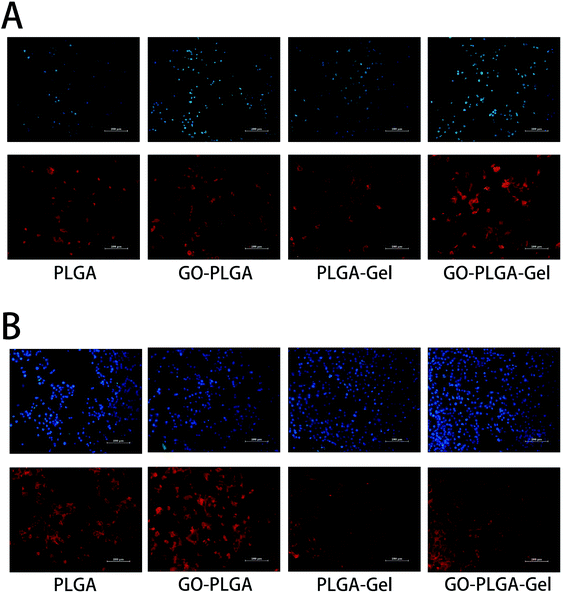
To visually investigate the effect of the GO and Gel for cell adhesion, the morphology of the MC3T3-E1 cells cultured on nanofibrous matrices at 1 and 4 days was observed using actin microfilaments (red) and nuclei (blue) fluorescent staining. As shown in Fig. 7, after 1 day post-seed, the results of the cytoskeleton and nuclear staining showed that there was no significant difference in cell the number of MC3T3-E1 cells cultured on the different nanofibrous matrices. However, cells on the nanofibrous matrices with Gel exhibited a greater spread with a better cytoskeleton than on nanofibrous matrices without Gel. This result indicated that the cell adhesion increased after blending with Gel. For PLGA–Gel nanofibrous matrices, gelatin functionalization was expected to be the major reason for the enhancement of cell adhesion because gelatin is a well-known cell adhesion mediator and its ability to increase cell attachment and proliferation has been previously described.44 After 4 days post-seed, compared to the pure PLGA groups (Fig. 6B), more adhered MC3T3-E1 cells were observed on the GO–PLGA, PLGA–Gel, and GO–PLGA–Gel matrices. The cell density ranking was as follows: GO–PLGA–Gel > PLGA–Gel > GO–PLGA > PLGA. This result is in accordance with the cell proliferation profiles. Moreover, the cells were fully extended on the GO–PLGA, PLGA–Gel, and GO–PLGA–Gel matrices, but more extended cells were observed on the matrices with Gel. Taken together, the data clearly indicate that the electrospun nanofibrous matrices containing a small amount of GO and Gel were beneficial for cell growth and cell–cell communication.
Alkaline phosphatase activity
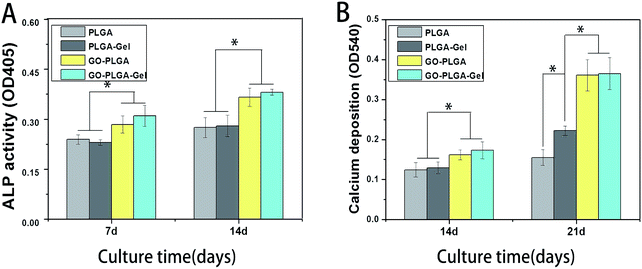
ALP enzyme activity, which is regarded as an early marker of osteoblast differentiation, was chosen to investigate the osteoinductive activity of the different nanofibrous matrices. Fig. 8A showed the ALP enzyme activities of MC3T3-E1 cells on different nanofibrous matrices at 7 and 14 days. A significantly higher ALP activity was detected in cells cultured on the GO–PLGA and GO–PLGA–Gel nanofibrous matrices than those on the PLGA and PLGA–Gel nanofibrous matrices (p < 0.05) that suggested the presence of GO stimulated an early stage of osteoblastic differentiation. Recent studies have demonstrated that GO could significantly increase the ALP expression of a variety of cells, such as MC3T3-E1 cells, human mesenchymal stem cells and human periodontal ligament stem cells.32,45–47 Zhao et al. found that GO coated quartz substrata could increase the ALP activity of MC3T3-E1 cells and promote osteogenic differentiation.48 In our case, GO-incorporated nanofibrous matrices apparently stimulated ALP expression of the MC3T3-E1 cells, which is in accordance with Zhao's work. Moreover, the OD values of ALP activity on the PLGA–Gel and GO–PLGA–Gel nanofibrous matrices did not exhibit significant difference with PLGA and GO–PLGA respectively, suggesting that Gel played little role in the early osteoblast differentiation of the MC3T3-E1 cells.Mineralization
The bioactivity of the bone implants was evaluated by examining the in vitro mineralization of the osteoblasts cultured in the bone implants. The capacity of minerals deposition is a late stage marker of osteogenic differentiation that can be used to conform that MC3T3-E1 cells entered into the mineralization phase to deposit mineralize ECM. After 14 and 21 days of culturing, the cells were stained with the alizarin red s that was utilized to determine calcium mineralization on the nanofibrous matrices quantitatively and qualitatively. Fig. 9 shows the optical images of the ARS staining for the PLGA, PLGA–Gel, GO–PLGA and GO–PLGA–Gel nanofibrous matrices. At day 14, the alizarin red s staining showed slight reddish dots on both GO-incorporated nanofibrous matrices but almost no positive stains were found over PLGA and PLGA–Gel nanofibrous matrices. Slightly enhanced staining was observed at day 21 for all nanofibrous matrices but the GO-incorporated nanofibrous matrices showed more intense ARS staining compared to the PLGA and PLGA–Gel matrices, indicated that GO can facilitates the calcium mineralization of MC3T3-E1 cells. A recent study reported that the GO-functionalized nanocomposites were able to elevate not only the ALP activity but also calcium deposition in MC3T3-E1 preosteoblasts.27 Quantitative cell mineralization was conducted after extracting the ARS with 10% CPC to evaluate the calcium-rich mineral deposits by MC3T3-E1 cells. As shown in Fig. 8B, the total calcium content was found to increase with time. At days 14 through 21, the total calcium content in the deposited minerals on the GO-incorporated nanofibrous matrices was significantly higher than that of the PLGA and PLGA–Gel matrices. Furthermore, after culturing for 21 days, the calcium mineral deposition in the PLGA–Gel nanofibrous matrices was greater than that in the PLGA matrices. We speculated that the negatively charged carboxylate groups on the surface of the gelatin could facilitate the binding of cells and calcium, which improved the nucleation of the HA. Therefore, it was concluded that the GO–PLGA–Gel nanofibrous matrices can promote the osteogenic differentiation of MC3T3-E1 cells.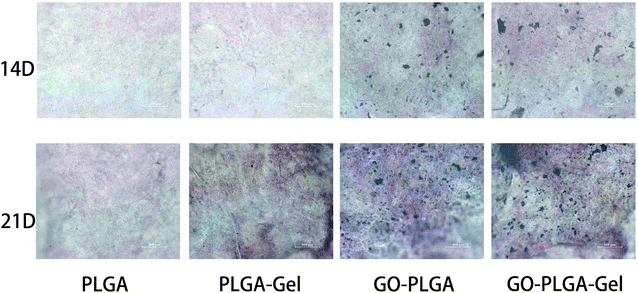 | ||
| Fig. 9 Alizarin red staining of MC3T3-E1 cells cultured on PLGA, PLGA–Gel, GO–PLGA and GO–PLGA–Gel nanofibrous matrices at 14 and 21 days. | ||
To better observe the effect of the different nanofibrous matrices on cell mineralization in cultured MC3T3-E1 cells, SEM images of the four kinds of nanofibrous matrices after 21 days incubation were acquired. The SEM images (Fig. 10) showed that the MC3T3-E1 cells were well coated on all nanofibrous matrices but cells growing on nanofibrous matrices with GO were more densely mineralized than those on matrices without GO. Compared to the PLGA matrices, the cells grown on the surface of the PLGA–Gel matrices had increased mineralized nodule formation. The SEM results also depicted the same trends observed from ARS staining (Fig. 8), demonstrating that the GO–PLGA–Gel nanofibrous matrices facilitate the calcium mineralization of MC3T3-E1 cells.
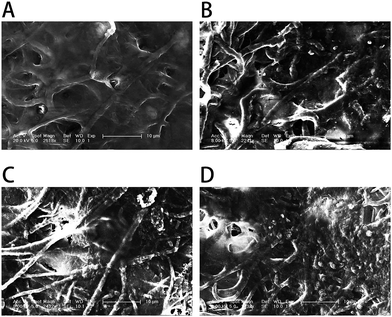 | ||
| Fig. 10 SEM images of MC3T3-E1 cells on (A) PLGA, (B) PLGA–Gel, (C) GO–PLGA and (D) GO–PLGA–Gel nanofibrous matrices after 21 days of mineralization. | ||
Quantitative real-time polymerase chain reaction
The expression of the osteogenesis-related genes during differentiation had characteristic changes that may be key markers in osteoblast differentiation. RUNX2 is an early differentiation marker observed at the early stage of differentiation, OPN expression is observed at the middle/late stage of differentiation.49 The expression of osteogenesis-related genes (RUNX2 and OPN) of the MC3T3-E1 cells after 7 days of culture on these nanofibrous matrices was quantitatively analysed using qRT-PCR and shown in Fig. 11. Among the genes, the expression levels of RUNX2 and OPN were higher in the GO–PLGA and GO–PLGA–Gel matrices, which is indicated that the GO alone or combined with Gel could enhanced the osteogenic differentiation of the MC3T3-E1 cells. It has been reported that GO could promote the osteogenic differentiation of MC3T3-E1 cells by increasing COL-I, BSP, Runx2 and OCN expression.47,50 It was interesting that the expression levels of OPN were higher in the GO–PLGA–Gel nanofibrous matrices than those in the GO–PLGA matrices, however, there was no significant difference in RUNX2 expression between the GO–PLGA–Gel and GO–PLGA nanofibrous matrices. We speculated that Gel improved the nucleation of HA that could better induce the osteogenic differentiation of MC3T3-E1 cells during the middle/late stages of differentiation.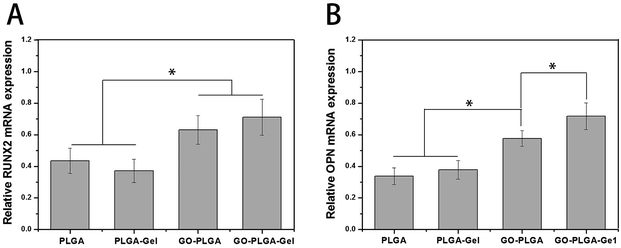 | ||
| Fig. 11 Quantitative real-time PCR analysis of osteogenesis-related gene expression of RUNX2 (A) and OPN (B) after MC3T3-E1 cells were cultured for 7 days. (p < 0.05, n = 3). | ||
The biodegradable polymeric implants for bone reconstruction have received significant attention because of their many advantages, including low immunogenicity, non-toxicity and absorption in the human body.51 Ideal bone tissue engineering scaffolds should mimic the extracellular matrix (ECM) and regulate cell function. Recently, many studies have demonstrated the application of electrospinning as a platform technology to generate artificial scaffolds composed of biocompatible polymers and various biomaterials for bone tissues repair. The primary advantage of the biodegradable electrospun scaffold in present study is that the electrospun scaffolds are highly porous, and have a high specific surface area and ECM-like nanotopography.52,53 The superior biodegradability of the PLGA and gelatin composite in the forms of a scaffold has been reported in many studies. Previous studies have demonstrated that the electrospun PLGA/gelatin nanofibrous scaffolds possessed higher cell adhesion and proliferation than the pure PLGA nanofibrous scaffolds.54,55 Therefore, the gelatin was utilized in this study to improve the biocompatibility of the PLGA electrospun scaffold. The results of this study showed that the protein adsorption and hydrophilicity of the nanofibrous matrices were increased by incorporation of gelatin due to its hydrophilic properties. The attachment and proliferation of MC3T3-E1 cells were increased significantly due to incorporation of gelatin. This might be due to the improved hydrophilicity of the nanofibrous matrices and higher protein adsorption. Furthermore, gelatin, a partially hydrolysed collagen, possesses the RGD sequences of collagen, making it highly effective for cell adhesion.44 However, our results revealed that the gelatin can efficiently improve the surface properties of PLGA nanofibrous matrices and promote the attachment and proliferation of cell, but the limited osteoinductive ability and poor mechanical properties of Gel-containing nanofibrous matrices should be considered when they were applied in bone tissue engineering.
To order to improve the mechanical properties, biocompatibility, and osteoinductive ability of the PLGA–Gel nanofibrous matrices, GO was employed in this study for modification of the nanofibrous matrices. Graphene and its derivatives that possess a higher availability and lower cell cytotoxicity than nanotubes, can promote the growth and osteogenesis differentiation of cells.27,32,45,50 Furthermore, GO–gelatin composites can mimic the proteins present in the extracellular matrix that can induce and assemble bone like apatite that is close to natural bone. The results of this study showed that the levels of ALP activity, calcium deposition and osteogenic gene expression of the MC3T3-E1 cells were increased significantly by incorporation of GO. More importantly, GO have greatly improved the mechanical properties of the PLGA–Gel nanofibrous matrices for clinical use. In addition, we found that the combination of Gel and GO had a synergistic effect in terms of hydrophilicity, protein adsorption and osteogenic induction. Thus, we can speculate that the GO–PLGA–Gel nanofibrous matrices may be successful bioactive polymeric scaffold for bone defect reparation. In our future study, a systematical investigation on bone tissue regeneration with polymeric scaffold containing Gel and GO will be undertaken in vivo.
Conclusions
This study was designed to develop a biomimetic scaffold with a three-dimensional porous structure similar to that of the natural ECM by electrospinning a blend of PLGA, 10 wt% gelatin, and 2 wt% grapheme oxide. Our results demonstrated that doping of GO and Gel into nanofibrous matrices caused slight increase in the hydrophilicity, and protein adsorption capacity, and significant improvement in mechanical properties. Moreover, the results suggest that Gel and GO are excellent candidates for proliferation and osteogenic differentiation of MC3T3-E1 cells, respectively. In particular, the GO and Gel showed the additive effect of promoting proliferation and osteogenic differentiation of MC3T3-E1 cells. Above all, GO–PLGA–Gel nanofibrous matrices have excellent biocompatibility, as well as suitable physicochemical and mechanical properties, and are promising biocomposite scaffolds for bone tissue engineering.References
- G. J. Lai, K. T. Shalumon, S. H. Chen and J. P. Chen, Carbohydr. Polym., 2014, 111, 288–297 CrossRef CAS PubMed.
- S. P. Nukavarapu, S. G. Kumbar, J. L. Brown, N. R. Krogman, A. L. Weikel, M. D. Hindenlang, L. S. Nair, H. R. Allcock and C. T. Laurencin, Biomacromolecules, 2008, 9, 1818–1825 CrossRef CAS PubMed.
- H. Liu, X. Y. Gu, M. Hu, Y. Hu and C. Y. Wang, RSC Adv., 2014, 4, 16751–16758 RSC.
- S. E. Kim, J. Wang, A. M. Jordan, L. T. Korley, E. Baer and J. K. Pokorski, ACS Macro Lett., 2014, 3, 585–589 CrossRef CAS PubMed.
- R. Murugan and S. Ramakrishna, Tissue Eng., 2007, 13, 1845–1866 CrossRef CAS PubMed.
- L. J. Zhang and T. J. Webster, Nano Today, 2009, 4, 66–80 CrossRef CAS.
- R. E. Scarber, A. D. Salaam, V. Thomas, G. M. Janowski and D. Dean, J. Biomater. Tissue Eng., 2013, 3, 440–447 CrossRef CAS.
- M. Ngiam, S. S. Liao, A. J. Patil, Z. Y. Cheng, C. K. Chan and S. Ramakrishna, Bone, 2009, 45, 4–16 CrossRef CAS PubMed.
- S. Pajoumshariati, S. K. Yavari and M. A. Shokrgozar, Ann. Biomed. Eng., 2016, 44, 1808–1820 CrossRef PubMed.
- M. F. Al Rez, H. Fouad, E. Laourine, M. Hild, D. Aibibu, C. Cherif, A. Mahmood, S. Abuelreich, M. Manikandan, S. W. Gosavi, M. Hashem, S. G. Ansari, F. S. Al-Mubaddel, Y. A. Elnakady and M. Alqahtani, Sci. Adv. Mater., 2015, 7, 2427–2435 CrossRef CAS.
- M. V. Jose, V. Thomas, K. T. Johnson, D. R. Dean and E. Nyalro, Acta Biomater., 2009, 5, 305–315 CrossRef CAS PubMed.
- T. Gao, N. Zhang, Z. Wang, Y. Wang, Y. Liu, Y. Ito and P. Zhang, Macromol. Biosci., 2015, 15, 1070–1080 CrossRef CAS PubMed.
- J. S. Choi, S. J. Lee, G. J. Christ, A. Atala and J. J. Yoo, Biomaterials, 2008, 29, 2899–2906 CrossRef CAS PubMed.
- Z. L. Wang, Y. Wang, P. B. Zhang and X. S. Chen, RSC Adv., 2015, 5, 96725–96732 RSC.
- A. Khojasteh, F. Fahimipour, M. B. Eslaminejad, M. Jafarian, S. Jahangir, F. Bastami, M. Tahriri, A. Karkhaneh and L. Tayebi, Mater. Sci. Eng., C, 2016, 69, 780–788 CrossRef CAS PubMed.
- M. Farokhi, F. Mottaghitalab, M. A. Shokrgozar, J. Ai, J. Hadjati and M. Azami, Mater. Sci. Eng., C, 2014, 35, 401–410 CrossRef CAS PubMed.
- L. Y. Cheng, X. M. Sun, B. Li, C. M. Hu, H. L. Yang, Y. G. Zhang and W. G. Cui, J. Mater. Chem. B, 2013, 1, 4428–4437 RSC.
- M. Mehrasa, M. A. Asadollahi, K. Ghaedi, H. Salehi and A. Arpanaei, Int. J. Biol. Macromol., 2015, 79, 687–695 CrossRef CAS PubMed.
- E. J. Chong, T. T. Phan, I. J. Lim, Y. Z. Zhang, B. H. Bay, S. Ramakrishna and C. T. Lim, Acta Biomater., 2007, 3, 321–330 CrossRef CAS PubMed.
- K. Su and C. M. Wang, Biotechnol. Lett., 2015, 37, 2139–2145 CrossRef CAS PubMed.
- J. Lee, G. Tae, Y. H. Kim, I. S. Park and S. H. Kim, Biomaterials, 2008, 29, 1872–1879 CrossRef CAS PubMed.
- D. A. Dikin, S. Stankovich, E. J. Zimney, R. D. Piner, G. H. B. Dommett, G. Evmenenko, S. T. Nguyen and R. S. Ruoff, Nature, 2007, 448, 457–460 CrossRef CAS PubMed.
- O. J. Yoon, C. Y. Jung, I. Y. Sohn, H. J. Kim, B. Hong, M. S. Jhon and N. E. Lee, Composites, Part A, 2011, 42, 1978–1984 CrossRef.
- O. J. Yoon, I. Y. Sohn, D. J. Kim and N. E. Lee, Macromol. Res., 2012, 20, 789–794 CrossRef CAS.
- S. L. Wu, X. D. Zhao, Z. G. Cui, C. T. Zhao, Y. Z. Wang, L. Du and Y. H. Li, Int. J. Nanomed., 2014, 9, 1413–1421 Search PubMed.
- Y. D. Ravichandran, T. Villaret and R. Rajesh, J. Indian Chem. Soc., 2015, 92, 649–651 CAS.
- J. Cheng, H. Y. Liu, B. J. Zhao, R. Shen, D. Liu, J. P. Hong, H. Wei, P. X. Xi, F. J. Chen and D. C. Bai, J. Bioact. Compat. Polym., 2015, 30, 289–301 CrossRef CAS.
- G. Y. Chen, D. W. P. Pang, S. M. Hwang, H. Y. Tuan and Y. C. Hu, Biomaterials, 2012, 33, 418–427 CrossRef CAS PubMed.
- Y. C. Shin, J. H. Lee, M. J. Kim, S. W. Hong, B. Kim, J. K. Hyun, Y. S. Choi, J.-C. Park and D.-W. Han, J. Biol. Eng., 2015, 9, 22 CrossRef PubMed.
- Y. C. Shin, J. H. Lee, L. H. Jin, M. J. Kim, Y. J. Kim, J. K. Hyun, T. G. Jung, S. W. Hong and D. W. Han, J. Nanobiotechnol., 2015, 13, 11 CrossRef PubMed.
- K. Zhang, H. Zheng, S. Liang and C. Gao, Acta Biomater., 2016, 37, 131–142 CrossRef CAS PubMed.
- Y. Luo, H. Shen, Y. X. Fang, Y. H. Cao, J. Huang, M. X. Zhang, J. W. Dai, X. Y. Shi and Z. J. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 6331–6339 CAS.
- C. Zhao, S. Pandit, Y. Fu, I. Mijakovic, A. Jesorka and J. Liu, RSC Adv., 2016, 6, 38124–38134 RSC.
- N. Zhang, T. Gao, Y. Wang, Z. Wang, P. Zhang and J. Liu, Mater. Sci. Eng., C, 2015, 46, 158–165 CrossRef CAS PubMed.
- M. Hazarika and T. Jana, Compos. Sci. Technol., 2013, 87, 94–102 CrossRef CAS.
- G. Zhao, Z. Schwartz, M. Wieland, F. Rupp, J. Geis-Gerstorfer, D. L. Cochran and B. D. Boyan, J. Biomed. Mater. Res., Part A, 2005, 74, 49–58 CrossRef CAS PubMed.
- L. Y. Yeo and J. R. Friend, J. Exp. Nanosci., 2006, 1, 177–209 CrossRef CAS.
- I. S. Chronakis, J. Mater. Process. Technol., 2005, 167, 283–293 CrossRef CAS.
- I. Armentano, M. Dottori, D. Puglia and J. M. Kenny, J. Mater. Sci.: Mater. Med., 2008, 19, 2377–2387 CrossRef CAS PubMed.
- R. L. Qi, X. Y. Cao, M. W. Shen, R. Guo, J. Y. Yu and X. Y. Shi, J. Biomater. Sci., Polym. Ed., 2012, 23, 299–313 CrossRef CAS PubMed.
- M. Patila, L. V. Pavlidis, A. Kouloumpis, K. Dimos, K. Spyrou, P. Katapodis, D. Gournis and H. Stamatis, Int. J. Biol. Macromol., 2016, 84, 227–235 CrossRef CAS PubMed.
- B. Cai, K. B. Hu, C. M. Li, J. Jin and Y. X. Hu, Appl. Surf. Sci., 2015, 356, 844–851 CrossRef CAS.
- H. Shen, M. Liu, H. X. He, L. M. Zhang, J. Huang, Y. Chong, J. W. Dai and Z. J. Zhang, ACS Appl. Mater. Interfaces, 2012, 4, 6317–6323 CAS.
- E. Rosellini, C. Cristallini, N. Barbani, G. Vozzi and P. Giusti, J. Biomed. Mater. Res., Part A, 2009, 91, 447–453 CrossRef PubMed.
- L. Jin, J. H. Lee, O. S. Jin, Y. C. Shin, M. J. Kim, S. W. Hong, M. H. Lee, J. C. Park and D. W. Han, J. Nanosci. Nanotechnol., 2015, 15, 7966–7970 CrossRef CAS PubMed.
- K. O. Park, J. H. Lee, J. H. Park, Y. C. Shin, J. B. Huh, J. H. Bae, S. H. Kang, S. W. Hong, B. Kim, D. J. Yang, D. W. Han and J. H. Yeum, Appl. Spectrosc. Rev., 2016, 51, 540–551 CrossRef CAS.
- Q. Zhou, P. S. Yang, X. L. Li, H. Liu and S. H. Ge, Sci. Rep., 2016, 6, 10 CrossRef PubMed.
- C. H. Zhao, X. Z. Lu, C. Zanden and J. H. Liu, Biomed. Mater., 2015, 10, 9 Search PubMed.
- T. L. Gao, W. W. Cui, Z. L. Wang, Y. Wang, Y. Liu, P. S. Malliappan, Y. Ito and P. B. Zhang, RSC Adv., 2016, 6, 20202–20210 RSC.
- J. Ruan, X. S. Wang, Z. Yu, Z. Wang, Q. Xie, D. D. Zhang, Y. Z. Huang, H. F. Zhou, X. P. Bi, C. W. Xiao, P. Gu and X. Q. Fan, Adv. Funct. Mater., 2016, 26, 1085–1097 CrossRef CAS.
- P. Y. Zhou, X. S. Cheng, Y. Xia, P. F. Wang, K. D. Zou, S. G. Xu and J. Z. Du, ACS Appl. Mater. Interfaces, 2014, 6, 20895–20903 CAS.
- C. Huang, R. Chen, Q. F. Ke, Y. Morsi, K. H. Zhang and X. M. Mo, Colloids Surf., B, 2011, 82, 307–315 CrossRef CAS PubMed.
- W. J. Li, C. T. Laurencin, E. J. Caterson, R. S. Tuan and F. K. Ko, J. Biomed. Mater. Res., 2002, 60, 613–621 CrossRef CAS PubMed.
- S. R. Son, R. A. Franco, S. H. Bae, Y. K. Min and B. T. Lee, J. Biomed. Mater. Res., Part B, 2013, 101, 1095–1105 CrossRef PubMed.
- M. Mehrasa, M. A. Asadollahi, B. Nasri-Nasrabadi, K. Ghaedi, H. Salehi, A. Dolatshahi-Pirouz and A. Arpanaei, Mater. Sci. Eng., C, 2016, 66, 25–32 CrossRef CAS PubMed.
Footnote |
| † Current Address: Department of Orthopaedic Surgery, the Second Hospital of Jilin University, Changchun, TX: 130021, PR China. |
| This journal is © The Royal Society of Chemistry 2017 |