Open Access Article
Open Access ArticleSynthesis, characterization and programmable toxicity of iron oxide nanoparticles conjugated with D-amino acid oxidase†
Riccardo Balzaretti *a,
Fabian Mederb,
Marco P. Monopolibc,
Luca Bosellib,
Ilaria Armenia
*a,
Fabian Mederb,
Marco P. Monopolibc,
Luca Bosellib,
Ilaria Armenia a,
Loredano Pollegioniad,
Giovanni Bernardini
a,
Loredano Pollegioniad,
Giovanni Bernardini ad and
Rosalba Gornatiad
ad and
Rosalba Gornatiad
aDepartment of Biotechnology and Life Sciences, University of Insubria, Varese, Italy. E-mail: riccardo.balzaretti@uninsubria.it
bCentre for Bionano Interactions, University College Dublin, Dublin, Ireland
cDepartment of Pharmacy and Medical Chemistry, Royal College of Surgeons in Ireland, 123 St. Stephen Green, Dublin 2, Ireland
dThe Protein Factory Research Center, Politecnico di Milano, University of Insubria, Milan, Varese, Italy
First published on 5th January 2017
Abstract
D-amino acid oxidase (DAAO) is an enzyme which generates reactive oxygen species (ROS) and it is believed to have potential uses as a novel therapeutic molecule if internalized by cancer cells or if they are localized close to their plasma membrane. When conjugated onto iron oxide nanoparticles (NPs), the enzyme can be magnetically directed to targeted locations with an increased efficacy. A subsequent injection of DAAO substrate D-alanine can initiate ROS production and induce apoptosis of cells surrounding the NP-DAAO complex. Here, we describe a platform for optimal bioconjugation using monodisperse γ-Fe2O3 NPs (∼10 nm) resulting in high DAAO loading, stable NP-DAAO dispersions and more than 90% enzymatic activity recovery, which is retained using the particles in human serum. Lastly, since the NP-DAAO system is designed for cancer therapy, we proved its efficacy in killing SKOV-3, U87 and HCT-116 cancer cells.
Nanobiotechnology has found widespread applications1 in biomedicine as diagnostic imaging tools,2,3 drug delivery systems,4,5 vaccines6,7 and in immunotherapy.8 NPs can be employed to produce a great variety of bioconjugates by grafting biomolecules onto a NP carrier. In oncology, for example, recent studies have shown that engineered NPs are capable of reaching specific targeted areas of the body and of releasing the therapeutic drug locally.9–12 This novel approach has the potential to increase the drug efficacy and to eliminate the side effects.13 To guide the bioconjugates through the body, one can use magnetic NPs that, after intravenous administration, can be directed to the cancerous tissue with an external magnetic field.14 Magnetic NPs alone are used as an effective anticancer agent during hyperthermic therapy in which NPs exposed to an alternating magnetic field produce heat that kills all the surrounding cells.15–19
D-Amino acid oxidase from Rhodotorula gracilis (RgDAAO, EC 1.4.3.3) is a flavoenzyme that utilizes D-amino acids to produce reactive oxygen species (ROS)20 and can be used to induce apoptosis. In particular the co-administration with D-alanine as substrate, gives the corresponding α-keto acid, ammonia and H2O2. Despite the benefit in using this enzyme in therapeutic applications, the main limitations are the low enzyme stability, the low efficacy in cellular internalization and the lack of specific molecular targeting. To overcome these limitations, in the past few years different attempts have been made in the development of NP-DAAO systems where the enzyme was covalently bound to the amino groups of APTES functionalized NPs. That resulted in an increased cellular internalization of the enzyme, a prolonged enzymatic activity and storage stability, and the possibility of targeting to the cancer mass with the help of an external magnetic field. However, while the APTES functionalization represents a good amino group donor, its activation via glutaraldehyde can affect the enzyme integrity21 leading to a loss of DAAO activity.22,23 Cappellini et al.24 reported a higher yield for the NP-DAAO synthesis when conducted via EDC/NHS instead of with glutaraldehyde. Furthermore, also the NP diameter and aggregation influence the amount of loaded enzyme.25
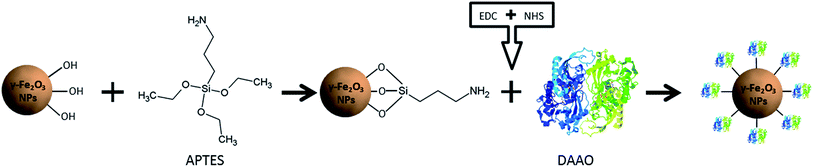
Here, we present a NP-DAAO system starting from the synthesis of γ-Fe2O3 NPs to obtain small and monodispersed γ-Fe2O3–APTES NPs to increase as much as possible the surface area available for the surface functionalization.
γ-Fe2O3 NP cores were synthetized following Geppert et al.26 (ESI†) and they were subsequently functionalized with APTES using a standard protocol and then conjugated with DAAO by coupling with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS), as summarized in Fig. 1. The RgDAAO enzyme was provided by the laboratory of Prof. Loredano Pollegioni of the Biotechnology and Life Sciences Department of Università degli Studi dell'Insubria, Varese. The γ-Fe2O3 NPs, NP-APTES and NP-APTES–DAAO were characterized by Dynamic Light Scattering (DLS), by Differential Centrifugal Sedimentation (DCS) and by Transmission Electron Microscopy (TEM) (ESI†).
The DLS characterization analysis (Fig. 2a) reports a diameter of about 38 nm for the synthetized γ-Fe2O3 NPs and about 47 nm for the APTES functionalized ones. After conjugation with the enzyme, the NP-DAAO complex acquires a larger diameter of approximately 185 nm.
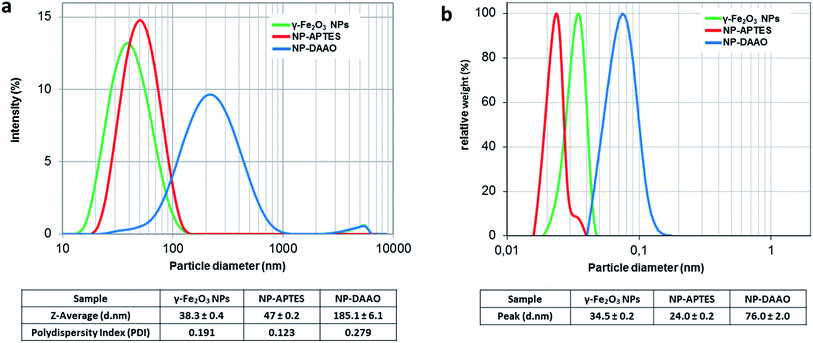 | ||
| Fig. 2 DLS (a) and DCS (b) analysis of γ-Fe2O3 NPs, NP-APTES and NP-DAAO. The polydispersity index is from cumulant fitting. | ||
The DCS characterization (Fig. 2b) is similar to the one obtained by DLS. The surface functionalization with APTES and DAAO results in a change in the particle sedimentation time. The NP-APTES and NP-DAAO have respectively an apparent size of 24 nm and 76 nm: this is due to a change in sample density as also reported elsewhere.27,28
At a glance, the TEM pictures (ESI†) do not suggest any notable changes in NP size as shown by DLS and DCS analysis. However, the TEM picture analyses (ESI†) reported that γ-Fe2O3 NPs, NP-APTES and NP-DAAO possess an average size of respectively 8.5, 10.3 and 11.0 nm. These results can be probably due to a slight NPs aggregation during DLS and DCS analysis.
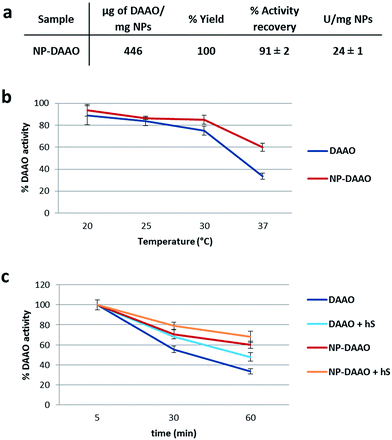
The amount of DAAO bound on the NP surface was determined by considering the difference between the starting amount of RgDAAO and the protein recovered in the supernatant at the end of the conjugation reaction. The quantification of conjugated DAAO was performed using the extinction coefficient at 455 nm (∼12.6 mM−1 cm−1)20 and an UV-Vis V-560 Spectrophotometer (JASCO, MD, USA). The amount of stock DAAO used for the conjugation to NP-APTES has been chosen starting from the ratio of μg of DAAO per NP-APTES surface area (calculated from the average particle diameter, assuming spherical particles) used by Cappellini et al.24 and then moving to DAAO saturation per NP-APTES. For one mg of NP-APTES particles up to 446 μg of RgDAAO was bound (Fig. 3a). Moreover, to determine the functionality of the NP-DAAO particles, we checked their enzymatic activity by measuring the absorbance increase accompanying the H2O2-induced oxidation of o-dianisidine. This reaction was initiated by the addition of the enzyme and the absorbance increase was monitored at 440 nm for 1 min.
The conjugation yield and the activity recovery of these NP-DAAO particles were of 100% and 91% respectively (Fig. 3a). Since the specific activity of the RgDAAO stock solution was 59 U mg−1, we estimated up to 24 U per mg of NP-APTES. The NPs here synthesized showed a smaller diameter compared to the ones used by Cappellini et al.24 and this allowed a 7 fold increase of the DAAO/NP (w/w) ratio.
Then, we detected the thermal stability of the NP-DAAO system, as enzymatic residual activity, after 1 hour incubation at different temperature (Fig. 3b) (ESI†). At 37 °C the NP-DAAO system is about 1.8 fold more stable compared to free DAAO.
Since this NP-DAAO system, thought for cancer therapy, has to be injected intravenously to perform its activity, we checked its stability also at 37 °C when exposed to 10% human serum (Sigma-Aldrich) (Fig. 3c). These results indicate that human serum has an active role in preserving the activity of both DAAO and NP-DAAO system.
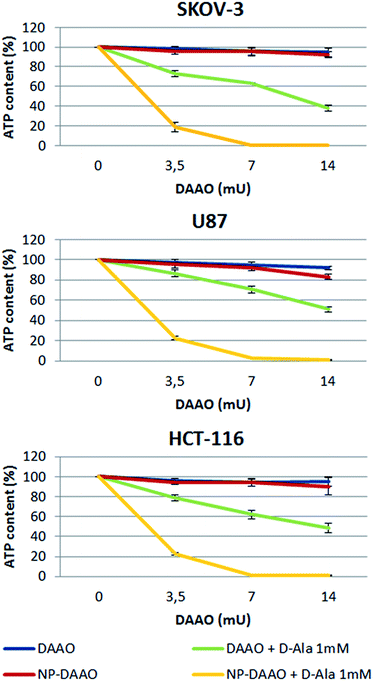
Lastly, we tested the efficacy of the NP-DAAO system on the three tumor cell lines SKOV-3, HCT116 and U87 (Sigma-Aldrich) (Fig. 4). In this experiment, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were exposed for 24 h to an increasing amount, expressed as DAAO units, of free DAAO or NP-DAAO with or without D-alanine (D-Ala). Hence, the cell viability was determined as ATP content by recording the luminescent signal after using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). The results reported in Fig. 4 show a very low cytotoxicity when cells are exposed to 7 mU of NP-DAAO particles without D-alanine. However, when D-Ala is added a full cell death was apparent. If compared to the NP-DAAO particles present in literature,23 the new particles result less cytotoxic whilst the effectiveness remains unchanged in the presence of the substrate: this can be explained by the lower amount of NP-DAAO particles required to reach the same DAAO activity (ESI†). We have performed preliminary experiments exposing human adipose stem cells (hASC), isolated from adipose tissue, to NP-DAAO. The results have shown that hASC are less responsive than tumor cell lines to the H2O2 produced by the NP-DAAO in presence of D-Ala.
000 cells were exposed for 24 h to an increasing amount, expressed as DAAO units, of free DAAO or NP-DAAO with or without D-alanine (D-Ala). Hence, the cell viability was determined as ATP content by recording the luminescent signal after using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). The results reported in Fig. 4 show a very low cytotoxicity when cells are exposed to 7 mU of NP-DAAO particles without D-alanine. However, when D-Ala is added a full cell death was apparent. If compared to the NP-DAAO particles present in literature,23 the new particles result less cytotoxic whilst the effectiveness remains unchanged in the presence of the substrate: this can be explained by the lower amount of NP-DAAO particles required to reach the same DAAO activity (ESI†). We have performed preliminary experiments exposing human adipose stem cells (hASC), isolated from adipose tissue, to NP-DAAO. The results have shown that hASC are less responsive than tumor cell lines to the H2O2 produced by the NP-DAAO in presence of D-Ala.
Conclusions
In conclusion, the new NP-DAAO particles represent a remarkable improvement of this NP-enzyme system.With a reduced amount of NPs required for the same enzymatic activity, we have obtained a safer and more efficient NP-DAAO system that might be used for cancer therapy, but also for biocatalysis. The new NPs, thanks to their smaller size, can bind about 7 times more enzyme than the previous ones28 thus increasing the efficacy and reducing the NP-DAAO system toxicity before external stimulation of ROS production (ESI†). Since smaller NPs might result more cytotoxic,29 in vivo experiments will be performed to assay their toxicity and half-life once injected into the blood stream.
It has been previously reported that enzymes conjugated to NPs may present a higher activity, a better specificity, a prolonged half-life, an enhanced thermal stability and, in some cases, even an enzymatic acceleration.30–32 In agreement with these reports, we have observed a prolonged storage half-life (ESI†) and an enhanced thermal stability (Fig. 3b). Conversely, we did not notice an increase of the enzymatic activity. In addition, the susceptibility of the NP-enzyme system to a magnetic field allows, in nanomedicine, its targeting to the desired area of the body, and, in biocatalysis, its efficient recovery enabling its reuse for several times and preventing the contamination of the final product.30
Acknowledgements
This study was partially funded and supported by QualityNano project. Riccardo Balzaretti and Ilaria Armenia are PhD students of the “Biotechnology, Biosciences and Surgical Technology” course at Università degli studi dell'Insubria. Fabian Meder acknowledges the support by the German Research Foundation (DFG) within the ‘Forschungsstipendium’ ME 4296/1-1. Marco P. Monopoli, acknowledges the financial support of the SFI Industry fellowship (15/IFA/3057) and Luca Boselli the EU H2020 Nanofacturing projects (grant agreement no. 646364).Notes and references
- A. G. Cattaneo, R. Gornati, E. Sabbioni, M. Chiriva-Internati, E. Cobos, M. R. Jenkins and G. Bernardini, J. Appl. Toxicol., 2010, 30, 730–744 CrossRef CAS PubMed.
- S. K. Nune, P. Gunda, P. K. Thallapally, Y.-Y. Lin, M. L. Forrest and C. J. Berkland, Expert Opin. Drug Delivery, 2009, 6(11), 1175–1194 CrossRef CAS PubMed.
- Y.-X. J. Wang, S. Xuan, M. Port and J.-M. Idee, Curr. Pharm. Des., 2013, 19, 6575–6593 CrossRef CAS PubMed.
- S. Onoue, S. Yamada and H.-K. Chan, Int. J. Nanomed., 2014, 9, 1025–1037 CrossRef CAS PubMed.
- L. Liu, Q. Ye, M. Lu, Y.-C. Lo, Y.-H. Hsu, M.-C. Wei, Y.-H. Chen, S.-C. Lo, S.-J. Wang, D. J. Bain and C. Ho, Sci. Rep., 2015, 5, 10881 CrossRef CAS PubMed.
- D. Boraschi and P. Italiani, Vaccines, 2015, 3, 930–939 CrossRef PubMed.
- Y. Fan and J. J. Moon, Vaccines, 2015, 3, 662–685 CrossRef PubMed.
- C. Petrarca, E. Clemente, V. Amato, P. Pedata, E. Sabbioni, G. Bernardini, I. Iavicoli, S. Cortese, Q. Niu, T. Otsuki, R. Paganelli and M. Di Gioacchino, Clin. Mol. Allergy, 2015, 13, 13 CrossRef PubMed.
- S. Barua and S. Mitragotri, Nano Today, 2014, 9, 223–243 CrossRef CAS PubMed.
- C. Castellini, S. Ruggeri, S. Mattioli, G. Bernardini, L. Macchioni, E. Moretti and G. Collodel, Syst. Biol. Reprod. Med., 2014, 60, 143–150 CrossRef CAS PubMed.
- T. Coccini, R. Gornati, F. Rossi, E. Signoretto, I. Vanetti, G. Bernardini and L. Manzo, J. Nanomed. Nanotechnol., 2014, 5, 227 Search PubMed.
- M. Masserini, ISRN Biochem., 2013, 238428 Search PubMed.
- J. R. Kanwar, G. Mahidhara and R. K. Kanwar, Nanomedicine and Cancer Therapies, 2013, vol. 2, pp. 1–18 Search PubMed.
- M. Liberatore, M. Barteri, V. Megna, P. D'Elia, S. Rebonato, A. Latini, F. De Angelis, F. Scaramuzzo, A. Francesca, M. E. De Stefano, N. A. Guadagno, S. Chondrogiannis, A. M. Maffione, D. Rubello, A. Pala and P. M. Colletti, Clinical Nuclear Medicine, 2015, 40(2), e104–e110 CrossRef PubMed.
- E. K. Lim, E. Jang, K. Lee, S. Haam and Y. M. Huh, Pharmaceutics, 2013, 5(2), 294–317 CrossRef CAS PubMed.
- J. H. Lee, K. J. Chen, S. H. Noh, M. A. Garcia, H. Wang, W. Y. Lin, H. Jeong, B. J. Kong, D. B. Stout, J. Cheon and H. R. Tseng, Angew. Chem., Int. Ed., 2013, 52, 1–6 CrossRef.
- M. H. Cho, E. J. Lee, M. Son, J. H. Lee, D. Yoo, J. W. Kim, S. W. Park, J. S. Shin and J. Cheon, Nat. Mat., 2012, 11, 1038–1043 CAS.
- C. R. Thomas, D. P. Ferris, J.-H. Lee, E. Choi, M. H. Cho, E. S. Kim, J. F. Stoddart, J.-S. Shin, J. Cheon and J. I. Zink, J. Am. Chem. Soc., 2010, 132, 10623–10625 CrossRef CAS PubMed.
- K. Katagiri, Y. Imai, K. Koumoto, T. Kaiden, K. Kono and S. Aoshima, Small, 2011, 7(12), 1683–1689 CrossRef CAS PubMed.
- L. Pollegioni, G. Molla, S. Sacchi, E. Rosini, R. Verga and M. S. Pilone, Appl. Microbiol. Biotechnol., 2008, 78(1), 1–16 CrossRef CAS PubMed.
- I. Migneault, C. Dartiguenave, M. J. Bertrand and K. C. Waldron, BioTechniques, 2004, 37(5), 790–802 CAS.
- H. C. Hsieh, I. C. Kuan, S. L. Lee, G. Y. Tien, Y. J. Wang and C. Y. Yu, Biotechnol. Lett., 2009, 31, 557–563 CrossRef CAS PubMed.
- A. Bava, R. Gornati, F. Cappellini, L. Caldinelli, L. Pollegioni and G. Bernardini, Nanomedicine, 2013, 8(11), 1797–1806 CrossRef CAS PubMed.
- F. Cappellini, C. Recordati, M. De Maglie, L. Pollegioni, F. Rossi, M. Daturi, R. Gornati and G. Bernardini, Future Sci. OA, 2015, 1, 4 Search PubMed.
- J. Conde, J. T. Dias, V. Grazú, M. Moros, P. V. Baptista and J. M. de la Fuente, Front. Chem., 2014, 2, 48 Search PubMed.
- M. Geppert, M. Hohnholt, L. Gaetjen, I. Grunwald, M. Bäumer and R. Dringen, J. Biomed. Nanotechnol., 2009, 5, 285–293 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. B. Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133(8), 2525–2534 CrossRef CAS PubMed.
- Z. Krpetić, A. M. Davidson, M. Volk, R. Lévy, M. Brust and D. L. Cooper, ACS Nano, 2013, 7(10), 8881–8890 CrossRef PubMed.
- M. Saji, M. Ilyas, C. Basheer, M. Tariq, M. Daud, N. Baig and F. Shehzad, Environ. Sci. Pollut. Res. Int., 2015, 22(6), 4122–4143 CrossRef PubMed.
- S. A. Ansari and Q. Husain, Biotechnol. Adv., 2012, 30(3), 512–523 CrossRef CAS PubMed.
- B. J. Johnson, W. Russ Algar, A. P. Malanoski, M. G. Ancona and I. L. Medintz, Nano Today, 2014, 9, 102–131 CrossRef CAS.
- S. Ding, A. A. Cargill, I. L. Medintz and J. C. Claussen, Curr. Opin. Biotechnol., 2015, 34, 242–250 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis of γ-Fe2O3 NPs, TEM analysis, DLS and DCS details, cell culture details, comparison with the previous NP-DAAO system. See DOI: 10.1039/c6ra25349k |
| This journal is © The Royal Society of Chemistry 2017 |