Deciphering the evolution of the functional genes and microbial community of the combined partial nitritation-anammox process with nitrate build-up and its in situ restoration†
Yuanyue Wangab,
Hongyan Wangab,
Junya Zhangab,
Li Yaoc and
Yuansong Wei*abd
aState Key Joint Laboratory of Environmental Simulation and Pollution Control, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, P. O. Box 2871, Beijing 100085, China. E-mail: yswei@rcees.ac.cn; Fax: +86-10-62849690; Tel: +86-10-62849690
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cSchool of Life Sciences, University of Science and Technology of China, Hefei 230026, Anhui, China
dInstitute of Energy, Jiangxi Academy of Sciences, Nanchang 330096, Jiangxi, China
First published on 18th November 2016
Abstract
The operation of a combined partial nitritation-anammox (CPNA) process was hampered by the overgrowth of nitrite-oxidizing bacteria (NOB), which was then successfully in situ restored by the combination of hydroxylamine (NH2OH) dosing and sludge retention time (SRT) control. To elucidate the microbial evolution on gene level and the microbial mechanism responding to the NO3− build-up and in situ restoration in the CPNA process, the qPCR and high-throughput sequencing were used to investigate the evolution of the N-transformation genes and microbial community in this paper. Results showed that the concentrations of amoA and hao were both decreased as well as the hzo along with the NO3− build-up, however, the nitrite-oxidation encoding gene nxrA was increased. Pearson correlation analysis demonstrated that the gene nxrA was significantly correlated with the NO3−. After restoration, the abundances of amoA, hao and hzo were increased to 26.58%, 43.56% and 26.17%, respectively, while the growth of nxrA was effectively limited. The obtained high abundance of narG gene (37.85%) as well as the significant correlation of hzo–narG and hzo–nirS illustrated the in situ restoring mechanism, i.e., the intensive denitrification of the accumulated NO3− to NO followed by the stimulation of the anammox by NO. Additionally, MiSeq sequencing showed a new microbial community was rebuilt after restoration and the Haliscomenobacter and Nitrosomonas were the dominant genera. The abundance of anammox genus Candidatus Kuenenia was also increased to 8.0%. The significant correlation between nirK and Ignavibacterium indicated that the filaments played an important role in the denitrification of the accumulated NO3−.
1. Introduction
The great advantages of anammox nitrogen removal technology have attracted lots of environmental researchers and engineers worldwide since it was discovered over two decades ago.1 In the following development, on one hand, the identification and isolation of the anammox microorganisms and the exploration of their cell structures and metabolism pathways, have led to a deep understanding of them,2–4 on the other hand, cultivating the anammox bacteria and developing many kinds of the anammox-based novel autotrophic nitrogen removal technologies with different descriptive terms has promoted the engineering application.5 Due to the extremely slow growth rate of the anammox microorganisms, it took 3.5 years for the first full-scale two-stage SHARON/ANAMMOX system in the world to start up in Rotterdam (NL) with a maximum nitrogen removal load of 9.5 kgN m−3 d−1.5 As summarized by Lackner et al.,6 the most commonly applied system was the one-stage combined partial nitritation-anammox (CPNA) process in the SBR configuration, and the steady increase in new plants over the past years has resulted in there being more than 100 operating installations worldwide by 2014.Though the engineering applications of the CPNA process are spreading all over the world, some of them were hampered by the problem of nitrate build-up that resulting in the deterioration of the overall nitrogen removal performance during their long-term operation, which signaled that the microbial community was unbalanced and too many nitrite-oxidizing bacteria (NOB) had accumulated,6 such as the systems at Zürich-Werdhölzli WWTP, Switzerland7 and Plettenberg WWTP, Germany.8 To deal with the nitrate build-up problem and restore the nitrogen removal of the CPNA process, several countermeasures had been proposed in the previous papers, such as the re-inoculation, reduction of the air flow rate, using the intermittent aeration and introducing the hydrocyclone to remove the NOB-containing floccular sludge, however, none of these methods were quite effective or time-efficient to solve the NOB overgrowth.9 Based on the previous researches that dosing the hydroxylamine (NH2OH), an intermediate in both nitrification and anammox, could not only promote the nitrite accumulation in the nitrifying system,10 but also stimulate the anammox activity,11 a novel in situ restoration strategy of combined NH2OH dosing and sludge retention time (SRT) control was developed in a lab-scale SBR,9 through which the nitrate build-up problem could be overcome in the short term.
However, during the researches of detecting the nitrate build-up and applying different measures to counteract it, most of the studies focused on the direct results of nitrogen removal performance and the changes of the activities of the ammonium-oxidizing bacteria (AOB), NOB and anammox,14 only a few papers conducted the direct detection of NOB by the fluorescence in situ hybridization (FISH) or quantitative real-time PCR (qPCR) technologies,7,8 and to the best of our knowledge, there are no studies to uncover the evolution of nitrogen removal functional genes (Fig. 1) in the CPNA process enduring the problem of nitrate build-up and its following restoration, and the change of the microbial community structure is still in a state of black box. Therefore, on the basis of our previous research that the nitrate build-up of the CPNA process was successfully in situ restored by the combination of NH2OH dosing and SRT control,9 the purpose of this study was to further understand the microbial mechanism of the deterioration by the nitrate build-up and the in situ restoration of the CPNA process through determining the nitrogen transformation functional genes by the qPCR assay, and investigating the changes of the microbial community using the 16S rRNA gene high-throughput sequencing.
 | ||
| Fig. 1 The nitrogen removal pathways and the involved functional genes. Anammox pathway I was proposed by van de Graaf,12 while the anammox pathway II was described by Kartal et al.13 | ||
2. Materials and methods
2.1. Setup of CPNA process and in situ restoration of the nitrate build-up
A CPNA process was operated in SBR system with the working volume of 80 L, and the Siemens PLC system combined a series of online sensors (DO, pH, ORP, conductivity and NH4+) were used to realize the automatic operation. The reactor was fed with the synthetic wastewater containing about 50% of real digested sludge liquid and 50% of tap water, NH4HCO3 was added to increase the concentration of ammonium, and NaHCO3 was supplied to secure the sufficient alkalinity for maintaining the pH.80 mgNH2OH L−1 of the stock solution was prepared with the hydroxylamine hydrochloride (NH2OH·HCl). Days from 237 to 305 were the first period of in situ restoration, only different volumes of stock solution were added obtaining the total NH2OH concentrations of 5, 10 and 20 mgNH2OH L−1 within each SBR cycle. In the second period of day 316 to 349, the combination of NH2OH dosing and 40 d of SRT control was carried out. The details could be referenced to Wang et al.9
The mole ratio of NO3−–N production to NH4+–N removal (MRNN) in each SBR cycle was used to determine the nitrate build-up, and could be calculated according to eqn (1).
 | (1) |
2.2. Chemical analysis
The pH, DO, conductivity and temperature in SBR were real-time monitored with the online sensors. Concentrations of ammonium, nitrite, nitrate, total nitrogen and mixed liquor suspended sludge (MLSS) were determined according to the standard methods.16 Aerobic and anaerobic batch tests were conducted to determine the activities of AOB, NOB and anammox, respectively.9 The parameter of nitrite accumulation rate was used to determine the activities of AOB and NOB comprehensively, and was calculated by dividing the nitrite production rate (the slope of the plot of the nitrite concentrations with time) by the MLSS.2.3. Sludge sampling, DNA extraction and quantitative real-time PCR
In the mixing state of the SBR operation, 8 sludge samples were collected for DNA extraction, which covered the three stages of CPNA process operation, i.e., deterioration by the nitrate build-up (day 186, 230), in situ restoration by NH2OH dosing and SRT control (day 251, 282, 307, 316, 349), and recovered stable operation (day 376). 2 mL of suspended sludge for each sample were centrifuged at 8000 rpm for 15 min, and the pellet was used for DNA extraction using the FASTDNA Spin Kit for Soil (MP Biomedicals, USA) in triplicate according to the manufacturer's instructions, and the resulting extracts were then merged together to average out the bias in sampling and extraction. Quality and concentration of the extracted DNA were determined through 1% agarose gel electrophoresis and NanoDrop 2000 (Thermo Scientific, USA), respectively, and then stored at −20 °C until use.9 nitrogen transformation functional genes including nitrification (amoA, hao, nxrA), denitrification (narG, nirS, nirK, norB, nosZ), anammox (hzo) as showed in Fig. 1 and bacterial 16S ribosomal RNA (rRNA) were quantified. Conventional PCR followed by TA cloning was conducted to obtain the standard plasmids, and then the standard plasmids were diluted to yield a series of tenfold concentrations and subsequently used for the establishment of qPCR standard curves. The R2 was higher than 0.99 for all the standard curves and the amplification efficiency based on the slope was between 91% and 104%. The 25 μL PCR reaction mixtures contained 12.5 μL SYBR Green qPCR SuperMix-UDG with Rox (Invitrogen, USA), 0.5 μL each of 10 μM forward and reverse primers, 10.5 μL DNA-free water, and 1.0 μL standard plasmid or DNA extract. The thermo cycling steps for qPCR amplification were as follows: (1) 50 °C, 2 min, (2) 95 °C, 5 min, (3) 95 °C, 20 s, (4) annealing temperature, 30 s, (5) 72 °C, 31 s, (6) plate read, go to (3)–(5), 39 more times, and (7) melt curve analysis: 60 °C to 95 °C, 0.2 °C per read. The reaction was conducted using an ABI Real-time PCR system 7500 (ABI, USA). Each gene was quantified in triplicate with a standard curve and negative control. The primers and annealing temperature used in this study were summarized in Table S1 in ESI.†
2.4. High-throughput sequencing
PCR primers 515F and 806R targeting the bacteria and archaea 16S V4 region were selected for the microbial community structure analysis,17 as the primers exhibit few biases and yield the accurate phylogenetic and taxonomic information. The reverse primer contains a 6 bp error-correcting barcode unique to each sample. The barcode was permuted for each sample and permitted the identification of individual sample within mixture in a single Illumina MiSeq sequencing run. DNA was amplified in triplicate for each sample following the protocol described previously.17 The size of PCR amplicons was determined by 2% agarose gels and further purified with a DNA purification kit (AxyPrepDNA, AXYGEN, USA), and concentrations were then determined by spectrometry using the QuantiFluor™-ST (Promega, USA). Amplicons from different samples were pooled to achieve equal mass concentrations in the final mixture, and then sent out to Majorbio Co., Ltd. (Shanghai, China) for small-fragment library construction and pair-end sequencing using Illumina MiSeq sequencing system (Illumina, USA).2.5. Post run analysis
Sequencing reads were assigned to each sample according to the unique 6 bp barcode of each sample. And then, barcode and primers were removed. Pairs of reads from the original DNA fragments were merged using FLASH,18 and then the raw reads were filtered by QIIME quality filters. PCR chimeras were filtered out using UCHIME.19 After the above filters were applied, the minimum number of selected sequences in the 7 samples was 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546. In order to analyze the different sample at the same sequencing depth, all samples were normalized by extracting the first 30
546. In order to analyze the different sample at the same sequencing depth, all samples were normalized by extracting the first 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 sequences from each sample for further analyses. The normalized sequences were uploaded to MG-RAST (http://www.metagenomics.anl.gov/, project = 12
546 sequences from each sample for further analyses. The normalized sequences were uploaded to MG-RAST (http://www.metagenomics.anl.gov/, project = 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 461). Taxonomic classification of the sequences of each sample was carried out using the RDP Classifier at a bootstrap cutoff of 50% suggested by the RDP.20 In addition, the alpha diversity including Shannon and Simpson diversity indices, species richness estimator of Chao1 and Ace, and diversity coverage were calculated in MOTHUR at cutoff level of 3%, while rarefaction curves were built using the relevant RDP pipeline modules as described elsewhere.21
461). Taxonomic classification of the sequences of each sample was carried out using the RDP Classifier at a bootstrap cutoff of 50% suggested by the RDP.20 In addition, the alpha diversity including Shannon and Simpson diversity indices, species richness estimator of Chao1 and Ace, and diversity coverage were calculated in MOTHUR at cutoff level of 3%, while rarefaction curves were built using the relevant RDP pipeline modules as described elsewhere.21
2.6. Statistical analysis
The results of chemical parameters and the genes copy concentrations were visualized through OriginPro 9.0 (OriginLab, USA). Pearson correlation coefficients were calculated using SPSS Statistics 20 (IBM, USA). An unconstrained ordination technique, principal component analysis (PCA) was conducted using Canoco 4.5 (Microcomputer Power, USA). A heat map of the top 10 genera in each sample based on reads log2 transformed was built using the HemI 1.0 (http://www.hemi.biocuckoo.org/).2.7. Mathematical models
The modified Boltzmann model (eqn (2))22 was employed to quantify the in situ restoration of the CPNA process. The curve fitting was conducted by OriginPro 9.0 (OriginLab, USA).
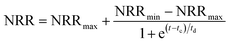 | (2) |
The exponential equations for the growth of functional genes were simulated by eqn (3).
| X = X0eμt | (3) |
3. Results and discussion
3.1. Nitrate accumulation and in situ restoration of the CPNA process
Once the growth of NOB was not under the effective control and accumulated in system, the nitrate build-up would occur causing the nitrogen removal deterioration of the CPNA process. Fig. 2 showed the operation of the CPNA process that could be divided into three stages, i.e., deterioration by nitrate build-up, in situ restoration and finally recovered stable operation. The average pH in SBR was kept in the range of 7.42–7.96 through all the stages (Fig. S1 in ESI†). Since the 11 days of doubling time of the anammox microorganisms were much longer than that of the general autotrophic bacteria,2 the daily sludge discharging for SRT control could not effectively wash out the accumulated NOB. In the initial period of deterioration of day 118 to 163 with the controlled 1200 mg L−1 of influent ammonium concentration, even though the build-up of nitrate gradually increased from 89.95 mg L−1 to 274.4 mg L−1, the obtained MRNN still did not exceed 23.92%, which was close to the acceptable value of 20%,7 thus the nitrogen removal rate (NRR) was maintained at 0.42 kgN m−3 d−1. However, later when the influent ammonium decreased to 800 mg L−1 from day 189 to 236, the nitrate concentration did not decrease according to the stoichiometric ratio of the CPNA equation but further increased to the highest 548.4 mg L−1 (day 235), and the MRNN also increased to the highest 86.99% on day 237. The activities of AOB and anammox were decreased to 0.37 and 0.20 gN gMLSS−1 d−1 on day 229, respectively, while the NOB activity increased to 0.91 gN gMLSS−1 d−1 on day 235 (Fig. S2 in ESI†). These results fully demonstrated the high population of NOB had been developed in system. | ||
| Fig. 2 The operational results of the CPNA process in the stages of deterioration, in situ restoration and recovered stable operation. | ||
In the first period of in situ restoration stage from day 237 to 305, different dosages of NH2OH were added into the system obtaining the total concentrations of 5, 10 and 20 mgNH2OH L−1 in each SBR cycle (Fig. 2), which were close to the concentrations used by Zekker et al.11 and Kindaichi et al.23 No sludge was discharged for accelerating the enrichment of anammox microorganisms. The nitrate concentration decreased sharply to 106.9 mg L−1 on day 305, and the MRNN also declined to 13.12%, which was less than the normal value of 15% for the stable CPNA process.15 These results showed an illusion that dosing NH2OH alone could restore the nitrate build-up problem, which was also proved by the optimized activities of AOB, NOB and anammox as well as the nitrite accumulation rate (Fig. S2 in ESI†), because it was unexpected to find that the nitrate build-up occurred again with the concentration increased to 287.90 mg L−1 as well as the resulted 38.6% of MRNN during the following days of 305 to 315 when the NH2OH dosing was stopped. This indicated that dosing NH2OH alone could not inhibit the NOB irreversibly. Therefore, in the second period of the in situ restoration from day 316 to 349, both NH2OH dosing and controlled 40 days of SRT control by sludge wasting were applied to inhibit and wash out the NOB simultaneously, through which the MRNN was decreased to around 15% again. The following days were regarded as the recovered stable operation section, since the MRNN was maintained stable under the normal operation condition without NH2OH dosing, the 0.59 kgN m−3 d−1 of NRR was recovered, indicating that the nitrate build-up problem of CPNA process was successfully in situ restored by the combination of NH2OH dosing and SRT control.
The modified Boltzmann model was applied to simulated the in situ restoration of the CPNA process as showed in Fig. S3 in ESI.† The regression of the experimental data yielded eqn (4) for the recovery of the NRR as follows:
| NRR = 0.61 − 0.40/[1 + exp(t − 36.6)/10.1], R2 = 0.88 | (4) |
The centre tc value provided by the modified Boltzmann model was 36.6 d, suggesting that the regained performance of the CPNA process needed a quite long period, which was extraordinary close to the second experimental in situ restoration stage from day 316 to 349 with the combination of NH2OH dosing and SRT control. However, the NRRmax predicated by the modified Boltzmann model was 0.61 kgN m−3 d−1, which was lower than actual nitrogen removal capacity (0.69 kgN m−3 d−1). This reason might be that lower R2 of the modeling and the co-existence of two pathways that supplied considerable nitrite for anammox, i.e., conventional partial nitrification by AOB, and partial denitrification of the high-strength accumulated nitrate to nitrite, hence higher NRR was obtained after the in situ restoration.
3.2. Response of functional genes
The Pearson correlation was calculated to investigated the interactions between the N-transformation genes, activities of AOB, NOB and anammox and the operational performance of the CPNA process. As showed in Table 1, the Pearson correlation coefficients between amoA–hao, nirS–narG, hzo–hao, hzo–narG and hzo–nirS all exceeded R = 0.92 (P < 0.01), the first two pairs of genes were connected by the intermediates of NH2OH and nitrite in nitritation and denitrification processes, respectively, while the hzo–hao and hzo–nirS cooperatively worked in the pathway I and II of the anammox process, respectively (Fig. 1). Under the circumstance of the build-up of high-strength nitrate in CPNA process, the high correlated hzo–narG demonstrated the high intensity of denitrification encoded by narG produced sufficient nitrite from nitrate reduction for the anammox process. The correlation coefficient of 0.838 (P < 0.01) between anammox activity and nirS was much higher than that between anammox activity and amoA and hao, indicating pathway II was the main anammox process in this study, which was also proved by the higher coefficient of 0.782 (P < 0.05) between NRR and nirS over than the 0.565 of coefficient between NRR and hao gene. Among all the N-transformation genes, only the nxrA gene was significant correlated with the nitrate concentration (R = 0.712, P < 0.05), this proved that the overgrowth of NOB in system was the main reason for the nitrate build-up.
| amoA | hao | nxrA | narG | nirS | nirK | norB | nosZ | hzo | AOB activity | NOB activity | Anammox activity | NO2−–N accumulation | MRNN | NRR | Effluent NO3−–N | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a Correlation is significant at the 0.01 level (2-tailed).b Correlation is significant at the 0.05 level (2-tailed). | ||||||||||||||||
| amoA | 1 | |||||||||||||||
| hao | 0.966a | 1 | ||||||||||||||
| nxrA | −0.217 | −0.326 | 1 | |||||||||||||
| narG | 0.846a | 0.878a | −0.054 | 1 | ||||||||||||
| nirS | 0.792b | 0.887a | −0.257 | 0.925a | 1 | |||||||||||
| nirK | 0.720b | 0.643 | −0.123 | 0.391 | 0.275 | 1 | ||||||||||
| norB | 0.658 | 0.729b | 0.155 | 0.773b | 0.835a | 0.329 | 1 | |||||||||
| nosZ | 0.157 | 0.243 | −0.574 | 0.299 | 0.422 | −0.277 | −0.075 | 1 | ||||||||
| hzo | 0.885a | 0.921a | −0.120 | 0.973a | 0.955a | 0.370 | 0.802b | 0.315 | 1 | |||||||
| AOB activity | 0.729b | 0.719b | −0.623 | 0.445 | 0.436 | 0.787b | 0.148 | 0.333 | 0.450 | 1 | ||||||
| NOB activity | 0.419 | 0.325 | −0.303 | 0.201 | 0.067 | 0.595 | −0.225 | 0.386 | 0.137 | 0.795b | 1 | |||||
| Anammox activity | 0.754b | 0.799b | −0.602 | 0.727b | 0.838a | 0.247 | 0.476 | 0.658 | 0.791b | 0.626 | 0.310 | 1 | ||||
| NO2−–N accumulation | 0.861a | 0.893a | −0.646 | 0.673 | 0.725b | 0.680 | 0.436 | 0.404 | 0.707b | 0.919a | 0.559 | 0.847a | 1 | |||
| MRNN | −0.648 | −0.716b | 0.446 | −0.598 | −0.749b | −0.364 | −0.700 | −0.102 | −0.654 | −0.426 | 0.099 | −0.745b | −0.706 | 1 | ||
| NRR | 0.427 | 0.565 | −0.531 | 0.555 | 0.782b | 0.015 | 0.605 | 0.491 | 0.592 | 0.305 | −0.099 | 0.794b | 0.602 | −0.861a | 1 | |
| Effluent NO3−–N | −0.685 | −0.730b | 0.712b | −0.501 | −0.590 | −0.572 | −0.379 | −0.181 | −0.536 | −0.744b | −0.259 | −0.745b | −0.876a | 0.865a | −0.678 | 1 |
The coefficient between NRR and anammox activity (R = 0.794, P < 0.05) was much higher compared with that of NRR–hzo (R = 0.592), indicating that the bacterial activities contributed much more than the gene concentrations to improving the nitrogen removal. The same conclusion could also be drawn from the comparison that the correlation coefficients of nitrate–AOB activity (R = −0.744, P < 0.05), nitrate–anammox activity (R = −0.745, P < −0.05) were higher than that of nitrate–hao (R = −0.73, P < 0.05), nitrate–hzo (R = −0.536). This result partly explained that though the concentration of the nitrite-oxidizing enzyme encoding gene nxrA was lower than other genes with 1 to 5 orders of magnitude (Fig. 3), the NOB activity was still comparable to those of AOB and anammox, thus able to lead to the build-up of nitrate. Our previous work had also detected the two kinds of NOB, i.e., Nitrobacter and Nitrospira, in this process of the nitrate build-up and the following in situ restoration by quantifying their 16S rRNA genes,9 the results showed that the abundance of Nitrobacter was relatively stable at 2.0% throughout this process while the abundance of Nitrospira fluctuated in the range of 5.21% to 2.34%. The similar situation had also occurred in the research by Langone et al.,24 where the primers targeting the 16S rRNA of Nitrobacter-like NOB yielded products of the correct size (225 nt), while no PCR products were obtained using the primers targeting the nxrA gene of Nitrobacter spp. The discrepancy of the NOB abundances based on the detections of 16S rRNA and nxrA gene might be attributed to that the nxrA gene containing cell numbers were too low and the other two of nxrB and nxrC genes functioned much in the nitrate build-up process.25
| Gene | Exponential growth fitting equation | R2 | P | Doubling time (d) |
|---|---|---|---|---|
| amoA | X = 4.5238 × 109e0.04148t | 0.96013 | 0.07761 | 16.71 |
| hao | X = 1.83409 × 1010e0.02658t | 0.97657 | 0.04818 | 26.08 |
| narG | X = 1.64034 × 1010e0.02638t | 0.99466 | 0.02369 | 26.28 |
| nirS | X = 1.92881 × 1010e0.01363t | 0.98766 | 0.02148 | 50.85 |
| nirK | X = 5.27902 × 107e0.01818t | 0.99721 | 0.0131 | 38.13 |
| norB | X = 2.33223 × 106e0.0219t | 0.90391 | 0.09308 | 31.65 |
| hzo | X = 6.85171 × 109e0.03444t | 0.98634 | 0.04256 | 20.13 |
PCA analysis was conducted to investigate the relationships between functional genes, bacterial activities and nitrogen removal performance (Fig. 4), and the results showed that the MRNN and the nitrate concentration in effluent were highly correlated with nxrA gene, while the NRR had a significantly positive relationship with the hzo and nirS genes. The anammox activity was influenced by the concentrations of amoA (R = 0.754, P < 0.05), hao (R = 0.799, P < 0.05) and narG (R = 0.727, P < 0.05), which all encoded the enzymes evolved in the nitrite production from ammonium oxidation and nitrate reduction, respectively. PCA also reflected the gene distribution along with the operation of CPNA process. Only the NOB contained nxrA gene contributed much more in the period of day 186 and 230, during which the nitrate build-up was occurred in system. The genes of hzo, nirS, hao, amoA, narG encoding the processes of nitrite production, nitric oxide production and anammox drew close on day 376 during the recovered stable operation, which all contributed to the nitrogen removal through anammox.
A series of stepwise regression models were built to provide a linear quantitative measure of gene association with the functional bacteria activities and nitrogen removal performance. All the functional genes as showed in Fig. 1 were used as the candidate variables, the obtained stepwise regression equations were showed in Table 3. The correlation coefficients between NRR and nirS, hzo were the highest compared with others as showed in Fig. 4, which was also demonstrated through the stepwise regression model that all other genes were excluded but only the nirS and hzo genes were included in the equation of NRR (R2 = 0.882). In the regression equation of nitrite accumulation rate (R2 = 0.939), the positive coefficient of hao gene was consistent with the encoded enzyme for nitrite production, the adverse impact of the nxrA on the nitrite accumulation was also reflected by its negative coefficient (−8.963 × 10−7).
| Equations | R2 | P |
|---|---|---|
| NRR = 3.986 × 10−11nirS − 1.923 × 10−11hzo − 0.87 | 0.882 | 0.005 |
| MRNN = −0.8045 × 10−10nirS + 43.853 | 0.561 | 0.032 |
| AOB activity = 4.401 × 10−0.09nirK − 1.039 × 10−6nxrA + 0.627 | 0.900 | 0.003 |
| Anammox activity = 3.396 × 10−11nirS − 6.919 × 10−0.08norB + 0.183 | 0.867 | 0.006 |
| Nitrite accumulation rate = 1.007 × 10−11hao − 8.963 × 10−7nxrA + 0.464 | 0.939 | 0.001 |
3.3. Evolution of the microbial community revealed by high-throughput sequencing
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 441 high quality sequences were obtained from 7 sludge samples, ranging from 30
441 high quality sequences were obtained from 7 sludge samples, ranging from 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 reads to 40
546 reads to 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 555 reads. To compare each other at the same sequencing depth, all sample sequences were normalized to 30
555 reads. To compare each other at the same sequencing depth, all sample sequences were normalized to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 reads for the downstream analysis. The rarefaction curves (Fig. S6 in ESI†) and the coverage values (Table 4) suggested that the sequencing depth in this study was enough to detect almost all of OTUs in the reactor. The alpha diversity indices for each sample at distance cutoff level of 0.03 were showed in Table 4. The diversity of microbial community was firstly increased along with the nitrate build-up then decreased in the following periods of in situ restoration and recovered operation, which was revealed by the OTUs and Chao1. The results of Shannon diversity index also demonstrated that the bacterial diversity in the CPNA process was firstly increased to 4.11 when the nitrate built up in system and then declined to finally 2.71 after the stable operation was recovered. This result indicated the unwanted inferring bacteria was effectively eliminated, the nitrogen removal functional microorganisms were selectively enriched and a more focused microbial community structure was obtained.
546 reads for the downstream analysis. The rarefaction curves (Fig. S6 in ESI†) and the coverage values (Table 4) suggested that the sequencing depth in this study was enough to detect almost all of OTUs in the reactor. The alpha diversity indices for each sample at distance cutoff level of 0.03 were showed in Table 4. The diversity of microbial community was firstly increased along with the nitrate build-up then decreased in the following periods of in situ restoration and recovered operation, which was revealed by the OTUs and Chao1. The results of Shannon diversity index also demonstrated that the bacterial diversity in the CPNA process was firstly increased to 4.11 when the nitrate built up in system and then declined to finally 2.71 after the stable operation was recovered. This result indicated the unwanted inferring bacteria was effectively eliminated, the nitrogen removal functional microorganisms were selectively enriched and a more focused microbial community structure was obtained.
| Sample ID | Sequences | 0.03 distance | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Raw | Clean | Normalized | OTUs | Ace | Chao1 | Coverage | Shannon | Simpson | |
| a OTUs, Ace, Chao1, Shannon and Simpson were all microbial community richness and diversity indices which were calculated using the relevant modules in MOTHUR. | |||||||||
| Day 186 | 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 628 628 |
34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 929 929 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
410 | 455 | 464 | 0.9976 | 4.01 | 0.0462 |
| Day 230 | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 645 645 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 921 921 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
453 | 511 | 504 | 0.9974 | 4.11 | 0.0387 |
| Day 251 | 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 506 506 |
39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 422 422 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
422 | 491 | 485 | 0.9971 | 3.84 | 0.0461 |
| Day 282 | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 994 994 |
34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130 130 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
412 | 461 | 464 | 0.9976 | 3.86 | 0.045 |
| Day 307 | 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 531 531 |
36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 769 769 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
362 | 458 | 453 | 0.9968 | 3.62 | 0.0552 |
| Day 349 | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 761 761 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
267 | 353 | 340 | 0.9975 | 2.98 | 0.1131 |
| Day 376 | 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 878 878 |
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 555 555 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
267 | 346 | 346 | 0.9972 | 2.71 | 0.1506 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 selected effective bacterial sequences in each sample were assigned to different taxa levels (from genus to phylum) using the RDP Classifier at 50% threshold. As shown in Fig. S7 in ESI,† Bacteroidetes, Proteobacteria, Chloroflexi, and Nitrospira were the dominant phyla in bacterial communities of the 7 samples in this study, which were similar to the previous studies towards the anammox-based processes.28,29 The abundance of phylum Bacteroidetes was continuously decreased from 32.8% on day 186 to 22.2% on day 230 during the deterioration, then increased gradually to 49.8% on day 376 in recovered stable operation. On the contrary, the abundance of phylum Chloroflexi was increased from 5.4% (day 186) to 18.3% (day 230), then decreased continuously to 4.4% (day 376).
546 selected effective bacterial sequences in each sample were assigned to different taxa levels (from genus to phylum) using the RDP Classifier at 50% threshold. As shown in Fig. S7 in ESI,† Bacteroidetes, Proteobacteria, Chloroflexi, and Nitrospira were the dominant phyla in bacterial communities of the 7 samples in this study, which were similar to the previous studies towards the anammox-based processes.28,29 The abundance of phylum Bacteroidetes was continuously decreased from 32.8% on day 186 to 22.2% on day 230 during the deterioration, then increased gradually to 49.8% on day 376 in recovered stable operation. On the contrary, the abundance of phylum Chloroflexi was increased from 5.4% (day 186) to 18.3% (day 230), then decreased continuously to 4.4% (day 376).For detailed analysis of the bacterial community structure during the deterioration and subsequent restoration stages, the top 10 abundant genera in each sample were selected (a total of 21 genera for all 7 samples) and compared their abundances with each other as shown in Fig. 5. The in situ restoration strategy increased the abundance of the genus Nitrosomonas for oxidizing ammonium to nitrite from 5.7% to finally increased to 13.8% on day 376. In the meantime, the members of genus Nitrospira, which acted as the NOB causing the nitrate build-up, bloomed to the highest abundance of 14.7% on day 251 even though the 5 mgNH2OH L−1 of hydroxylamine had already dosed, but after restoration its abundance was decreased to only 3.4% on day 349. Candidatus Kuenenia was the dominant anammox bacterium in the CPNA process of this study, the abundance of which was increased from 2.8% in deterioration to 8.0% on day 349. These results were consistent with the recovery of AOB and anammox activities that realizing the high rate of nitrite accumulation and anammox nitrogen removal (Fig. S2 in ESI†).
Pearson correlation analysis (Table S2 in ESI†) showed that the amoA and hao genes encoding the ammonium oxidation were significantly correlated with the abundances of Nitrosomonas genus (R2 = 0.933, P < 0.01 and R2 = 0.766, P < 0.05, respectively), while the nxrA gene in NOB was correlated with the genera of Chthonomonas (R2 = 0.755, P < 0.05), Parcubacteria genera incertae sedis (R2 = 0.826, P < 0.05) and Phycisphaera (R2 = 0.867, P < 0.05), and the abundances of which were all decreased after restoration (Fig. 5a). The increase of anammox gene hzo was reflected by the decrease of the abundance of Subdivision3 genera incertae sedis (from 0.36% to 0.05%) as the significant negative correlation was calculated between each other (R2 = −0.806, P < 0.05). But it was unexpected to find that there was no significant correlation relationship between the hzo gene and the Candidatus Kuenenia, and this could be explained by the relative low abundance of Candidatus Kuenenia detected by aligning the OTU to the RDP database.
High abundance of Haliscomenobacter in the reactor became the dominant population after restoration. Haliscomenobacter is known as the filamentous activated sludge bacterium which is associated with O2-deficient situations.30 The abundance of the genus Haliscomenobacter was increased from 4.0% (day 230) to 21.1% (day 376). Several similar observations of the proliferation of such filamentous bacteria in other anammox reactors had been reported,31,32 and the positive effect of the filamentous bacteria on conferring the structural integrity or forming the initial framework by providing a stabilizing backbone for the small anammox aggregates was speculated in the CPNA process. The highest 7.0% of the abundance of Ignavibacterium, another typical filamentous bacterium, was detected on day 251, which was much lower than the 32% from Li et al.33 The Acidobacteria GP4 is a common genus that had been identified from many fields,34 the abundance of which was kept increasing from 0.12% (day 186) to 11.3% (day 349), and then decreased to 2.6% after restoration. Researchers had pointed out that in system with low COD concentration, the filamentous microorganisms had a higher specific growth rate than the floc-forming bacteria, and could perform the denitrification function.35 This might be the reason for the massive proliferation of filamentous bacteria, i.e., Haliscomenobacter and Ignavibacterium, in the CPNA process, where the high concentration of nitrate was accumulated. Additionally, the Pearson correlation analysis showed that the denitrifying gene nirK had a significant correlation relationship with the Ignavibacterium (R2 = 0.802, P < 0.05) (Table S2 in ESI†).
The principal component analysis (PCA) in Fig. 5b showed the evolution of the bacterial community on the genus level along with the operation of the CPNA process. It was found that the bacterial communities of day 349 and 376 were quite different from those before, indicating that the new bacterial community of the CPNA process was rebuilt after the in situ restoration. Most of the genera contributed to the bacterial community in the processes of deterioration and the first period of in situ restoration as detected by the samples of day 186, 230, 251, and 282, while only the genera of Candidatus Kuenenia, Haliscomenobacter, GP4 and Byssovorax contributed in the recovered stable operation (day 376). These results not only were consistent with the reduction of the Shannon diversity index in Table 4 that the elimination of other unwanted bacteria was effective by the in situ restoration strategy to make the bacterial community more concentrated on nitrogen removal, but also demonstrated that the high contribution of the anammox genera Candidatus Kuenenia in CPNA process was restored as well as the proliferation of the filamentous bacterium of Haliscomenobacter to carry out the reduction of nitrate to the nitrite for the anammox microorganism.
4. Conclusions
The combination of NH2OH dosing and SRT control could in situ restore the nitrate build-up problem of the CPNA process. Along with the nitrate build-up, the concentrations of amoA, hao and hzo were all decreased, however, after restoration their abundances were recovered to 26.58%, 43.56% and 26.17%, respectively. Pearson correlation analysis and PCA result showed that the overgrowth of NOB (nxrA) was the main reason for the nitrate build-up, while the intensive denitrification of the accumulated nitrate to nitric oxide (narG, nirS) and the stimulation of anammox (hzo) by nitric oxide was the mechanism of removing the accumulated nitrate and recovering the nitrogen removal. The microbial diversity index was decreased after restoration and a new microbial community was rebuilt with the Haliscomenobacter and Nitrosomonas as the dominant genera (21.1% and 13.8%, respectively). Candidatus Kuenenia was the dominant anammox bacterium with its abundance increased to 8.0%. The significant correlation between the denitrifying gene nirK and the filament Ignavibacterium (R2 = 0.802, P < 0.05) illustrated that the massive proliferation of filaments might be attributed to their obligate denitrifying function in conquering the nitrate build-up.Acknowledgements
This study was supported by the National Water Pollution Control and Management Technology Major Project (No. 2012ZX07203-002; No. 2015ZX07203-005), the National Natural Science Foundation of China (No. 21607167), and the Project for Jiangxi-CAS Collaborative Innovation Team-building (2014SYXTCX-02).References
- A. Mulder, A. A. Vandegraaf, L. A. Robertson and J. G. Kuenen, FEMS Microbiol. Ecol., 1995, 16, 177–183 CrossRef CAS.
- M. Strous, J. J. Heijnen, J. G. Kuenen and M. S. M. Jetten, Appl. Microbiol. Biotechnol., 1998, 50, 589–596 CrossRef CAS.
- M. Strous, E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartol-Mavel, P. Wincker, V. Barbe, N. Fonknechten, D. Vallenet, B. Segurens, C. Schenowitz-Truong, C. Medigue, A. Collingro, B. Snel, B. E. Dutilh, H. J. M. Op den Camp, C. van der Drift, I. Cirpus, K. T. van de Pas-Schoonen, H. R. Harhangi, L. van Niftrik, M. Schmid, J. Keltjens, J. van de Vossenberg, B. Kartal, H. Meier, D. Frishman, M. A. Huynen, H. W. Mewes, J. Weissenbach, M. S. M. Jetten, M. Wagner and D. Le Paslier, Nature, 2006, 440, 790–794 CrossRef PubMed.
- M. Strous, J. A. Fuerst, E. H. M. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen and M. S. M. Jetten, Nature, 1999, 400, 446–449 CrossRef CAS PubMed.
- W. R. van der Star, W. R. Abma, D. Blommers, J. W. Mulder, T. Tokutomi, M. Strous, C. Picioreanu and M. C. van Loosdrecht, Water Res., 2007, 41, 4149–4163 CrossRef CAS PubMed.
- S. Lackner, E. M. Gilbert, S. E. Vlaeminck, A. Joss, H. Horn and M. C. M. van Loosdrecht, Water Res., 2014, 55, 292–303 CrossRef CAS PubMed.
- A. Joss, N. Derlon, C. Cyprien, S. Burger, I. Szivak, J. Traber, H. Siegrist and E. Morgenroth, Environ. Sci. Technol., 2011, 45, 9735–9742 CrossRef CAS PubMed.
- N. Jardin and J. Hennerkes, Water Sci. Technol., 2012, 65, 447–455 CrossRef CAS PubMed.
- Y. Wang, Y. Wang, Y. Wei and M. Chen, Biochem. Eng. J., 2015, 98, 127–136 CrossRef CAS.
- W. F. Harper Jr, A. Terada, F. Poly, X. Le Roux, K. Kristensen, M. Mazher and B. F. Smets, Biotechnol. Bioeng., 2009, 102, 714–724 CrossRef PubMed.
- I. Zekker, K. Kroon, E. Rikmann, T. Tenno, M. Tomingas, P. Vabamaee, S. E. Vlaeminck and T. Tenno, Biodegradation, 2012, 23, 739–749 CrossRef CAS PubMed.
- A. A. van de Graaf, P. de Bruijn, L. A. Robertson, M. S. M. Jetten and J. G. Kuenen, Microbiology, 1997, 143, 2415–2421 CrossRef CAS.
- B. Kartal, N. C. G. Tan, E. Van de Biezen, M. J. Kampschreur, M. C. M. Van Loosdrecht and M. S. M. Jetten, Appl. Environ. Microbiol., 2010, 76, 6304–6306 CrossRef CAS PubMed.
- B. Wett, G. Nyhuis, I. Takács and S. Murthy, Proceedings of the Water Environment Federation, 2010, 2010, 5917–5926 CrossRef.
- A. O. Sliekers, N. Derwort, J. L. C. Gomez, M. Strous, J. G. Kuenen and M. S. M. Jetten, Water Res., 2002, 36, 2475–2482 CrossRef CAS PubMed.
- APHA, Standard methods for the examination of water and wastewater, Washington, 21st edn, 2005 Search PubMed.
- J. G. Caporaso, C. L. Lauber, W. A. Walters, D. Berg-Lyons, C. A. Lozupone, P. J. Turnbaugh, N. Fierer and R. Knight, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4516–4522 CrossRef CAS PubMed.
- T. Magoč and S. L. Salzberg, Bioinformatics, 2011, 27, 2957–2963 CrossRef PubMed.
- R. C. Edgar, B. J. Haas, J. C. Clemente, C. Quince and R. Knight, Bioinformatics, 2011, 27, 2194–2200 CrossRef CAS PubMed.
- Q. Wang, G. M. Garrity, J. M. Tiedje and J. R. Cole, Appl. Environ. Microbiol., 2007, 73, 5261–5267 CrossRef CAS PubMed.
- J. Zhang, X. Cai, L. Qi, C. Shao, Y. Lin, J. Zhang, Y. Zhang, P. Shen and Y. Wei, Appl. Microbiol. Biotechnol., 2015, 99, 7321–7331 CrossRef CAS PubMed.
- R. C. Jin, Q. Q. Zhang, G. F. Yang, B. S. Xing, Y. X. Ji and H. Chen, Bioresour. Technol., 2013, 142, 162–170 CrossRef CAS PubMed.
- T. Kindaichi, S. Okabe, H. Satoh and Y. Watanabe, Water Sci. Technol., 2004, 49, 61–68 CAS.
- M. Langone, J. Yan, S. Haaijer, H. Op Den Camp, M. Jetten and G. Andreottola, Front. Microbiol., 2014, 5(28), 1–12 Search PubMed.
- D. Y. Sorokin, S. Lücker, D. Vejmelkova, N. A. Kostrikina, R. Kleerebezem, W. I. C. Rijpstra, J. S. S. Damsté, D. Le Paslier, G. Muyzer and M. Wagner, ISME J., 2012, 6, 2245–2256 CrossRef CAS PubMed.
- J. Pérez, T. Lotti, R. Kleerebezem, C. Picioreanu and M. C. M. van Loosdrecht, Water Res., 2014, 66, 208–218 CrossRef PubMed.
- D. Kaelin, R. Manser, L. Rieger, J. Eugster, K. Rottermann and H. Siegrist, Water Res., 2009, 43, 1680–1692 CrossRef CAS PubMed.
- X. R. Li, B. Du, H. X. Fu, R. F. Wang, J. H. Shi, Y. Wang, M. S. M. Jetten and Z.-X. Quan, Syst. Appl. Microbiol., 2009, 32, 278–289 CrossRef CAS PubMed.
- L. Wang, P. Zheng, T. Chen, J. Chen, Y. Xing, Q. Ji, M. Zhang and J. Zhang, Bioresour. Technol., 2012, 123, 78–85 CrossRef CAS PubMed.
- G. Gaval and J.-J. Pernelle, Water Res., 2003, 37, 1991–2000 CrossRef CAS PubMed.
- Z. R. Chu, K. Wang, X. K. Li, M. T. Zhu, L. Yang and J. Zhang, Chem. Eng. J., 2015, 262, 41–48 CrossRef CAS.
- H. Park, A. Rosenthal, K. Ramalingam, J. Fillos and K. Chandran, Environ. Sci. Technol., 2010, 44, 6110–6116 CrossRef CAS PubMed.
- Z. Li, Y. Ma, D. Hira, T. Fujii and K. Furukawa, Bioresour. Technol., 2011, 102, 5702–5708 CrossRef CAS PubMed.
- T. Zhang, M. F. Shao and L. Ye, ISME J., 2012, 6, 1137–1147 CrossRef CAS PubMed.
- A. M. P. Martins, K. Pagilla, J. J. Heijnen and M. C. M. van Loosdrecht, Water Res., 2004, 38, 793–817 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra23865c |
| This journal is © The Royal Society of Chemistry 2016 |



