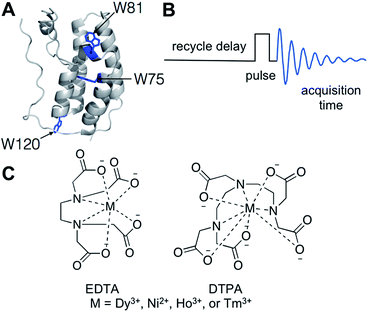
Paramagnetic relaxation enhancement for protein-observed 19F NMR as an enabling approach for efficient fragment screening†
Laura M. L. Hawka,
Clifford T. Geea,
Andrew K. Urickab,
Haitao Hub and
William C. K. Pomerantz*a
aDepartment of Chemistry, University of Minnesota, 207 Pleasant St. SE, Minneapolis, MN 55455, USA. E-mail: wcp@umn.edu
bDiscovery Chemistry Research & Technologies, Lilly Research Laboratories, Eli Lilly and Company, Lilly Corporate Center, Indianapolis, IN 46285, USA
First published on 29th September 2016
Abstract
Protein-observed 19F (PrOF) NMR is an emerging tool for ligand discovery. To optimize the efficiency of PrOF NMR experiments, paramagnetic relaxation enhancement through the addition of chelated Ni(II) was used to shorten longitudinal relaxation time without causing significant line broadening. Thus enhancing relaxation time leads to shorter experiments without perturbing the binding of low- or high-affinity ligands. This method allows for time-efficient screening of potential ligands for a wide variety of proteins in the growing field of fragment-based ligand discovery.
Introduction
Biomolecular NMR is a widespread tool for evaluating the structure and dynamics of biopolymers in isolation and in complexes. In the context of small molecule discovery, biomolecular NMR of proteins labelled with NMR active nuclei (e.g., 1H–15N HSQC NMR) has proven to be a valuable structure-based method for identifying binding sites and quantifying affinities of small molecule ligands.1,2 New fast pulsing methods such as SOFAST-HMQC have decreased experiment time considerably; however, screening large compound libraries for protein binding by NMR can be a time- and material-intensive process.3 We recently reported a protein-observed 19F NMR (PrOF NMR) method for small molecule screening using a fluorine-labelled protein domain, KIX.4 In that study, 508 small molecules were screened in mixtures of six requiring five minutes per experiment or 510 minutes of total experiment time and 20 mg of protein. Although this approach gives a modest time enhancement over SOFAST 1H–15N HMQC NMR on small to medium sized proteins,5 the length and material consumption can still be limiting.In NMR experiments, the recycle delay, during which the spectrometer is idle while the magnetization relaxes after a radio frequency pulse, comprises up to 80% or more of the total experiment time (Fig. 1B).6 Shorter recycle delays allow more scans per unit time, leading to more efficient instrument use. One strategy employed to shorten recycle delays is the addition of a paramagnetic metal to decrease the longitudinal relaxation time (T1) of 1H, 13C, and 15N nuclei in labelled proteins while minimizing unfavourable line-broadening from decreasing transverse relaxation time (T2).6–11 The application of paramagnetic additives to 19F NMR has also been described in the context of NMR with molecular oxygen.12 However, the specialized NMR tubes required would not be ideal for routine small molecule screening. These studies did not explore if the beneficial effects of paramagnetic metal additives are retained in the presence of bound ligands of varying affinity, a question we examine herein.
In this work, we explore the application of paramagnetic relaxation enhancement (PRE) to PrOF NMR, utilizing a solvent-accessible side-chain labelling scheme. This labelling scheme is well-suited to studying protein–protein interaction interfaces, which contain a high proportion of aromatic amino acids13 that can readily be biosynthetically replaced with their fluorinated counterparts. The appended fluorine atoms on the side chains can be more solvent exposed than backbone amides, and thus more accessible to paramagnetic additives in the bulk solvent. We demonstrate that PRE can reduce experiment times significantly by shortening the recycle delay with minimal line broadening from T2 effects, ultimately leading to a marked enhancement in sensitivity. In the context of low-complexity molecules commonly used in fragment-based ligand discovery screening, we further show improvements by using this method for rapidly obtaining dissociation constants for weak-binding fragments. In addition to fragment screening, this method may also be useful for proteins available in limited abundance such as GPCRs,14,15 as the signal to noise increases by the square root of the number of scans. Importantly, this approach is also compatible with proteins containing metal binding hexahistidine affinity tags and with various fluorinated amino acids.
NMR studies of fluorinated biomolecules have been applied to diverse biophysical processes, including protein folding and ligand binding.16,17 The high sensitivity (83% that of proton), 100% natural abundance, absence of background in biological systems, and large chemical shift range all serve to make 19F NMR an attractive tool for characterizing biological systems. Several recent studies have screened for ligand binding by PrOF NMR.4,18–21 Although the 19F NMR spectra can be readily acquired,5 library screening by this method nevertheless requires a significant amount of experiment time. Shortened experiments would increase throughput in a small molecule NMR screen where the number of experiments may be large.
Results and discussion
To find a metal that would shorten T1 while minimally affecting T2, the effects of several metals on 19F T1 and T2 times were compared. To minimize interactions between the metal and protein or small molecule, the metals were chelated, initially with the common chelator EDTA. For subsequent work we moved to DTPA, which forms a more stable metal chelate (Fig. 1C).22,23 We initially tested nickel, dysprosium, holmium, and thulium chelates to determine their effect on the relaxation properties of small molecule 4-fluorophenylalanine (4FF).16,17 In addition to changing the sample's relaxation properties, the three lanthanides tested also cause lanthanide-induced shifts in the NMR spectrum, which have been utilized in 2D 1H–1H and 1H–15N systems to resolve resonance overlap.23 Because of the environmental sensitivity of the 19F nucleus, most PrOF NMR spectra of folded proteins show strong resonance dispersion and would not significantly benefit from lanthanide-induced shift reagents. Chelated Ni(II) showed the strongest effect on T1 of the metals tested (Fig. S1A†), was the most economical, and did not introduce the additional complications of a shift reagent. Therefore, it was selected for further study.To determine the optimal chelated Ni(II) concentration for protein studies, we explored the effect of a range of concentrations of chelated Ni(II) on the relaxation properties of 4FF (Fig. S1A†) as well as 5-fluoroindole (Fig. S1B†), the side chain present in proteins that have been 19F-labelled at their tryptophan residues. The T1 values of both of these molecules at Ni(II) concentrations up to 320 mM for 4FF and 200 mM for 5-fluoroindole were determined by inversion recovery experiments.  values were determined from the peakwidth at half height. Because of the importance of peakwidth for resolving spectral resonances,
values were determined from the peakwidth at half height. Because of the importance of peakwidth for resolving spectral resonances,  was used rather than T2. For both 4FF and 5-fluoroindole, the highest Ni(II) concentration tested led to a more than a 100-fold decrease in T1 values (Table 1).
was used rather than T2. For both 4FF and 5-fluoroindole, the highest Ni(II) concentration tested led to a more than a 100-fold decrease in T1 values (Table 1).  also decreased with increasing chelated Ni(II) concentration, leading to approximately a 5-fold increase in peak width at 200 mM chelated Ni(II). Concentrations of 20–50 mM provide an optimal balance between T1 reduction and
also decreased with increasing chelated Ni(II) concentration, leading to approximately a 5-fold increase in peak width at 200 mM chelated Ni(II). Concentrations of 20–50 mM provide an optimal balance between T1 reduction and  reduction and agree well with the 50 mM Ni(II) used by Cai et al. in 1H, 15N, and 13C experiments.6
reduction and agree well with the 50 mM Ni(II) used by Cai et al. in 1H, 15N, and 13C experiments.6
 values for fluorine resonances of small molecules and protein Brd4 at increasing Ni(II) concentrationsa
values for fluorine resonances of small molecules and protein Brd4 at increasing Ni(II) concentrationsa
| Compound | Ni-DTPA (mM) | T1 (ms) | (ms) |
|---|---|---|---|
a Small molecules used in this study were 5-fluoroindole and 4-fluorophenylalanine (4FF). T1 and  values were also measured for 5-fluorotryptophan residues within the protein Brd4 with and without the addition of Brd4 ligand, JQ1, that binds near W81. values were also measured for 5-fluorotryptophan residues within the protein Brd4 with and without the addition of Brd4 ligand, JQ1, that binds near W81. |
|||
| 5-Fluoroindole | 0 | 4890 | 208 |
| 20 | 528 | 107 | |
| 50 | 202 | 80 | |
| 4FF | 0 | 1870 | 77 |
| 20 | 216 | 97 | |
| 40 | 113 | 80 | |
| W75 (+JQ1) | 0 | 508 (552) | 7.5 (7.1) |
| 20 | 269 (399) | 7.2 (6.2) | |
| 40 | 294 (346) | 6.4 (6.9) | |
| W120 (+JQ1) | 0 | 721 (685) | 8.4 (6.1) |
| 20 | 146 (212) | 8.0 (7.2) | |
| 40 | 86 (156) | 8.4 (8.1) | |
| W81 (+JQ1) | 0 | 837 (772) | 8.8 (7.0) |
| 20 | 196 (289) | 8.6 (5.4) | |
| 40 | 192 (149) | 8.6 (2.9) | |
We then moved to testing the effects of Ni-DTPA on a model protein, the first domain of the bromodomain-containing protein Brd4, hereafter referred to as Brd4. Bromodomains are epigenetic regulatory domains, inhibition of which has therapeutic implications for cancer, heart disease, and inflammation,24,25 and are often targeted in fragment-screening campaigns.26–28 For these studies, Brd4 was biosynthetically 19F-labelled using the amino acid precursor 5-fluoroindole at all three tryptophan sites (W75, W81, and W120; Fig. 1A) due to the enrichment of aromatic amino acids at protein–protein interaction interfaces.13,29 When small molecules or ε-acetylated histone substrates bind to Brd4, we have observed a significant perturbation of the W81 resonance, which is located in the binding pocket. The resonance corresponding to W75, which is located beneath the binding pocket, experiences minor perturbations due to the environmental sensitivity of fluorine. The resonance corresponding to W120, which is located 36 Å from the binding site, generally shows minimal perturbation. T1 measurements were carried out on 5-fluorotryptophan (5FW)-labelled Brd4 solutions titrated with Ni-DTPA without significantly affecting resonance width (Fig. S2†).
As expected, increasing concentrations of Ni-DTPA led to decreases in the T1 (Table 1 and Fig. S3†). The T1 of the fluorine appended to W120 decreases the most rapidly and continues decreasing as more Ni-DTPA is added, suggesting that this fluorine is most accessible to the paramagnetic additive (80% reduction in T1 at 20 mM Ni-DTPA), with W81 being intermediate in accessibility (75% reduction). The fluorine on W75 (50% reduction) is least accessible, and its T1 value is less affected by Ni-DTPA. While the level of accessibility to paramagnetic additives consistent with T1 measurements differs from the level of solvent exposure determined by the program GetArea,30 the accessibility of the appended fluorine atom may differ from that of the residue as a whole. The extent of T1 reduction upon addition of Ni-DTPA correlates with the apparent solvent exposure of the 5-position of the indole ring of the labelled tryptophans (Fig. S4†). These results are consistent with solvent accessible nuclei experiencing the strongest PRE effects.
The fluorines appended to W75 and W81 reach their T1 minima at 20 mM metal; doubling the metal concentration to 40 mM had little effect on T1 values. Therefore, 20 mM Ni-DTPA was used for further experiments to minimize the Q-damping effects resulting from additional ions in solution and concomitant loss of sensitivity.31
In the absence of Ni-DTPA, experiments were carried out with a recycle delay of 1 second (1.2 times the measured T1 value of W81, the residue most affected by ligand binding in the binding pocket), with a total experiment time of five minutes (280 scans) at modest protein concentrations of 50 μM. With 20 mM Ni-DTPA present, the recycle delay was shortened to 0.24 seconds for a consistent value of 1.2 times the measured T1 of W81. In this case, the experiment time was reduced to two minutes (350 scans) at the same protein concentration with similar signal-to-noise for the W81 resonance (11.8 with Ni-DTPA vs. 10.9 without), a 60% reduction in experiment time (Fig. 2). The signal-to-noise ratio may be further improved if Q-damping effects could be mitigated. The T1 value of W120 (0.15 s) is more reduced than that of W81 (0.20 s) by the addition of 20 mM Ni-DTPA. Consequently, the relative intensity of W120 increases with the addition of 20 mM Ni-DTPA because the net magnetization has more fully relaxed back to the ground state before application of the next pulse. In contrast, W75 has a longer T1 value (0.27 s) than W81 and thus its resonance is truncated by the short recycle delay of this experiment. Because W75 is farther from the binding pocket, the experiment was optimized for maximizing signal-to-noise per unit time of W81. Because binding sites must be accessible to their binding partners, amino acid side chains located at protein–protein interaction interfaces should also be accessible to paramagnetic additives and thus subject to substantial PRE effects, allowing for shortened recycle delays and experiment times.
The chemical shift of W75 of Brd4 resonance is slightly perturbed upon titration of Ni-DTPA, moving 0.04 ppm in the presence of 20 mM Ni-DTPA and 0.07 ppm in the presence of 40 mM Ni-DTPA, which are above the threshold for significance of 0.03 ppm.19 However, binding is non-specific and the change in chemical shift does not reach a saturation point up to 200 mM Ni-DTPA (Fig. S5†). Similarly, the chemical shift of fluorine resonances can change substantially upon titration of deuterated solvent, underscoring the large environmental sensitivity of the fluorine nucleus.32 Protein stability does not undergo large perturbations upon titration of Ni-DTPA, as measured by differential scanning fluorimetry, a thermal shift assay. Changes in melting temperature were ≤1.2 °C by this method (Fig. S6†). To assess the functional effect of Ni-DTPA on binding, fluorescence anisotropy was used to quantitate the binding affinity between a fluorescently labelled BET bromodomain ligand BI-BODIPY19,33 and Brd4 (Fig. S7†). Binding was minimally perturbed, with only minor perturbations to the dissociation constant upon increasing concentrations of Ni-DTPA (Table S1†). These results reduce concerns over the perturbing nature of the paramagnetic additive.
Having optimized conditions for running a time-efficient NMR experiment on a fluorinated protein, we next evaluated the time-enhancement effects of these conditions on ligand binding of both fragments and potent molecules. To this end, we tested two Brd4 inhibitors, compounds 1 and 2 (Fig. 3A), uncovered in our small molecule discovery NMR fragment screens. Both 1 and 2 bind to 5FW-Brd4 (Fig. 3B). For molecules like 1 and 2 that bind in the fast exchange regime, Kd values can be obtained by plotting changes in chemical shift against ligand concentration and fitting the resulting data to a binding isotherm that accounts for ligand depletion.16 For both fragments, the resonance for W81 was the most perturbed, W75 was slightly perturbed, and W120 remained unaffected (Fig. 3C and D). Fitting of the data for 1 to a binding isotherm yielded a Kd value of 81 ± 8 μM in the absence of paramagnetic metal and 87 ± 9 μM in the presence of 20 mM Ni-DPTA based on the chemical shift perturbation of W81. For 2, the Kd values were 390 ± 60 μM without paramagnetic additive and 440 ± 30 μM with 20 mM Ni-DTPA. Additional bromodomain ligands were shown to bind to 5FW-Brd4 in the presence and absence of Ni-DTPA (Fig. S8†). These experiments were acquired at a higher magnetic field strength (564 MHz) allowing for a one-minute experiment time. We conclude that moderate concentrations of Ni-DTPA minimally perturb the binding of low-affinity molecules to this fluorinated protein and can thus be applied for fragment-based screens.
An experimental time reduction of one to two minutes allows for facile Kd determinations of fragments. Affinity determinations are valuable in fragment screening for prioritizing hits. We found that because low-affinity fragments have a short residence time on the protein, the 19F-labelled residues are subject to PRE effects similar to those in the absence of fragment, allowing a full titration (nine samples) to be carried out in less than 20 minutes of experiment time. Without the advantage of pulse sequences like those used for 2D heteronuclear experiments such as SOFAST-HMQC which result in seven minute experiment times for similar sized proteins,5,34 these 19F experiments represent some of the fastest PrOF NMR experiments at the modest concentrations used.
We next sought to determine whether this method was compatible with more potent binders with longer residence times on the protein. We previously showed 19F NMR spectra of 5FW-Brd4 exhibit slow exchange kinetics with known ligand (+)-JQ1 (Kd = 77 nM),35 consistent with a long residence time on the protein. In our hands, the spectra acquired with one equivalent of (+)-JQ1 are qualitatively similar in the presence and absence of Ni-DTPA (Fig. S9†). However, the presence of this tight binding ligand limits the interaction of the paramagnetic additive and the labelled residue. Therefore, the T1 experiences a smaller reduction in the presence of (+)-JQ1, consistent the r−6 distance dependence of relaxation times as described by the Solomon–Bloembergen equations.6 Despite the changes of T1 values, the W81 resonance is similarly perturbed by (+)-JQ1 in the presence and absence of Ni-DTPA, and spectra remain consistent with a tight-binding molecule in slow exchange.
Finally, to assess the generality of using PRE to shorten PrOF NMR experiments, we tested the compatibility of Ni-DTPA with three additional 19F-labelled protein constructs. We tested proteins with hexahistidine affinity tags due to their ability to chelate metals. Encouragingly, spectra of hexahistidine-tagged 5FW-Brd4 recorded in the presence of 40 mM Ni-DTPA had linewidths similar to those of 5FW-Brd4 without its affinity tag, indicating that the Ni-DTPA is compatible with the presence of a hexahistidine tag despite this sequence's high affinity for Ni(II) (Fig. S10†). Ni-DTPA was also tested and is compatible with 3-fluorotyrosine (3FY)-labelled Brd4 (Fig. S11†) and 3FY-labelled KIX (Fig. S12†). Because 3FY KIX shows larger perturbations in its NMR spectrum than 5FW Brd4 upon addition of Ni-DPTA, the capacity of KIX for binding small molecules in the presence of Ni-DTPA was tested by both fluorescence anisotropy and PrOF NMR. Fluorescence anisotropy showed a small but significant dose-dependent increase in the Kd of the interaction between a fluorescein-labelled peptide of the transcriptional activation domain of MLL and the KIX protein at increasing concentrations of Ni-DPTA (Fig. S13 and Table S2†). For 20 mM Ni-DTPA, the increases in Kd were within two-fold. Using PrOF NMR, the Kd measured for the binding of KIX ligand naphthol-AS-E phosphate36 in the presence of 20 mM Ni-DTPA was 187 ± 50 μM, similar to the previously reported Kd of 115 ± 15 μM (Fig. S14†).18
In addition to the proteins studied here, Cai et al. also measured PRE effects on the 15N-labeled protein SUMO-1 using a DO2A (1,7-dicarboxymethyl-1,4,7,10-tetraazacyclododecane) chelator for Ni(II) rather than DTPA.6 PRE effects from chelated Ni(II) have also been applied to 13C-ribosome nascent chains.10 Finally, the 13C, 15N-labelled intrinsically disordered protein α-synuclein, which is known to bind metals, has also been subjected to PRE conditions with chelated Ni(III) with minimal changes observed in its spectra.7 These results support a broader use with proteins.
Conclusions
We were able to shorten T1 times of our 19F-labelled proteins without significantly changing resonance width by adding Ni-DTPA. The T1 time of each residue was reduced by at least 50%, with the actual value depending on residue accessibility to the paramagnetic additive. The T1 of the binding site residue was reduced by ∼75%. By shortening the recycle delay based on these T1 times, experiment time decreases 60% while conserving the signal-to-noise ratio of the binding site residue. The addition of Ni-DTPA does not significantly perturb ligand binding, making it a useful additive for shortening the total time for screening compounds by PrOF NMR and obtaining dissociation constants by ligand titration. For fragment screening, a two minute experiment would allow for ∼700 mixtures of small molecules to be screened in 24 h of spectral acquisition time. Using six molecules per mixture, this affords libraries of over 4000 compounds to be tested in a day for a protein target of this size. Additional gains in sensitivity may be possible by using neutral Ni(II) chelates.6,7,37 Other metals may also have favourable properties. Fe(III) has shown promise in some experiments,38 although T2 effects may be too substantial for PrOF NMR experiments.39 In addition to efficient screening of large compound libraries, this method can reduce protein concentration needs, or allow for the use of non-cryogenically cooled NMR probes for PrOF NMR screening.Experimental
General NMR methods
Unless otherwise noted, spectra were acquired on a 500 MHz Bruker Avance III HD equipped with a 5 mM Prodigy TCI cryoprobe at 25 °C. All experiments were performed in 50 mM Tris, 100 mM NaCl at pH 7.4 with 5% D2O added for lock. Spectra were referenced to TFA at −76.55 ppm. All processing was done using TopSpin.T1 and  determination
determination
Inversion recovery experiments were carried out with 100 μM 5FW-Brd4 or 5 mM small molecule (4-fluorophenylalanine and 5-fluoroindole). 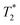 measurements for small molecules were taken on a 500 MHz Bruker Avance III with a BBFO probe. 19F NMR spectra were acquired at 470 MHz. The 90° pulsewidth was calibrated for each experiment. Variable delay values ranged from 50 ms to 8 seconds, with the small molecules requiring 4 transients and the protein 40–80 transients. The recycle delay time was set to at least five times the measured T1 value.
measurements for small molecules were taken on a 500 MHz Bruker Avance III with a BBFO probe. 19F NMR spectra were acquired at 470 MHz. The 90° pulsewidth was calibrated for each experiment. Variable delay values ranged from 50 ms to 8 seconds, with the small molecules requiring 4 transients and the protein 40–80 transients. The recycle delay time was set to at least five times the measured T1 value.  values were determined by measuring the peakwidth at half height of the 1H-decoupled 19F resonances for small molecules and undecoupled 19F resonances for proteins. Peakwidth was converted to
values were determined by measuring the peakwidth at half height of the 1H-decoupled 19F resonances for small molecules and undecoupled 19F resonances for proteins. Peakwidth was converted to  using the equation
using the equation  .
.
PrOF NMR experiments
Experiments were carried out with 50 μM 5FW-Brd4. Spectra were taken with a spectral width of 10 ppm, acquisition time of 0.05 seconds, and calibrated pulsewidths. Recycle delays were 1 second in the absence of Ni-DTPA and 0.24 seconds with 20 or 40 mM Ni-DTPA. JQ1 was added from a 10 mM stock solution in ethylene glycol. Fragments were added from 100 mM stock solutions in DMSO. Napththol-AS-E-phosphate was added from a solution of 40 mM DMSO. Line broadening of 10 Hz was applied during processing. Spectra of 5FW-Brd4 with compounds 3 and 4 were taken on a 600 MHz Bruker Avance III spectrometer with a quadruple resonance HFCN CryoProbe. 19F NMR spectra were acquired at 564 MHz. Changes in chemical shift upon titration of small molecules was fit to eqn (1) to determine Kd, where [L]0 and [P]0 are the total concentrations of ligand and protein, respectively, Δobs is the observed change in chemical shift, and Δmax is the maximum change in chemical shift.
 | (1) |
Synthesis of DTPA-chelated metals
Metal (Ho(III)Cl3·6H2O (Sigma Aldrich), Ni(II)SO4·6H2O (Alfa Aesar), DyI3 (Sigma Aldrich), or TmCl3 (Sigma Aldrich)) was dissolved to 0.25 mM in H2O with two equivalents of DTPA. The pH was raised with NaOH until all the DTPA went into solution (pH ∼ 7). The reaction was stirred for an additional two hours. At this point, a xylenol orange test was negative for free metal.40 Water was removed by lyophilization and the complex was resuspended at 0.5 to 1 M in H2O for use in NMR experiments.Protein expression and purification
5FW-Brd4, was expressed and purified as previously described.41 Briefly, E. coli containing plasmids for both pRARE with a chloramphenicol resistance selectable gene and Brd4 in a pNIC-BSA2 plasmid with a kanamycin selectable gene were cultured in LB containing 100 mg L−1 kanamycin and 35 mg L−1 chloramphenicol at 37 °C, then pelleted by centrifugation and resuspended in defined media containing 60 mg L−1 5-fluoroindole and no tryptophan. 3FY-Brd4 and 3FY-KIX were expressed in a similar manner, but with unlabelled tryptophan and 80 mg L−1 3-fluorotyrosine in place of tyrosine. For all proteins, after a 60 minute recovery period, the culture was cooled to 20 °C and expression was induced with 1 mM IPTG. The bacterial pellet was lysed with sonication and purified on a Ni-NTA affinity column, eluted with an imidazole gradient. The hexahistidine tag was removed by cleavage with TEV protease. After buffer exchange, the protein was stored in 50 mM Tris, 100 mM NaCl, pH 7.4.Differential scanning fluorimetry
Differential scanning fluorimetry was performed on a CFX384 Real-Time PCR Detection System (BioRad). Purified unlabelled Brd4 (18 μM final concentration in 50 mM phosphate buffer (pH 7.4) with 150 mM NaCl) was assayed, in replicates of 12, in a 384-well plate with increasing Ni-DTPA concentrations. SYPRO Orange Protein Gel Stain (1:5000; Sigma-Aldrich) was used as the fluorescent probe at 5× concentration. Protein stability was investigated using 0.5 °C increments and 15 s incubations per increment from 24 to 99 °C. The inflection point of the transition curve/melting temperature (Tm) was calculated using the Boltzmann equation within the Protein Thermal Shift Software (v.1.1; Applied Biosystems).Fluorescence anisotropy
Fluorescence anisotropy experiments were performed to quantitate the binding of BI-BODIPY33 and unlabelled Brd4 as well as Fl-MLL18 and unlabelled KIX. Emission and excitation were measured at 485 and 535 nm, respectively, on a Tecan Infinite 500 in a 384-well plate (Corning 4511). All experiments were carried out in triplicate, with at least two independent measurements. Data were fit in GraphPad Prism 6. Kd values were determined from eqn (2), where c is the maximum anisotropy level, b is the minimum anisotropy value, and [L]0 and [P]0 have the same meanings as eqn (1).
 | (2) |
Acknowledgements
This work was supported by the NSF CAREER award (CHE-1352019) and NIH training grants 5T32GM008347-23 (A. K. U.) and T32-GM08700 (C. T. G.).References
- S. B. Shuker, P. J. Hajduk, R. P. Meadows and S. W. Fesik, Science, 1996, 274, 1531–1534 CrossRef CAS PubMed
.
- M. Vogtherr and K. Fiebig, in Modern Methods of Drug Discovery, Birkhäuser Basel, Basel, 2003, pp. 183–202 Search PubMed
.
- M. Pellecchia, I. Bertini, D. Cowburn, C. Dalvit, E. Giralt, W. Jahnke, T. L. James, S. W. Homans, H. Kessler, C. Luchinat, B. Meyer, H. Oschkinat, J. Peng, H. Schwalbe and G. Siegal, Nat. Rev. Drug Discovery, 2008, 7, 738–745 CrossRef CAS PubMed
.
- C. T. Gee, E. J. Koleski and W. C. K. Pomerantz, Angew. Chem., Int. Ed. Engl., 2015, 54, 3735–3739 CrossRef CAS PubMed
.
- M. J. Harner, B. A. Chauder, J. Phan and S. W. Fesik, J. Med. Chem., 2014, 57, 9687–9692 CrossRef CAS PubMed
.
- S. Cai, C. Seu, Z. Kovacs, A. D. Sherry and Y. Chen, J. Am. Chem. Soc., 2006, 128, 13474–13478 CrossRef CAS PubMed
.
- F.-X. Theillet, A. Binolfi, S. Liokatis, S. Verzini and P. Selenko, J. Biomol. NMR, 2011, 51, 487–495 CrossRef CAS PubMed
.
- K. Yamamoto, S. Vivekanandan and A. Ramamoorthy, J. Phys. Chem. B, 2011, 115, 12448–12455 CrossRef CAS PubMed
.
- A. Eletsky, O. Moreira, H. Kovacs and K. Pervushin, J. Biomol. NMR, 2003, 26, 167–179 CrossRef CAS PubMed
.
- S. H. S. Chan, C. A. Waudby, A. M. E. Cassaignau, L. D. Cabrita and J. Christodoulou, J. Biomol. NMR, 2015, 63, 151–163 CrossRef CAS PubMed
.
- K. Takeuchi, D. P. Frueh, S. G. Hyberts, Z.-Y. J. Sun and G. Wagner, J. Am. Chem. Soc., 2010, 132, 2945–2951 CrossRef CAS PubMed
.
- J. L. Kitevski-LeBlanc, F. Evanics and R. S. Prosser, J. Biomol. NMR, 2009, 45, 255–264 CrossRef CAS PubMed
.
- A. A. Bogan and K. S. Thorn, J. Mol. Biol., 1998, 280, 1–9 CrossRef CAS PubMed
.
- J. J. Liu, R. Horst, V. Katritch, R. C. Stevens and K. Wüthrich, Science, 2012, 335, 1106–1110 CrossRef CAS PubMed
.
- A. Manglik, T. H. Kim, M. Masureel, C. Altenbach, Z. Yang, D. Hilger, M. T. Lerch, T. S. Kobilka, F. S. Thian, W. L. Hubbell, R. S. Prosser and B. K. Kobilka, Cell, 2015, 161, 1101–1111 CrossRef CAS PubMed
.
- K. E. Arntson and W. C. K. Pomerantz, J. Med. Chem., 2015, 59, 5158–5171 CrossRef PubMed
.
- E. N. G. Marsh and Y. Suzuki, ACS Chem. Biol., 2014, 9, 1242–1250 CrossRef CAS PubMed
.
- W. C. Pomerantz, N. Wang, A. K. Lipinski, R. Wang, T. Cierpicki and A. K. Mapp, ACS Chem. Biol., 2012, 7, 1345–1350 CrossRef CAS PubMed
.
- A. K. Urick, L. M. L. Hawk, M. K. Cassel, N. K. Mishra, S. Liu, N. Adhikari, W. Zhang, C. O. Dos Santos, J. L. Hall and W. C. Pomerantz, ACS Chem. Biol., 2015, 10, 2246–2256 CrossRef CAS PubMed
.
- X. Ge, C. A. MacRaild, S. M. Devine, C. O. Debono, G. Wang, P.
J. Scammells, M. J. Scanlon, R. F. Anders, M. Foley and R. S. Norton, J. Med. Chem., 2014, 57, 6419–6427 CrossRef CAS PubMed
.
- E. W. W. Leung, H. Yagi, J. R. Harjani, M. D. Mulcair, M. J. Scanlon, J. B. Baell and R. S. Norton, Chem. Biol. Drug Des., 2014, 84, 616–625 CAS
.
- K. L. Nash, D. Brigham, T. C. Shehee and A. Martin, Dalton Trans., 2012, 41, 14547–14556 RSC
.
- M. Sattler and S. W. Fesik, J. Am. Chem. Soc., 1997, 119, 7885–7886 CrossRef CAS
.
- R. K. Prinjha, J. Witherington and K. Lee, Trends Pharmacol. Sci., 2012, 33, 146–153 CrossRef CAS PubMed
.
- E. Nicodeme, K. L. Jeffrey, U. Schaefer, S. Beinke, S. Dewell, C.-W. Chung, R. Chandwani, I. Marazzi, P. Wilson, H. Coste, J. White, J. Kirilovsky, C. M. Rice, J. M. Lora, R. K. Prinjha, K. Lee and A. Tarakhovsky, Nature, 2010, 468, 1119–1123 CrossRef CAS PubMed
.
- L. J. Martin, M. Koegl, G. Bader, X.-L. Cockcroft, O. Fedorov, D. Fiegen, T. Gerstberger, M. H. Hofmann, A. F. Hohmann, D. Kessler, S. Knapp, P. Knesl, S. Kornigg, S. Muller, H. Nar, C. Rogers, K. Rumpel, O. Schaaf, S. Steurer, C. Tallant, C. R. Vakoc, M. Zeeb, A. Zoephel, M. Pearson, G. Boehmelt and D. McConnell, J. Med. Chem., 2016, 59, 4462–4475 CrossRef CAS PubMed
.
- A. M. Taylor, A. Côté, M. C. Hewitt, R. Pastor, Y. Leblanc, C. G. Nasveschuk, F. A. Romero, T. D. Crawford, N. Cantone, H. Jayaram, J. Setser, J. Murray, M. H. Beresini, G. de Leon Boenig, Z. Chen, A. R. Conery, R. T. Cummings, L. A. Dakin, E. M. Flynn, O. W. Huang, S. Kaufman, P. J. Keller, J. R. Kiefer, T. Lai, Y. Li, J. Liao, W. Liu, H. Lu, E. Pardo, V. Tsui, J. Wang, Y. Wang, Z. Xu, F. Yan, D. Yu, L. Zawadzke, X. Zhu, X. Zhu, R. J. Sims, A. G. Cochran, S. Bellon, J. E. Audia, S. Magnuson and B. K. Albrecht, ACS Med. Chem. Lett., 2016, 7, 531–536 CrossRef CAS PubMed
.
- J.-L. Yu, T.-T. Chen, C. Zhou, F.-L. Lian, X.-L. Tang, Y. Wen, J.-K. Shen, Y.-C. Xu, B. Xiong and N.-X. Zhang, Acta Pharmacol. Sin., 2016, 37, 984–993 CrossRef CAS PubMed
.
- A. M. Watkins and P. S. Arora, ACS Chem. Biol., 2014, 9, 1747–1754 CrossRef CAS PubMed
.
- R. Fraczkiewicz and W. Braun, J. Comput. Chem., 1998, 19, 319–333 CrossRef CAS
.
- A. E. Kelly, H. D. Ou, R. Withers and V. Dötsch, J. Am. Chem. Soc., 2002, 124, 12013–12019 CrossRef CAS PubMed
.
- P. E. Hansen, H. D. Dettman and B. D. Sykes, J. Magn. Reson., 1985, 62, 487–496 CAS
.
- Z. Zhang, N. Kwiatkowski, H. Zeng, S. M. Lim, N. S. Gray, W. Zhang and P. L. Yang, Mol. BioSyst., 2012, 8, 2523–2524 RSC
.
- M. J. Harner, A. O. Frank and S. W. Fesik, J. Biomol. NMR, 2013, 56, 65–75 CrossRef CAS PubMed
.
- N. K. Mishra, A. K. Urick, S. W. J. Ember, E. Schönbrunn and W. C. Pomerantz, ACS Chem. Biol., 2014, 9, 2755–2760 CrossRef CAS PubMed
.
- J. L. Best, C. A. Amezcua, B. Mayr, L. Flechner, C. M. Murawsky, B. Emerson, T. Zor, K. H. Gardner and M. Montminy, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17622–17627 CrossRef CAS PubMed
.
- M. W. Voehler, G. Collier, J. K. Young, M. P. Stone and M. W. Germann, J. Magn. Reson., 2006, 183, 102–109 CrossRef CAS PubMed
.
- A. A. Kislukhin, H. Xu, S. R. Adams, K. H. Narsinh, R. Y. Tsien and E. T. Ahrens, Nat. Mater., 2016, 15, 662–668 CrossRef CAS PubMed
.
- Z.-X. Jiang, Y. Feng and Y. B. Yu, Chem. Commun., 2011, 47, 7233–7235 RSC
.
- D. J. Averill, J. Garcia, B. N. Siriwardena-Mahanama, S. M. Vithanarachchi and M. J. Allen, J. Visualized Exp., 2011, 53, e2844 Search PubMed
.
- C. T. Gee, K. E. Arntson, A. K. Urick, N. K. Mishra, L. M. L. Hawk, A. J. Wisniewski and W. C. K. Pomerantz, Nat. Protoc., 2016, 11, 1414–1427 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra21226c |
| This journal is © The Royal Society of Chemistry 2016 |