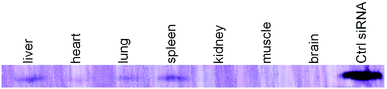Open Access Article
Open Access ArticleEfficient in vivo siRNA delivery by stabilized D-peptide-based lipid nanoparticles†
Tsogzolmaa Ganbold‡
,
Gerile Gerile‡,
Hai Xiao‡ and
Huricha Baigude *
*
School of Chemistry & Chemical Engineering, Inner Mongolia University, 235 West College Road, Hohhot, Inner Mongolia Autonomous Region 010020, PR China. E-mail: hbaigude@imu.edu.cn; Fax: +86-471-4993165
First published on 30th January 2017
Abstract
RNA interference (RNAi) has shown great potential for clinical treatment of a variety of diseases. As an effective post-transcriptional regulation, RNAi induces degradation of specific target mRNA, leading to a decreased level of disease related gene product, which could be resistant to conventional small molecule therapeutics. Because of the relatively small size of short interfering RNA (siRNA) as well as it's robustness in the design of efficient 20–22 nucleotide sequences targeting any known gene, delivery of siRNA in vitro and in vivo has achieved tremendous success by the design and fabrication of various carrier materials including peptides, lipids, and polymers. While siRNA delivery efficiency of these materials proved to be appreciable, their tissue specificity, biocompatibility and cytotoxicity still need much improvement. We report a novel siRNA delivery material designed by optimization of our previously reported peptidomimetic based lipoplex. The novel lipoplex, which bears a D-amino acid based dipeptide head group, possesses an ideal balance of increased stability in a tissue environment, and enhanced efficiency of tissue specific delivery as well as minimized cytotoxicity. The novel lipoplex carrying siRNA efficiently targeted mouse liver, initiated RNAi and knocked down the apoB dose dependently, which is confirmed by both RT-qPCR and Western blotting. Moreover, the novel lipoplex does not induce any apparent cytotoxicity, as confirmed by the measurement of liver enzyme levels in serum.
1. Introduction
RNA interference (RNAi) has shown great potential for the clinical treatment of a variety of diseases. Since the first proof of RNAi activity triggered by synthetic short RNA mimicking naturally occurring short interference RNA (siRNA) in the cells,1–3 tremendous effort has been made to realize the clinical potential of RNAi, especially for targeting therapeutic pathways that have little to no application for conventional small molecule based drug screening.4,5 The development of RNAi based therapeutics has shown enormous progress, with genetic diseases, cancer6–10 and virus infection11,12 as the major focus in clinical trials. Although promising outcomes have been reported for RNAi based clinical trials of diseases such as ATTR amyloidosis,13,14 no siRNA drug has been approved by the FDA so far. One of the main obstacles for the application of RNAi in clinical trials is the lack of an efficient and safe delivery system.15 Therefore, development of the siRNA delivery system is one of the most urgent tasks for the clinical application of RNAi.Clinically evaluated non-viral siRNA delivery systems comprise of small molecule conjugated siRNA, as well as polymers covalently or non-covalently complexed siRNA,16 and lipid nanoparticles.17 For example, previous studies have shown that cholesterol conjugated siRNA targeting liver apoB efficiently accumulated in mouse liver and induced a 60% reduction.5 More recently, multivalent N-acetyl galactosamine (GalNAc) conjugated siRNA has been reported.18–20 Combined with enhanced stabilization chemistry on the siRNA sequence, GalNAc conjugated siRNA efficiently targeted hepatocyte through binding to asialoglycoprotein receptors (ASGPR) on the hepatocytes. Although small molecule conjugated siRNA could eliminate cellular interference of heavily loaded carrier materials, the requirement for the high dosage of siRNA may affect the clinical feasibility, especially for cost efficiency of a heavily modified siRNA sequence. The higher dosage of siRNA may also raise the possibility of increased immune responses.
Polymeric nanoparticles, either based on a naturally occurring polymer such as chitosan and curdlan, or chemically synthesized polymers, have been tested for their efficiency as well as biocompatibility as siRNA delivery materials.16,21,22 Non-covalent approach for polymeric siRNA delivery depends on protonated amine groups, which could facilitate the endosomal escape of siRNA/polymer complex through proton sponge activity that breaks endosomal membrane through increased compartmental pH value. Tissue targeting features could be obtained by coupling targeted ligands such as galactose,23 GalNAc,24 transferrin,7 and siRNA which can also be covalently conjugated to the polymer backbone, through cleavable bonds such as disulfide bonds. The first confirmation of RNAi in a human was achieved by using a polymeric nanoparticle based on cyclodextrin containing a cationic polymer.7 A dynamic poly conjugate (PDC) bearing covalently conjugated siRNA as well as hydrolysable GalNAc ligands showed remarkable liver targeted siRNA delivery efficiency.25 A recent study demonstrated that a pH-responsive polymer based nanoplatform efficiently targeted a xenograft tumor and induced RNAi activity.21 Nonetheless, uncertainty of cytotoxicity as well as cellular clearance pathway raise concerns about the clinical safety of cationic polymer based nanoparticles.
Lipid nanoparticles (LNP) are the most widely investigated siRNA delivery material. In most cases, cationic lipid materials were used for the formulation of siRNA. Various cationic lipid molecules have been designed, formulated and tested for their siRNA delivery efficiency. With appropriate formulation with the fusogenic lipids and other ingredients, cationic lipids provided LNP with much higher siRNA delivery efficiency compared to other delivery materials. For example, stable nucleic acid lipid particles (SNALP) containing three siRNA sequences targeting viral genes protected rhesus macaques that were exposed to a lethal dose of the ebola virus.26 Parisina, a LNP targeting TTR has shown promising delivery efficiency in phase III clinical trials.13 A SNALP containing CSN5 siRNA efficiently targeted the orthotopic xenograft model of liver cancer, remarkably inhibiting tumor growth.27
However, artificial cationic head groups in the lipid components of LNP may potentially induce adverse effects for siRNA delivery. Therefore, cytotoxicity of LNP needs to be thoroughly investigated for assurance in clinical applications. On the other hand, lipid molecules containing natural amino acids may have enhanced feasibility for a biocompatible siRNA delivery system. Previously, we reported peptidomimetic based lipid nanoparticles for successful siRNA delivery in vivo.28 DoGo3 lipids showed enhanced biocompatibility while exhibiting high potential for liver targeted siRNA delivery at higher doses. In this study, in order to increase the siRNA delivery efficacy of DoGo lipids, we synthesized novel D-amino acid incorporated peptidomimetics, and evaluated their siRNA delivery efficiency in vitro and in vivo.
2. Experimental section
Chemicals and general methods
N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochlorides (EDCI), Boc-D-Orn-OH, Boc-D-Glu-OH, Boc-D-Asp-OH and oleylamine were purchased from Aladdin (Pudong, Shanghai, China). Dichloromethane (DCM) was distilled after drying over 4 Å molecular sieve. 1H NMR and 13C NMR were recorded on a Brucker Avance III 500 NMR spectrometer. MS was recorded on Finnigan LCQ Advantage MAX.Synthesis of D-DoGo1
D-DoGo1 was synthesized according to the previously reported method.28 Briefly, Boc-D-Glu-OH (1.0 g, 4.0 mmol) and oleylamine (2.1 g, 8.1 mmol) were dissolved in DCM (10 mL). Then EDCI reagent (1.7 g, 8.9 mmol) was added, and the mixture was stirred for 24 h under nitrogen atmosphere at room temperature. The solvent was removed under reduced pressure and the resulting syrup was dissolved in dichloromethane, followed by washing sequentially with 5% critic acid, distilled water, saturated sodium chloride and drying over anhydrous sodium sulfate. The product was purified by silica gel chromatography using dichloromethane and methanol (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as eluent to give D-DoGo1 (2.3 g). Yield: 61%. 1H NMR (DMSO-d6, ppm): 0.838–0.851 (t, 6H, –CH3), 1.235–1.550 (m, 48H, –CH2–), 1.971–1.980 (t, 4H, –NHCOCH2–, –NHCOCH2CH2–), 2.066–2.084 (m, 8H, –CH2CH
1) as eluent to give D-DoGo1 (2.3 g). Yield: 61%. 1H NMR (DMSO-d6, ppm): 0.838–0.851 (t, 6H, –CH3), 1.235–1.550 (m, 48H, –CH2–), 1.971–1.980 (t, 4H, –NHCOCH2–, –NHCOCH2CH2–), 2.066–2.084 (m, 8H, –CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2–), 2.982–3.058 (m, 4H, –CH2–NH–), 3.092 (t, 1H, –OCCH(NH–)CH2–), 5.319–5.350 (m, 4H, –CH
CHCH2–), 2.982–3.058 (m, 4H, –CH2–NH–), 3.092 (t, 1H, –OCCH(NH–)CH2–), 5.319–5.350 (m, 4H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 7.738–7.807 (m, 4H, –CONH–, –NH2). MS (ESI) calc. 746.20; found: 769.32 (M + Na+). Anal. calcd. for C46H87N3O4: C, 74.04; H, 11.75; N, 5.63. Found: C, 73.94; H, 11.05; N, 5.99.
CH–), 7.738–7.807 (m, 4H, –CONH–, –NH2). MS (ESI) calc. 746.20; found: 769.32 (M + Na+). Anal. calcd. for C46H87N3O4: C, 74.04; H, 11.75; N, 5.63. Found: C, 73.94; H, 11.05; N, 5.99.
Synthesis of D-DoGo2
Deproteced D-DoGo1 (1.2 g, 1.9 mmol), Di-Boc-D-Orn-OSu (0.8 g, 1.9 mmol) and triethylamine (0.26 mL, 1.9 mmol) was stirred in dichloromethane (6 mL) at room temperature for 24 h. The resulting compound, D-DoGo2, was purified in identical method described above. Yield: 1.5 g, 83%. 1H NMR (DMSO-d6, ppm): 0.838–0.852 (t, 6H, –CH3), 1.232–1.371 (m, 48H, –CH2–), 1.581–1.594 (m, 2H, NH2–CH2–CH2–), 1.692–1.719 (m, 2H, NH2–C(CO)H–CH2–), 1.786–2.062 (m, 4H, –NHCOCH2–, –NHCOCH2CH2–), 2.076–2.183 (m, 8H, –CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2–), 2.797 (t, 2H, NH2–CH2–), 2.994–3.133 (m, 4H, NHCOCH2–), 3.849 (m, 1H, –NHCOC(NH2)H–), 4.202–4.257 (m, 1H, OCCH(NH–)CH2–), 5.319–5.350 (m, 4H, –CH
CHCH2–), 2.797 (t, 2H, NH2–CH2–), 2.994–3.133 (m, 4H, NHCOCH2–), 3.849 (m, 1H, –NHCOC(NH2)H–), 4.202–4.257 (m, 1H, OCCH(NH–)CH2–), 5.319–5.350 (m, 4H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 7.795–8.596 (m, 7H, –CONH–, –NH2). MS (ESI) calc. 959.80; found: 960.87 (M + H+). Anal. calcd. for C56H105N5O7: C, 70.03; H, 11.02; N, 7.29. Found: C, 70.83; H, 10.92; N, 7.09.
CH–), 7.795–8.596 (m, 7H, –CONH–, –NH2). MS (ESI) calc. 959.80; found: 960.87 (M + H+). Anal. calcd. for C56H105N5O7: C, 70.03; H, 11.02; N, 7.29. Found: C, 70.83; H, 10.92; N, 7.09.
Synthesis of D-DoAo1
D-DoAo1 was synthesized according to the previously reported method. Briefly, a mixture of Boc-D-Asp-OH (1.0 g, 4.3 mmol) and oleylamine (2.3 g, 8.6 mmol) was stirred in DCM (10 mL) under nitrogen atmosphere. Then EDCI reagent (1.6 g, 8.6 mmol) was added, and the mixture was stirred at room temperature for 24 h. The solvent was removed and the resulting syrup was dissolved in dichloromethane, followed by washing sequentially with 5% critic acid, distilled water, saturated sodium chloride and drying over anhydrous sodium sulfate. The product was purified by silica gel chromatography using dichloromethane and methanol (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as eluent to give D-DoAo1 (1.9 g). Yield: 61%. 1H NMR (DMSO-d6, ppm): 0.852–0.865 (t, 6H, –CH3), 1.290–1.368 (m, 44H, –CH2–), 1.938–1.995 (m, 8H, –CH2CH
1) as eluent to give D-DoAo1 (1.9 g). Yield: 61%. 1H NMR (DMSO-d6, ppm): 0.852–0.865 (t, 6H, –CH3), 1.290–1.368 (m, 44H, –CH2–), 1.938–1.995 (m, 8H, –CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2–), 2.141–2.187 (m, 2H, –CH2CH2CH
CHCH2–), 2.141–2.187 (m, 2H, –CH2CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2–), 2.338–2.376 (m, 2H, –CH2CH2CH
CHCH2–), 2.338–2.376 (m, 2H, –CH2CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2–), 2.978–3.055 (m, 6H, NH2–C(CO)H–CH2–, –NHCOCH2–), 3.410–3.489 (t, 1H, –OCCH(NH2)CH2–), 5.290–5.353 (m, 4H, –CH
CHCH2–), 2.978–3.055 (m, 6H, NH2–C(CO)H–CH2–, –NHCOCH2–), 3.410–3.489 (t, 1H, –OCCH(NH2)CH2–), 5.290–5.353 (m, 4H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 7.807–7.856 (m, 4H, –CONH–, –NH2). MS (ESI): calc.: 731.65; found: 754.75 (M + Na+). Anal. calcd. for C45H85N3O4: C, 73.82; H, 11.70; N, 5.74. Found: C, 72.80; H, 11.45; N, 5.24.
CH–), 7.807–7.856 (m, 4H, –CONH–, –NH2). MS (ESI): calc.: 731.65; found: 754.75 (M + Na+). Anal. calcd. for C45H85N3O4: C, 73.82; H, 11.70; N, 5.74. Found: C, 72.80; H, 11.45; N, 5.24.
Synthesis of D-DoAo2
D-DoAo1 (1.9 g, 2.9 mmol), Di-Boc-D-Orn-OSu (1.2 g, 2.9 mmol) and triethylamine (0.4 g, 2.9 mmol) was stirred in dichloromethane (6 mL) at room temperature for 24 h. The resulting compound, D-DoAo2, was purified in identical method described above. Yield: 2.4 g, 84%. 1H NMR (DMSO-d6, ppm): 0.876–0.890 (t, 6H, –CH3), 1.087–1.486 (m, 44H, –CH2–), 1.582–1.627 (m, 6H, H2NCH2CH2–, –OCNHCH2CH2–), 1.693–1.740 (m, 2H, H2NCH2CH2CH2–), 1.938–1.994 (m, 8H, –CH2CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CHCH2–), 2.425–2.474 (t, 4H, NH2–CH2–, –OC(NH)CHCH2–), 2.790–2.802 (m, 2H, –CH2CONHCH2–), 2.899–2.956 (m, 2H, –OCNHCH2–), 3.068 (m, 1H, –NHCOC(NH2)H–), 4.572–4.586 (m, 1H, OCCH(NH–)CH2–), 5.311–5.359 (m, 4H, –CH
CHCH2–), 2.425–2.474 (t, 4H, NH2–CH2–, –OC(NH)CHCH2–), 2.790–2.802 (m, 2H, –CH2CONHCH2–), 2.899–2.956 (m, 2H, –OCNHCH2–), 3.068 (m, 1H, –NHCOC(NH2)H–), 4.572–4.586 (m, 1H, OCCH(NH–)CH2–), 5.311–5.359 (m, 4H, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–), 7.861–8.622 (m, 7H, –CONH–, –NH2). MS (ESI): calc.: 945.79; found: 946.78 (M + H+). Anal. calcd. for C55H103N5O7: C, 69.80; H, 10.97; N, 7.40. Found: C, 69.89; H, 11.12; N, 7.66.
CH–), 7.861–8.622 (m, 7H, –CONH–, –NH2). MS (ESI): calc.: 945.79; found: 946.78 (M + H+). Anal. calcd. for C55H103N5O7: C, 69.80; H, 10.97; N, 7.40. Found: C, 69.89; H, 11.12; N, 7.66.
Design of apoB siRNA
SiRNA (targeting mouse apoB) was custom synthesized by Takara Biotechnology Co. Ltd. (Dalian, Liaoning, China) according to the previous reports.29 The sequences are: apoB sense strand: 5′-GUCAUCACACUGAAUACCAAU-3′, apoB antisense strand: 5′-AUUGGUAUUCAGUGUGAUGACAC-3′. For biodistribution study, a 3′-biotin conjugated antisense strand was custom synthesized by the same company. The sequence is: 5′-AUUGGUAUUCAGUGUGAUGACAC-biotin-3′.Enzyme stability test
SiRNA (1 μL, 50 μM) and D- or L-DoAo2 (3 μL, 2 mg mL−1) were mixed at a N/P ratio of 3. Trypsin (6 μL, 5% solution) was added and the mixture was incubated at 37 °C for 2 h. The resulting complexes were subjected to agarose gel (1%) electrophoresis analysis.Cell culture, cytotoxicity measurement and in vitro siRNA transfection
Human cancer cell lines (HepG2, HeLa, A549, ATCC, Manassas, VA, USA) were maintained at 37 °C (with 5% CO2) in DMEM supplemented (10% fetal bovine serum), 100 μg mL−1 streptomycin and 100 U mL−1 penicillin. For cytotoxicity assay, cells were plated onto 96-well plates at 5000 cells per well in 100 μL of culture medium. Twenty-four hours after plating, lipoplex stock solutions were added to the cells in triplicate and incubated as described above. After a 24 h of incubation, cell viability was assayed using the MTT test. For in vitro transfection experiment, siGenome Non-Targeting siRNA Control, siGenome Human GAPDH siRNA (Dharmacon, Lafayette, CO, USA) were used. Cells (2.5 × 105 cells per well) were seeded into 12-well plates at the day before transfection. Cells were transfected with 0.5 mL per well of siRNA (25 nM) complexed to D-DoAo2 for 24 h. To determine mRNA levels in cell culture after siRNA treatment, total RNA was extracted with TRIZOL (Invitrogen, Carlsbad, CA, USA). The expression of mRNA was measured using iScriptTM Reverse Transcription Supermix for RT-qPCR and iTaqTM Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) for quantitative PCR. The sequences of primers used for RT-qPCR were as following: human β-actin, forward: 5′-ccaaccgcgagaagatga-3′, reverse: 5′-ccagaggcgtacagggatag-3′, human GAPDH, forward: 5′-agccacatcgctcagacac-3′, reverse: 5′-gcccaatacgaccaaatcc-3′.Preparation and characterization of D-DoAo2 lipoplex
Stock solutions of D-DoAo2, cholesterol and mPEG-OLA (Nanocs Inc., New York, NY, USA) were prepared in chloroform and mixed in 39%, 60% and 1% respectively. The dry film was then hydrated by adding 278 mM sucrose, followed by extruding through a membrane with 100 nm pore size for 20 times by using Avanti Mini-Extruder (Avanti Polar Lipids, Alabaster, Alabama, USA). The resulting lipoplex stock solution had total lipid concentration of 4.34 mg mL−1. The size distribution of D-DoAo2 lipoplex was measured on Malvern Zetasizer Nano S instrument. Zeta potential was measured on Malvern ZEN 1690 instrument. To generate siRNA–D-DoAo2 complex, the obtained lipoplex solution (200 μL) was mixed with an equal volume of a solution of siRNA in 278 mM sterile sucrose. After incubating at room temperature for 15 min, the siRNA–lipoplex formulations were administered intravenously through tail vein injection at the indicated doses.Organ distribution of siRNA/D-DoAo2 lipoplex
C57BL/6 mice (Vital River Laboratories, Beijing, China) were housed in plastic cages under controlled conditions and maintained according to the Guide for the Care Use of Laboratory Animals established by Inner Mongolia University (IMU, Hohhot, China). All animal experiments were performed in compliance with Institutional Guidelines for the Care and Use of Laboratory Animals established by Inner Mongolia University (Hohhot, Inner Mongolia Autonomous Region, China) after approval from Inner Mongolia University Animal Care and Use Committee.A six-week-old male C57BL/6 mouse was injected via tail vein with DdoAo2 lipoplex complexed with a 3′-biotin labeled siRNA. 12 h after the injection, total RNAs from liver, heart, lung, spleen, kidney, muscle, and brain were extracted. 300 ng of total RNAs were run on 14% denaturing gel, and the RNAs were transferred to a membrane, which was subsequently detected for biotin using Biotin Chromogenic Detection Kit (Thermo scientific, MA, USA) following the manufacturer's instruction.
In vivo siRNA delivery by D-DoAo2 lipoplex
Eight-week-old male C57BL/6 mice were maintained under a 12 h dark cycle. Mice were injected via the lateral tail vein with sucrose or D-DoAo2 complexes of apoB siRNA or its mm siRNA. Dosage either of 2.0 mg kg−1 or 4.0 mg kg−1 siRNA was delivered in a final volume of 0.15 mL. For analysis of target mRNA level, liver was dissected immediately after sacrifice of the mice and homogenized in a mortar. The total RNA was prepared with Trizol reagent and used for quantitative RT-PCR.Measurement of apoB mRNA and protein level
To determine apoB mRNA level in liver tissue after siRNA treatment, tissue samples were collected from three regions of the liver. Total RNA was extracted with Trizol and treated with DNase I before quantification. apoB mRNA levels were determined by qPCR. Mouse apoB primers were: forward, 5′-ttccagccatgggcaactttacct-3′; reverse, 5′-tactgcagggcgtcagtgacaaat-3′. Mouse β-actin primers were: forward, 5′-ctaaggccaaccgtgaaaag-3′, reverse, 5′-accagaggcatacagggaca-3′. The mice plasma was used for analysis of apoB protein level. Anti-apolipoprotein B antibody and anti-fibronectin antibody (Abcam, Cambridge, MA, USA) were used for analysis of serum apoB protein levels by Western blot. Liver enzyme levels (AST/ALT) were analyzed using an ELISA kit (R&D systems, Minneapolis, MN, USA).3. Results and discussion
Synthesis of lipid conjugated D-dipeptides
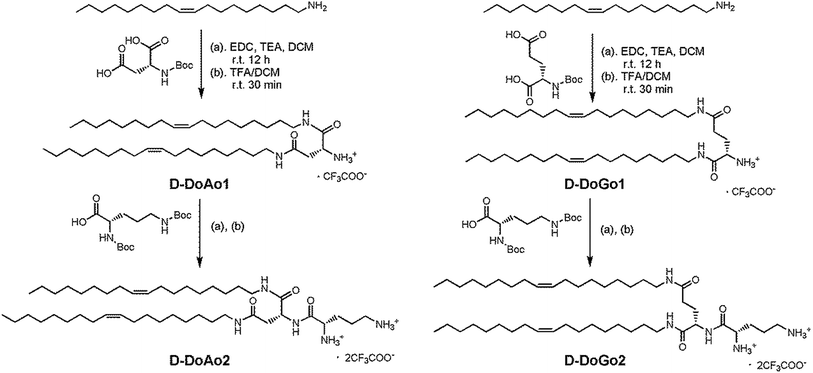
We synthesized 2 lipid conjugated dipeptides using D-aspartic acid and D-glutamic acid as well as D-ornithine coupled to oleylamine, respectively (Scheme 1). The rationales for such design are (1) to simplify the chemical process for lipid synthesis, and (2) to enhance in vivo siRNA delivery efficacy by stabilizing the head group of amphipathic lipids. The resulting lipids are denoted as D-DoAo2 and D-DoGo2. The goal of such design is to investigate the bio-stability, biocompatibility as well as siRNA delivery efficiency of D-peptide containing lipids after further formulation. The synthesis applied conventional liquid phase peptide coupling method, using EDCI as coupling reagent, which facilitated smooth reaction and gave appreciable yields for all 4 reactions (Tables 1 and 2). D-Amino acids showed equal reactivity as L-amino acids. Thus, lipid functionalized dipeptides bearing free alpha amine and free delta amine were successfully synthesized. Both D-DoAo2 and D-DoGo2 showed excellent solubility in aqueous medium, facilitating further formulation.| Lipids | Boc-D-Asp-OH g (mmol) | Boc-D-Glu-OH g (mmol) | Oleylamine g (mmol) | EDC g (mmol) | Yield g (%) |
|---|---|---|---|---|---|
| D-DoAo1 | 1.0 (4.3) | — | 2.3 (8.6) | 1.6 (8.6) | 1.9 (61%) |
| D-DoGo1 | — | 1.0 (4.0) | 2.1 (8.1) | 1.7 (8.9) | 2.3 (79%) |
| Lipids | D-DoAo1 g (mmol) | D-DoGo1 g (mmol) | Boc-D-Orn(Boc)-OSu g (mmol) | TEA g (mmol) | Yield g (%) |
|---|---|---|---|---|---|
| D-DoAo2 | 1.9 (2.9) | — | 1.2 (2.9) | 0.4 (2.9) | 2.4 (84%) |
| D-DoGo2 | — | 1.2 (1.9) | 0.8 (1.9) | 0.26 (1.9) | 1.5 (83%) |
SiRNA binding efficiency and enzyme resistance assay
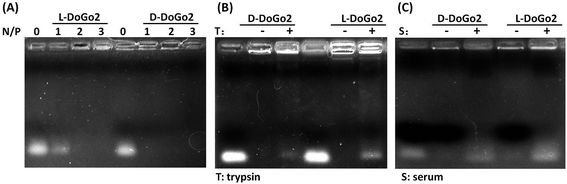
Electrostatic interaction between the carrier and the nucleic acid therapeutic is one of the pre-requirements for non-viral cationic molecule-based nucleic acid delivery systems. As demonstrated in our previous study, lipid conjugated short peptidomimetics can efficiently bind and encapsulate short nucleic acids such as siRNA. To test whether or not di-oleyl functionalized D-dipeptides electrostatically interact and bind with siRNA, we complexed L- and D-DoGo2 with siRNA at different amine (N) to phosphate (P) ratio and performed electrophoresis analysis. All lipids (D-DoGo2 and D-DoAo2) readily bind to siRNA at N/P ratio above 2 (Fig. 1A). D-Peptides are well known for their remarkable stability in bio-environment such as serum and extracellular matrix, which is a preferable characteristic for drug delivery agents, especially for the systemic drug delivery systems. Therefore, incorporation of D-amino acids in lipid conjugated dipeptides may strengthen their stability by enhancing their resistance to enzymatic hydrolysis in the biosystem. To test this, we complexed siRNA with L- and D-DoAo2, respectively, and examined their stability by incubation with trypsin at 37 °C, followed by an electrophoresis mobility shift analysis. At N/P ratios above 2 which gave complete binding and retardation of siRNA mobility, D-DoAo2 complexed siRNA showed apparently less electrophoresis mobility, while L-DoAo2 complexed siRNA appeared as bright band, indicating that D-DoAo2 is more resistant to enzymatic hydrolysis by trypsin, hence it may bear higher stability in bio-environment (Fig. 1B). D-DoGo2 also showed similar protease resistance (data not shown). Based on these results, we focused on in vitro and in vivo evaluation of D-DoAo2 in the following studies.Cell penetrating and in vitro siRNA delivery
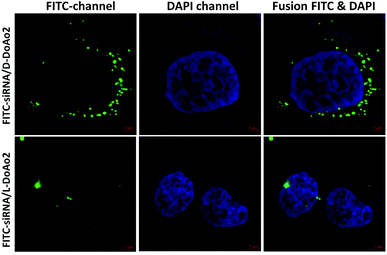
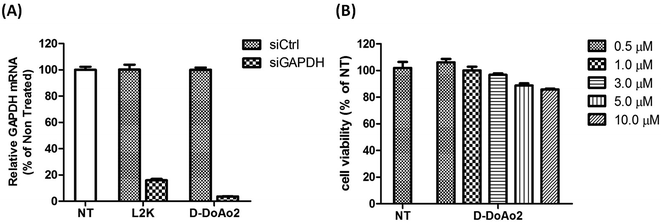
Next we investigated the cell penetrating ability of D-DoAo2 based lipoplexes by complexing them with an FITC-labeled siRNA, respectively. To do this, we treated HepG2 cells with either FITC-siRNA only or FITC-siRNA/D-DoAo2 complexes for 4 hours. Confocal images of HepG2 cells treated with D-DoAo2 based lipoplex carrying FITC-siRNA showed intense fluorescence signal scattered in the cytosol, while L-DoAo2 based lipoplex treated cells showed less fluorescence signal aggregated in cytosol, indicating that D-DoAo2 based lipoplex delivered siRNA into cells in high efficiency (Fig. 2). This is consistent with the observation that D-DoAo2 binds with siRNA more efficiently than L-DoAo2 does (Fig. 1A), which may contribute to the enhanced intracellular delivery of siRNA. D-DoAo2 based lipoplex not only delivered siRNA into the cells, but also induced efficient RNAi activity, as confirmed by a subsequent RT-qPCR measurement on the endogenous target gene GAPDH in HepG2 cells, which exhibited 90% knock down of GAPDH mRNA, while showing no apparent cytotoxicity (Fig. 3).Formulation and characterization of D-DoAo2 lipoplex
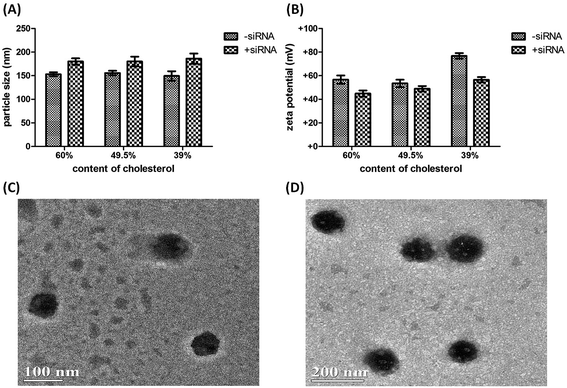
Since the physicochemical properties are important parameters for in vivo application of lipoplex, especially for the lipoplex as carriers for siRNA, we next performed formulation of the D-DoAo2 based lipoplex by incorporating cholesterol and PEGylated lipids (mPEG-OLA) at different ratios, followed by the measurement of particle size and zeta potential. Particle size of the lipoplexes measured by DLS showed little difference between particles containing 60%, 49.5% and 39% cholesterol, with about 150 nm being the average size (Fig. 4A). Addition of siRNA increased particle size by about 50 nm. However, zeta potential of three different formulations showed significantly different values: incorporation of 60% cholesterol in the lipoplex showed +57 mV; complexing of siRNA with the lipoplex decreased the zeta potential to +45 mV, which is the lowest in all three formulations hence represents a more closely bound nanoparticle with an increased stability (Fig. 4B).30 Based on these observations, we further investigated the in vivo siRNA delivery efficiency of the lipoplex formulated with 60% cholesterol, 39% D-DoAo2 and 1% mPEG-OLA.Tissue distribution of D-DoAo2 lipoplex
Since the D-DoAo2 based lipoplex formulation has a particle size of a preferable for sustained blood circulation and organ uptake, we next investigated the biodistribution of D-DoAo2/siRNA lipoplex. To do this, we labeled a siRNA by conjugating biotin at the 3′ end of the antisense strand of apoB siRNA. Since siRNA conjugated biotin can be detected on the membrane, we analyzed total RNA from different organs of mouse systematically administered biotin-siRNA/D-DoAo2 lipoplex 12 h after the intravenous (i.v.) injection. Although biotin molecule exists in the cells, there are no reports of naturally occurring biotin containing small RNAs. Furthermore, a positive control siRNA coupled with biotin clearly indicated the existence of antisense strand of injected siRNA in the organs, including liver, heart, lung and spleen (Fig. 5).In vivo siRNA delivery of D-DoAo2 lipoplex
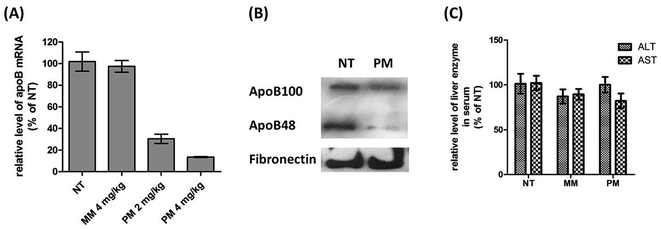
Since D-DoAo2 lipoplex can efficiently target mouse liver, we next examined whether or not it can penetrate the tissue, deliver the siRNA to hepatocytes and induce in vivo RNAi. To do this, we complexed an unmodified siRNA targeting mouse liver apoB with D-DoAo2 lipoplex, and administered the complex to mice through i.v. injection. After 24 h, mice were sacrificed, liver total RNA was extracted, and the level of apoB mRNA in the control group and test group was analyzed by RT-qPCR. At a siRNA dose of 2.0 mg kg−1, apoB mRNA level in the D-DoAo2/siRNA treated group decreased by 60% compared to the control group, indicating that D-DoAo2 lipoplex complexed apoB siRNA entered the hepatocytes and induced RNAi in vivo (Fig. 6A). At an increased dose of 4.0 mg kg−1 siRNA, 90% knock down of target apoB mRNA was achieved by a single i.v. injection. Moreover, a three consecutive administration of D-DoAo2/siRNA lipoplex at a dose of 4.0 mg kg−1 induced significant decrease of serum apoB protein level (Fig. 6B). Remarkably, D-DoAo2 carried unmodified siRNA and triggered RNAi in the target tissue at a clinically feasible dose. Moreover, the administration of D-DoAo2 lipoplex in the mice did not elevate serum level of liver enzyme (ALT, AST), indicating that D-DoAo2 lipoplex do not have apparent liver toxicity (Fig. 6C).4. Conclusions
Lipid functionalized dipeptides containing D-amino acids have been successfully synthesized. Digestion by trypsin as well as subsequent electrophoresis analysis revealed that D-DoAo2 has excellent resistance to enzymic hydrolysis, which may contribute to enhanced stability and better siRNA protection during in vivo systemic delivery. Increased enzyme resistance of D-DoAo2 did not induce elevated cytotoxicity, as demonstrated by MTT assay on HepG2 cells. D-DoAo2 showed efficient siRNA delivery to HepG2 cells only, as confirmed by fluorescence microscopic analysis. D-DoAo2 lipoplex delivered siRNA to HepG2 cells and efficiently knocked down endogenous GAPDH gene. Formulation of D-DoAo2 with cholesterol resulted in lipoplexes with approximate diameters of 150 nm to 200 nm, which efficiently delivered unmodified siRNA to mouse liver, lung and spleen. In liver, apoB gene expression was dose dependently silenced by siapoB delivered by D-DoAo2 lipoplex. The novel D-amino acids containing lipoplex carrying siRNA achieved superior RNAi in vivo at a lower dose (4.0 mg kg−1) compared to the previously reported DoGo3 lipid.28 Our results collectively demonstrated that D-DoAo2 lipoplex has enhanced siRNA delivery efficacy, which is a promising in vivo siRNA delivery agent for systemic siRNA.Acknowledgements
This research has been supported by the National Natural Science Foundation of China (21364006, 21375058 and 81560568).References
- S. M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber and T. Tuschl, Nature, 2001, 411, 494–498 CrossRef CAS PubMed.
- Y. L. Chiu and T. M. Rana, RNA, 2003, 9, 1034–1048 CrossRef CAS PubMed.
- A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806–811 CrossRef CAS PubMed.
- A. Akinc, A. Zumbuehl, M. Goldberg, E. S. Leshchiner, V. Busini, N. Hossain, S. A. Bacallado, D. N. Nguyen, J. Fuller, R. Alvarez, A. Borodovsky, T. Borland, R. Constien, A. de Fougerolles, J. R. Dorkin, K. Narayanannair Jayaprakash, M. Jayaraman, M. John, V. Koteliansky, M. Manoharan, L. Nechev, J. Qin, T. Racie, D. Raitcheva, K. G. Rajeev, D. W. Sah, J. Soutschek, I. Toudjarska, H. P. Vornlocher, T. S. Zimmermann, R. Langer and D. G. Anderson, Nat. Biotechnol., 2008, 26, 561–569 CrossRef CAS PubMed.
- J. Soutschek, A. Akinc, B. Bramlage, K. Charisse, R. Constien, M. Donoghue, S. Elbashir, A. Geick, P. Hadwiger, J. Harborth, M. John, V. Kesavan, G. Lavine, R. K. Pandey, T. Racie, K. G. Rajeev, I. Rohl, I. Toudjarska, G. Wang, S. Wuschko, D. Bumcrot, V. Koteliansky, S. Limmer, M. Manoharan and H. P. Vornlocher, Nature, 2004, 432, 173–178 CrossRef CAS PubMed.
- J. A. McCarroll, T. Dwarte, H. Baigude, J. Dang, L. Yang, R. B. Erlich, K. Kimpton, J. Teo, S. M. Sagnella, M. C. Akerfeldt, J. Liu, P. A. Phillips, T. M. Rana and M. Kavallaris, Oncotarget, 2015, 6, 12020–12034 CrossRef PubMed.
- M. E. Davis, J. E. Zuckerman, C. H. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, 1067–1070 CrossRef CAS PubMed.
- J. Shen, H. C. Kim, H. Su, F. Wang, J. Wolfram, D. Kirui, J. Mai, C. Mu, L. N. Ji, Z. W. Mao and H. Shen, Theranostics, 2014, 4, 487–497 CrossRef CAS PubMed.
- J. Shen, R. Xu, J. Mai, H. C. Kim, X. Guo, G. Qin, Y. Yang, J. Wolfram, C. Mu, X. Xia, J. Gu, X. Liu, Z. W. Mao, M. Ferrari and H. Shen, ACS Nano, 2013, 7, 9867–9880 CrossRef CAS PubMed.
- J. Shen, H. C. Kim, C. Mu, E. Gentile, J. Mai, J. Wolfram, L. N. Ji, M. Ferrari, Z. W. Mao and H. Shen, Adv. Healthcare Mater., 2014, 3, 1629–1637 CrossRef CAS PubMed.
- H. J. Unwalla and J. J. Rossi, Virol. J., 2010, 7, 33 CrossRef PubMed.
- D. V. Morrissey, J. A. Lockridge, L. Shaw, K. Blanchard, K. Jensen, W. Breen, K. Hartsough, L. Machemer, S. Radka, V. Jadhav, N. Vaish, S. Zinnen, C. Vargeese, K. Bowman, C. S. Shaffer, L. B. Jeffs, A. Judge, I. MacLachlan and B. Polisky, Nat. Biotechnol., 2005, 23, 1002–1007 CrossRef CAS PubMed.
- T. Coelho, D. Adams, A. Silva, P. Lozeron, P. N. Hawkins, T. Mant, J. Perez, J. Chiesa, S. Warrington, E. Tranter, M. Munisamy, R. Falzone, J. Harrop, J. Cehelsky, B. R. Bettencourt, M. Geissler, J. S. Butler, A. Sehgal, R. E. Meyers, Q. Chen, T. Borland, R. M. Hutabarat, V. A. Clausen, R. Alvarez, K. Fitzgerald, C. Gamba-Vitalo, S. V. Nochur, A. K. Vaishnaw, D. W. Sah, J. A. Gollob and O. B. Suhr, N. Engl. J. Med., 2013, 369, 819–829 CrossRef CAS PubMed.
- J. S. Butler, A. Chan, S. Costelha, S. Fishman, J. L. Willoughby, T. D. Borland, S. Milstein, D. J. Foster, P. Goncalves, Q. Chen, J. Qin, B. R. Bettencourt, D. W. Sah, R. Alvarez, K. G. Rajeev, M. Manoharan, K. Fitzgerald, R. E. Meyers, S. V. Nochur, M. J. Saraiva and T. S. Zimmermann, Amyloid, 2016, 23, 109–118 CrossRef CAS PubMed.
- M. L. Bobbin and J. J. Rossi, Annu. Rev. Pharmacol. Toxicol., 2016, 56, 103–122 CrossRef CAS PubMed.
- K. A. Howard, U. L. Rahbek, X. Liu, C. K. Damgaard, S. Z. Glud, M. O. Andersen, M. B. Hovgaard, A. Schmitz, J. R. Nyengaard, F. Besenbacher and J. Kjems, Mol. Ther., 2006, 14, 476–484 CrossRef CAS PubMed.
- M. Jayaraman, S. M. Ansell, B. L. Mui, Y. K. Tam, J. Chen, X. Du, D. Butler, L. Eltepu, S. Matsuda, J. K. Narayanannair, K. G. Rajeev, I. M. Hafez, A. Akinc, M. A. Maier, M. A. Tracy, P. R. Cullis, T. D. Madden, M. Manoharan and M. J. Hope, Angew. Chem., Int. Ed., 2012, 51, 8529–8533 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967–977 CrossRef CAS PubMed.
- J. K. Nair, J. L. Willoughby, A. Chan, K. Charisse, M. R. Alam, Q. Wang, M. Hoekstra, P. Kandasamy, A. V. Kel'in, S. Milstein, N. Taneja, J. O'Shea, S. Shaikh, L. Zhang, R. J. van der Sluis, M. E. Jung, A. Akinc, R. Hutabarat, S. Kuchimanchi, K. Fitzgerald, T. Zimmermann, T. J. van Berkel, M. A. Maier, K. G. Rajeev and M. Manoharan, J. Am. Chem. Soc., 2014, 136, 16958–16961 CrossRef CAS PubMed.
- S. Matsuda, K. Keiser, J. K. Nair, K. Charisse, R. M. Manoharan, P. Kretschmer, C. G. Peng, A. V. Kel'in, P. Kandasamy, J. L. Willoughby, A. Liebow, W. Querbes, K. Yucius, T. Nguyen, S. Milstein, M. A. Maier, K. G. Rajeev and M. Manoharan, ACS Chem. Biol., 2015, 10, 1181–1187 CrossRef CAS PubMed.
- X. Xu, J. Wu, Y. Liu, M. Yu, L. Zhao, X. Zhu, S. Bhasin, Q. Li, E. Ha, J. Shi and O. C. Farokhzad, Angew. Chem., Int. Ed., 2016, 55, 7091–7094 CrossRef CAS PubMed.
- S. Yu, X. Gao, H. Baigude, X. Hai, R. Zhang, B. Shen, Z. Li, Z. Tan and H. Su, ACS Appl. Mater. Interfaces, 2015, 7, 5089–5096 CAS.
- Y. Wu, J. Cai, J. Han and H. Baigude, ACS Appl. Mater. Interfaces, 2015, 7, 21521–21528 CAS.
- K. G. Rajeev, J. K. Nair, M. Jayaraman, K. Charisse, N. Taneja, J. O'Shea, J. L. Willoughby, K. Yucius, T. Nguyen, S. Shulga-Morskaya, S. Milstein, A. Liebow, W. Querbes, A. Borodovsky, K. Fitzgerald, M. A. Maier and M. Manoharan, ChemBioChem, 2015, 16, 903–908 CrossRef CAS PubMed.
- D. B. Rozema, D. L. Lewis, D. H. Wakefield, S. C. Wong, J. J. Klein, P. L. Roesch, S. L. Bertin, T. W. Reppen, Q. Chu, A. V. Blokhin, J. E. Hagstrom and J. A. Wolff, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12982–12987 CrossRef CAS PubMed.
- T. W. Geisbert, A. C. Lee, M. Robbins, J. B. Geisbert, A. N. Honko, V. Sood, J. C. Johnson, S. de Jong, I. Tavakoli, A. Judge, L. E. Hensley and I. Maclachlan, Lancet, 2010, 375, 1896–1905 CrossRef CAS.
- Y. H. Lee, A. D. Judge, D. Seo, M. Kitade, L. E. Gomez-Quiroz, T. Ishikawa, J. B. Andersen, B. K. Kim, J. U. Marquardt, C. Raggi, I. Avital, E. A. Conner, I. MacLachlan, V. M. Factor and S. S. Thorgeirsson, Oncogene, 2011, 30, 4175–4184 CrossRef CAS PubMed.
- H. Xiao, A. Altangerel, G. Gerile, Y. Wu and H. Baigude, ACS Appl. Mater. Interfaces, 2016, 8, 7638–7645 CAS.
- H. Baigude, J. McCarroll, C. S. Yang, P. M. Swain and T. M. Rana, ACS Chem. Biol., 2007, 2, 237–241 CrossRef CAS PubMed.
- J. D. Clogston and A. K. Patri, Methods Mol. Biol., 2011, 697, 63–70 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: MS and 1H NMR spectra of D-DoAo2 and D-DoGo2. See DOI: 10.1039/c6ra25862j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |