An affinity triggered MRI nanoprobe for pH-dependent cell labeling†
Susana I. C. J. Palmaa,
Alexandra R. Fernandesbc and
Ana C. A. Roque *a
*a
aUCIBIO, REQUIMTE, Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, 2829-516 Caparica, Portugal. E-mail: cecilia.roque@fct.unl.pt
bUCIBIO, REQUIMTE, Departamento de Ciências da Vida, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, 2829-516 Caparica, Portugal
cCQE, Centro de Química Estrutural, Instituto Superior Técnico, Universidade de Lisboa, 1490-001 Lisboa, Portugal
First published on 28th November 2016
Abstract
The pH-sensitive affinity pair composed by neutravidin and iminobiotin was used to develop a multilayered Magnetic Resonance Imaging (MRI) nanoprobe responsive to the acidic pH of tumor microenvironment. The multilayer system was assembled on meso-2,3-dimercaptosuccinic acid-coated iron oxide magnetic nanoparticles (MNP), which convey negative MRI contrast enhancement properties to the nanoprobe. The outer stealth PEG-layer is altered in acidic media due to the disruption of interactions between neutravidin–iminobiotin. As a consequence, the positively charged inner layer is exposed and enhances interactions with cells. The nanoprobe uptake by HCT116 cells cultured in vitro under acidic conditions had a 2-fold increase compared to the uptake at physiological pH. The uptake difference is particularly clear in T2-weighted MRI phantoms of cells incubated with the nanoprobes at both pH conditions. This work sets the proof-of-concept of a MNP-based MRI nanoprobe targeting acidic tumor microenvironment through the use of a specific bio-recognition interaction that is pH-sensitive. This tumor targeting strategy is potentially applicable to the generality of tumors since the typical hypoxic conditions and high glycolysis rate in cancer cells create an acidic environment common to the majority of cancer types.
Introduction
Nanotechnology research triggered the development of colloidal iron oxide magnetic nanoparticles (MNP) for magnetic resonance imaging (MRI), delivery of therapeutics, hyperthermia or theranostics of several diseases.1–4 Cancer, in particular, has been widely studied due to the importance of early detection and the need of targeted treatments. Tumor targeted MNP-based systems are valuable approaches for that purpose as they combine the inherent MRI contrast enhancement properties of superparamagnetic iron oxide nanoparticles (for diagnosis) with the versatility for surface functionalization with biologically or chemically active moieties (for targeted therapy).A possible tumour-targeting strategy consists in creating nanoparticles activated by tumour physicochemical characteristics.5 In this context, the pH difference between the extracellular medium of tumours and healthy tissues can be used to engineer tumour-targeted nanoparticles.6,7 Acidic extracellular tumour pH mainly results from the high rate of glycolytic metabolism and poor perfusion typically found in tumours, as 90% of the pyruvate generated by glycolysis is converted to lactic acid, and co-transported outside the cell with H+ ions. This process, associated with decreased blood flow rate and poor lymphatic drainage in the tumour cells, leads to the accumulation of H+ ions in the extracellular medium thereby causing acidity (pH ranging approximately between 6.5 and 7.0) compared with healthy tissues and blood (pH around 7.4).5,8,9
One of the strategies for pH-activation of nanoparticles relies on the hypothesis that the nanoparticles maintain stealth during blood circulation and passively accumulate at tumour sites. Here, they can be activated by the acidic environment, and be transformed into a more cell-interactive form for enhanced tumour cell internalization, cytotoxicity or release of cargo. For example, MNP with a glycol-chitosan (GC) coating generated a 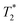 -weighted Magnetic Resonance (MR) contrast agent with enhanced cellular interactions and MRI contrast at tumour pH both in vitro and in vivo due to the pH-titrable charge of GC, which becomes positive under acidic conditions.10 Mok et al.11 reported a dual therapeutic and MR imaging MNP nanosystem for chlorotoxin-mediated tumour-targeted delivery of siRNA. This system makes use of the acid-hydrolysable linkage between citraconic anhydride and primary amines to block the cytotoxic effect of polyethylenimine (PEI) and reduce cellular interactions at physiological pH. At acidic conditions, due to citraconic anhydride removal, the positive charges of PEI are unblocked promoting cytotoxicity, chlorotoxin is exposed (for receptor-mediated tumour cellular uptake) and siRNA delivered to the intracellular medium. Saha et al.12 developed a pH-sensitive MR contrast agent using melamine-dendron functionalized MNP. In this system, large R2 values are provided at low pH in physiological salt conditions, but decrease for higher pH. A sharp inflection at pH values just below the pKa of melamine monomer (∼5) is observed due to the pH-dependent transient and reversible clustering of magnetic cores modulated by the interplay between surface charge at different pHs and ionic strength. A more complex nanosystem was recently reported by Ling et al.,13 who developed a multifunctional pH-sensitive nanosystem composed of self-assembled ultrasmall MNP, a fluorescent tag, a photodynamic therapeutic moiety and pH-sensitive ligands. The authors engineered polymeric pH-sensitive ligands based on a protonable imidazole group and used them to fabricate magnetic nanogrenades that upon exposure to acidic extratumoral pH switch charge from negative to positive and swell, promoting cellular uptake. Once inside the cell, the system disassembles and activates T1-weighted MRI contrast and photoactivity for therapeutic effect.
-weighted Magnetic Resonance (MR) contrast agent with enhanced cellular interactions and MRI contrast at tumour pH both in vitro and in vivo due to the pH-titrable charge of GC, which becomes positive under acidic conditions.10 Mok et al.11 reported a dual therapeutic and MR imaging MNP nanosystem for chlorotoxin-mediated tumour-targeted delivery of siRNA. This system makes use of the acid-hydrolysable linkage between citraconic anhydride and primary amines to block the cytotoxic effect of polyethylenimine (PEI) and reduce cellular interactions at physiological pH. At acidic conditions, due to citraconic anhydride removal, the positive charges of PEI are unblocked promoting cytotoxicity, chlorotoxin is exposed (for receptor-mediated tumour cellular uptake) and siRNA delivered to the intracellular medium. Saha et al.12 developed a pH-sensitive MR contrast agent using melamine-dendron functionalized MNP. In this system, large R2 values are provided at low pH in physiological salt conditions, but decrease for higher pH. A sharp inflection at pH values just below the pKa of melamine monomer (∼5) is observed due to the pH-dependent transient and reversible clustering of magnetic cores modulated by the interplay between surface charge at different pHs and ionic strength. A more complex nanosystem was recently reported by Ling et al.,13 who developed a multifunctional pH-sensitive nanosystem composed of self-assembled ultrasmall MNP, a fluorescent tag, a photodynamic therapeutic moiety and pH-sensitive ligands. The authors engineered polymeric pH-sensitive ligands based on a protonable imidazole group and used them to fabricate magnetic nanogrenades that upon exposure to acidic extratumoral pH switch charge from negative to positive and swell, promoting cellular uptake. Once inside the cell, the system disassembles and activates T1-weighted MRI contrast and photoactivity for therapeutic effect.
As an alternative to chemically engineered pH-dependent materials, specific bio-recognition interactions can be explored to derive pH-sensitivity. The complex formed by biotin and avidin (or its analogues) is the strongest known non-covalent interaction (Kd = 10−15 M)14 between a protein and ligand, and once formed is not affected by extreme conditions. However, the guanido-version of biotin (iminobiotin) binds to avidin and its derivatives in a pH-dependent fashion. At pH 9.5–11.0, the avidin–iminobiotin complex binds tightly (Kd = 3.4 × 10−10 M) but the bond strength decreases with pH until the weaker binding is reached at pH 4 (Kd = 10−3 M).15 Due to the reversible binding property, this affinity pair has been utilized in bioseparation applications,16–18 in the production of thin films decomposable by pH19 and in the development of layer-by-layer acidity-triggered quantum-dot nanoprobes for in vivo tumour imaging by fluorescence.20
In this work, we explore the neutravidin–iminobiotin pH-dependent affinity interaction to develop an affinity-triggered MNP-based MRI nanoprobe for preferential labelling of tumour cells. The system consists of a multilayer-coated magnetic nanoprobe with a pH-removable PEG outer-layer (Fig. 1). At the acidic tumour microenvironment, the outer-layer tends to dissociate from the nanoprobe, rendering cell–MNP interactions more favourable, and cancer cells visible by MRI. The multilayer system was deposited onto meso-2,3-dimercaptosuccinic acid (DMSA)-functionalized MNP and characterized regarding its size and surface charge after depositing each coating step. The pH responsiveness of the final MNPs was first evaluated in buffer solutions at different pHs and then in in vitro cultures of human colorectal carcinoma cells (HCT116 cell line) at acidic and physiological pH. Cell phantoms were imaged by MRI to evaluate the efficacy of the particles to provide differential contrast depending on the pH of the cultures.
Results and discussion
Multilayer MNP assembly
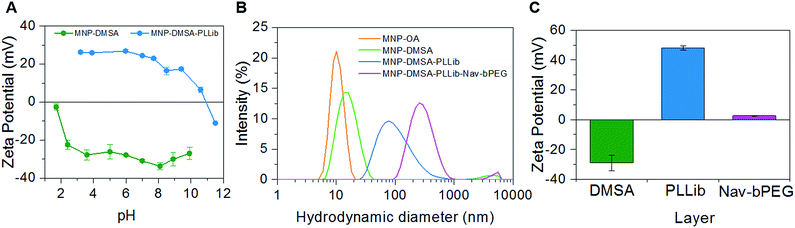
Hydrophobic iron oxide magnetic nanoparticles coated with oleylamine and oleic acid (MNP–OA) were synthesized by the thermal decomposition method and transferred to aqueous phase by replacement of the oleic acid and oleylamine moieties at their surface by DMSA molecules.21 This process provided the negatively charged template nanoparticles (MNP–DMSA) for further functionalization with the pH-responsive layer system. The assembly of the multi-layered magnetic nanoprobes was followed by assessing, at each layering step, particles size and surface charge (Fig. 2 and Table 1).| Nanoparticles | dh (nm) | pdI | Z-Ave (nm) | Zeta potential (mV) |
|---|---|---|---|---|
| MNP–OA | 10 ± 0.3 | 0.14 ± 0.06 | 12 ± 3.2 | n. a. |
| MNP–DMSA | 16 ± 0.1 | 0.19 ± 0.01 | 15 ± 0.1 | −29.1 ± 5.2 |
| MNP–DMSA–PLLib | 139 ± 10.1 | 0.22 ± 0.01 | 97 ± 0.8 | 48.1 ± 1.4 |
| MNP–DMSA–PLLib–Nav–bPEG | 302 ± 15.6 | 0.25 ± 0.013 | 261 ± 7.9 | 2.6 ± 0.1 |
The first layer to be added onto MNP–DMSA was PLL, which was previously functionalized with iminobiotin on 28% of its free amine groups (36 mol (ib) per mol (PLL)). The deposition of iminobiotin-modified PLL (PLLib) onto MNP–DMSA by electrostatic adsorption yielded positively charged nanoprobes (Fig. 2 and Table 1) with anchoring points for neutravidin (Nav) (1.6 μmol (ib) per mg (MNP)). These nanoprobes are composed by aggregates of multiple MNP–DMSA, as supported by the increase in hydrodynamic diameter (Fig. 2B and Table 1). As can be seen in Fig. 2A, there was a complete reversal of the particles surface charge due to the presence of amino groups from PLLib. The isoelectric point of the particles shifted from pH 1.5 to pH 11. The colloidal suspension of MNP–DMSA–PLLib was very stable at pH 7.4 and low salt concentrations (1 mM PBS), with a clean size distribution and relatively low PdI (Fig. 2B and Table 1). However, we observed that physiological salt conditions (10 mM PBS, 150 mM NaCl) caused flocculation (after ∼20 h). This phenomenon was not observed upon addition of the PEG shell, which helped to stabilize the multilayer clustered nanoparticles (Fig. 2B and S1†), through inter-particle steric interactions provided by the electrically neutral hydrophilic chains of the polymer.
The PEG shell consisted of a neutravidin–PEG conjugate (Nav–bPEG). PEG was functionalized with biotin in one of the chain terminus (bPEG) for subsequent binding to Nav (Kd = 10−15 M). The deposition of the Nav–bPEG layer onto MNP–DMSA–PLLib resulted in the neutralization of the particles surface charge at pH 7.4 (Fig. 2C and Table 1), which shows that the inner cationic PLLib layer was shielded by the neutral PEG chains bound to the particles. Neutralization of surface charge with PEG is important because PEG is known to provide stealth properties to nanoparticles, due to its hydrophilicity, flexibility, and neutral charge in biological fluids. PEG-coated nanomaterials usually have longer circulation times in the blood stream and escape more effectively to the monophagocytic system.22–24 The measured hydrodynamic diameter had a 3-fold increase after deposition of Nav–bPEG (Fig. 2B and Table 1), which resulted in a final particle size of 300 nm. Besides the specific molecular interaction between Nav–bPEG and MNP–DMSA–PLLib, further aggregation of the nanoprobes may be caused by inter-particle bridges established by the PEG chains network.
pH-Dependent MNP response
The pH sensitivity of the prepared multilayer nanoparticles was assessed by exposing the particles to different pH conditions by means of dialysis (20 h) to PBS at pH 5, 6.5, 7.4 and 8.5. The release profile of Nav–bPEG shell was determined (Fig. 3A) as well as the modifications observed in samples' surface charge, size and polydispersity (Fig. 3B and C).The strength of Nav–ib binding is maximal between pH 9.5 and pH 11 (Kd = 10−10) and lowers with the pH until maximal dissociation of ib from Nav at pH 4 (Kd = 10−3).15 In MNP–DMSA–PLLib–Nav–bPEG, a specific response to pH was observed, which is attributed to the pH-dependent bio-recognition interaction between ib and Nav. After 20 h of dialysis, the quantification of Nav–bPEG in the dialysates revealed that the fraction of Nav–bPEG released from the particles to the dialysate significantly decreases with increasing pH, (Fig. 3A). This was expected given the nature of the Nav–ib affinity interaction. The most significant difference (p < 0.0001) was observed between pH 5 and pH 8.5. At pH 8.5, due to the stronger affinity between the ib on the particles and the Nav from Nav–bPEG, most Nav–bPEG was kept bound to the particles. On the other hand, at pH 5, close to the lower limit of affinity, maximal dissociation of Nav–bPEG from the particles was triggered. The release of Nav–bPEG was incomplete for all tested pH conditions. In particular, at pH 5 and pH 6.5 respectively 65% and 45% of the particles total Nav–bPEG was found in the dialysates, and consequently a fraction of Nav–bPEG remained bound to the particles. Accordingly, MNP–DMSA–PLLib–Nav–bPEG did not equal the zeta potential of MNP–DMSA–PLLib subjected to the same acidic pH treatments (Fig. 3B). Still, releasing the PEG layer and exposing PLLib cationic groups, yielded a change in the zeta potential from neutral (at pH 7.4) to positive (at pH 6.5), and increase to +10 mV at pH 5 (Fig. 3B). Samples become heterogeneous as a result of the structural alterations caused by the partial release of Nav–bPEG shell from particles at acidic pHs, and this is reflected in the average size and polydispersity index (Fig. 3C and S2†). Under acidic conditions, the (at least partial) exposure of positive charges may responsible by particle flocculation due to the interaction with buffer salt (interparticle salt bridges), similar to what was observed for MNP–DMSA–PLLib (PdI around 1). MNP–DMSA–PLLib–Nav–bPEG samples thus become polydisperse, meaning that populations of large and smaller particles co-exist in the sample (Fig. S2†) and the average hydrodynamic size of the sample increases, as it is represented in Fig. 3C.
pH-Dependent in vitro MNP–cell interactions and MR imaging
After observing the pH-dependent erosion of Nav–bPEG shell in saline buffers and the resultant alterations in particles charge and sizes, in vitro interactions with cells were studied. To mimic the tumoral environment and compare the results with physiological conditions, assays with the human colorectal carcinoma HCT116 cell line were performed in acidified culture medium and in standard non-modified Dulbecco's modified Eagle's culture medium (DMEM).Compared to untreated cells, cells incubated with MNP–DMSA–PLLib–Nav–bPEG at 10 μg Fe per ml for 5 h did not present significant loss of viability (Fig. 4). Indeed, PEG-based materials are commonly employed as nanoparticle coatings for biocompatibility purposes due to its hydrophilicity, low immunogenicity and low toxicity.24–26 Interestingly, at pH 6.5, the particles tended to be slightly more cytotoxic than at pH 7.4 (Fig. 4), which is in accordance with the observed cationic character of the particles at pH 6.5 due to Nav–bPEG erosion. Incubation of the same cell type with MNP–DMSA–PLLib at the same concentration and pH resulted in very low cellular viability (Fig. S3†), confirming the cytotoxic effect of highly cationic MNP.
Microscopy images of Prussian blue-stained cellular preparations show that upon incubation with cells, in either physiological (pH 7.4) or acidic (pH 6.5) conditions, MNP–DMSA–PLLib–Nav–bPEG are found adhered to the cell surface and internalized (Fig. S4†). Internalization was found to be mediated by an endocytosis pathway (Fig. 5A and B). The internalized MNP co-localize with the lysosomes in preparations incubated at 37 °C but remain mainly attached to the cell membrane and in the extracellular medium in preparations incubated at 4 °C (Fig. 5A and B and S5†), when energy-dependent processes in the cell (like endocytosis) are blocked. These results are in accordance with other works where multilayer nanoparticles with PEG shell were employed.27 Also, Calero and collaborators demonstrated that an active energy-dependent transport was implicated in DMSA–SPION internalization within breast cancer cells and that once inside the cells, the co-localization of the internalized nanoparticles with the fluorescence from LysoTracker, indicated that the DMSA–SPION were accumulated in the endosome/lysosome fraction.28
ICP-AES cellular iron quantification demonstrated that cell labelling with MNP–DMSA–PLLib–Nav–bPEG presents a pH-dependent behaviour (Fig. 5C) since after 5 h of incubation with the particles at 10 μg Fe per ml, significantly higher iron content was detected in cells cultured in acidic medium (22.82 ± 8.15 pg Fe per cell) than in cells cultured in physiological conditions (11.79 ± 3.22 pg Fe per cell) (p < 0.05). Approximately, a 2-fold increase in the cellular iron content was promoted by the acidic conditions compared to the physiological ones, in accordance to the findings of Poon et al.20 for HeLa, KB and A549 cancer cells treated with nanoprobes with a similar ib/Nav-based nanosystem. These results suggest that besides having a complex architecture, the developed MRI nanoprobe may be able to interact preferentially with cells in the acidic tumour microenvironment.
When a nanomaterial is put in contact with a biological environment, a protein corona rapidly forms around it that, overall, will affect the interaction of the material with the tissues or cells.29,30 Generally, neutral and anionic nanoparticles show lower interactions with medium proteins than cationic ones, that interact strongly with proteins, and undergo nonspecific binding, in some cases, leading to cell lysis.31–33 Since culture medium supplemented with fetal bovine serum (FBS) was used in this work, it is likely that the medium proteins also play a role in the way MNPs interact with cells at different pH values. At acidic pH, a conjunction of factors, namely pH-dependent surface charge and aggregation, contribute to enhance MNP interactions with cells.
Superparamagnetic iron oxide nanoparticles with core sizes between 6 and 20 nm are negative MRI contrast agents,1 i.e., have the ability to shorten the transversal relaxation time (T2) of water protons in their vicinity, which translates in a darkening effect of MR images in the areas were the nanoprobes are present. MNP–DMSA, which are the basis of this multilayer system, were previously shown to possess superparamagnetic and T2 MRI contrast agent properties.21,34 T2-Weighted MR images of agarose dispersions of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells incubated with the MNP–DMSA–PLLib–Nav–bPEG show a clear contrast difference relative to unlabeled cells. Furthermore, these results reflect a preferential labelling of cells in acidic conditions compared to those in physiological medium (Fig. 6A). Labeled cells are detectable as hypointense regions in the images, which are more intense in the acidic than in the physiological sample due to the higher content of MNPs in the cells labeled at pH 6.5. Correspondingly T2 of cells in acidic medium (T2 = 56.75 ± 0.48 ms) is shorter than in physiological medium (T2 = 63.90 ± 5.48 ms) (Fig. 6B). Similar results were reported by Crayton et al.,10 which employed a biopolymeric coating with pH-titrable charges to obtain pH-dependent in vitro T2 relaxation times.
000 cells incubated with the MNP–DMSA–PLLib–Nav–bPEG show a clear contrast difference relative to unlabeled cells. Furthermore, these results reflect a preferential labelling of cells in acidic conditions compared to those in physiological medium (Fig. 6A). Labeled cells are detectable as hypointense regions in the images, which are more intense in the acidic than in the physiological sample due to the higher content of MNPs in the cells labeled at pH 6.5. Correspondingly T2 of cells in acidic medium (T2 = 56.75 ± 0.48 ms) is shorter than in physiological medium (T2 = 63.90 ± 5.48 ms) (Fig. 6B). Similar results were reported by Crayton et al.,10 which employed a biopolymeric coating with pH-titrable charges to obtain pH-dependent in vitro T2 relaxation times.
The multilayer pH-sensitive magnetic nanoprobe MNP–DMSA–PLLib–Nav–bPEG overall presented a good labelling performance in vitro. The observed selectivity for labelling HCT116 cells in acidic compared to physiological medium confirms that the ib–Nav pH-dependent and biologically derived affinity pair is suited to confer pH-sensitivity in the tested range of pH values and that immobilization within the multi-layered architecture system does not affect its robustness. Regarding translation to in vivo application of the presented magnetic nanoprobe, the challenge of reducing particle size still remains, as for extravasation of the nanoprobes in the tumor and escape clearance from circulation by the reticulo-endothelial system, hydrodynamic diameters lower than 100 nm are preferable.35 In the present formulation blood half-time would be short, and this would reduce the tumor-accumulation efficiency of the system. Compared to a similar architecture system,20 the magnetic nanoprobe presented here has the advantage of allowing imaging by MRI, which is well established for human-scale diagnostics in the clinic unlike other imaging modalities such as fluorescence. Moreover, it offers the possibility (not explored in this work) of magnetic targeting using an external magnet to direct the nanoprobe to desired areas after systemic injection. Furthermore, given the availability of free amine groups in the PLLib layer, other nanoprobes could be engineered using the developed system as template. For example, reporter and/or specific targeting molecules could be attached to create a combined targeting strategy for a pH-sensitive multimodal drug delivery and imaging system.
Since acidity is characteristic of most tumour tissues, the proposed iminobiotin/neutravidin based MNP multilayer architecture could contribute to surpass some of the issues associated with ligand/receptor mediated tumour targeting strategies in vivo. These include the heterogeneity among cancer cell populations and the heterogeneous expression of receptors or antigens on cancer cell membranes,8 which limit the efficacy of nanoprobes decorated with specific ligands targeting for one biomarker.
Conclusions
In this work, a proof-of-concept of an affinity triggered T2 MRI contrast agent for cancer cell labelling was presented. The developed nanoprobe is activated by acidic pH so that preferential interaction with cells is promoted under those conditions, leading to higher contrast in MRI compared to the one in physiological conditions. While most strategies to produce iron oxide MNP sensitive to pH rely on the use of chemically engineered polymers with pH-sensitive bonds or chemical groups, this work demonstrated the feasibility of using a biologically-derived affinity interaction (iminobiotin/neutravidin) to impart pH-sensitivity to the nanoprobes. It was shown that the PEGylated layer of MNP–DMSA–PLLib–Nav–bPEG is able to shield the cationic charges of the underlying PLLib molecules at physiological pH (pH 7.4) and expose them at acidic pH. Upon exposure to different pH conditions, the proportion of Nav–bPEG released from the particles significantly increases with decreasing pH. Although non-specific interactions within the nanosystem cannot be neglected, the weaker strength of Nav–ib affinity interaction at acidic pH contributes to trigger the partial release of Nav–bPEG from the nanoparticles in acidic environment and to enhance cell–MNP interactions. Approximately 2-fold increase in cell-associated iron was promoted in acidic culture conditions compared to the observed in physiological conditions. For cells cultured at pH 7.4, PEG shell also contributes to decrease the cytotoxicity of the nanoprobes. The difference in cell-associated iron content resulted in clear hypointensity differences between cells cultured in acidic medium (T2 = 56.75 ± 0.48 ms) and cells cultured in physiological medium (T2 = 63.90 ± 5.48 ms). As acidity is a characteristic of most of cancer tissues, the proposed nanosystem architecture is envisaged as a general tumour targeting approach, expected to provide MR labelling of tumoral tissues disregarding the type of cancer.Experimental
Materials
All materials were purchased from Sigma Aldrich unless otherwise specified.Production of multi-layer functionalized magnetic nanoparticles
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–30
000–30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da) was functionalized with NHS-activated iminobiotin (Thermo Scientific) on approximately 30% of its primary amines by incubation in aqueous conditions (borate buffer 50 mM, pH 8) for 2 h at 4 °C. Prior to use, the modified PLL was dialyzed against water in a MWCO 10 kDa dialysis membrane, with four complete water changes, to remove unreacted iminobiotin and reaction leftovers. The biotinylation yield is the ratio between the number of primary amines in PLLib after the reaction with NHS–iminobiotin and the number of primary amines in native PLL. The Kaiser test was used to estimate the amount of primary amines, as described previously.38
000 Da) was functionalized with NHS-activated iminobiotin (Thermo Scientific) on approximately 30% of its primary amines by incubation in aqueous conditions (borate buffer 50 mM, pH 8) for 2 h at 4 °C. Prior to use, the modified PLL was dialyzed against water in a MWCO 10 kDa dialysis membrane, with four complete water changes, to remove unreacted iminobiotin and reaction leftovers. The biotinylation yield is the ratio between the number of primary amines in PLLib after the reaction with NHS–iminobiotin and the number of primary amines in native PLL. The Kaiser test was used to estimate the amount of primary amines, as described previously.38For the deposition of PLLib layer, MNP–DMSA (at 0.7 mg ml−1) were added dropwise to an equal volume of PLLib solution (at 1.25 mg ml−1) under magnetic stirring (600 rpm) and left incubating under gentle magnetic stirring (200 rpm) for 2 h at room temperature. Dialysis in a MWCO 50 kDa membrane against MilliQ water (four complete water changes) was used to wash the particles prior to further use. Filtration near the flame through a syringe filter with 0.2 μm diameter pore was performed to ensure sterility of the particles for the following steps and in vitro testing.
All the materials, including buffers and water, were autoclaved prior to use and the reactions were performed near the flame to maximize the sterility condition of the produced particles.
Characterization of magnetic nanoparticles
Nanoparticle hydrodynamic diameter (dh) and zeta potential were characterized using a Zetasizer Nano ZS (Malvern). The mean value of the intensity-weighted size distribution measured at pH 7 in low salt conditions (water for MNP–DMSA and MNP–DMSA–PLLib or 1 mM phosphate buffer for MNP–DMSA–PLLib–Nav–bPEG) was considered as the dh of the pristine nanoparticles. The Z-average was considered instead when characterizing the size of the particles after dialysis to PBS with 150 mM NaCl at different pH values. Zeta potential of the pristine nanoparticles was measured in low salt conditions. Variation of zeta potential of MNP–DMSA and MNP–DMSA–PLLib with pH was measured in a 10 mM KNO3 solution (HNO3 or KOH solutions were used for pH adjustment). The amine groups on MNP–DMSA–PLLib were quantified through the Kaiser test38 and from this characterization, the amount of PLL and ib on the particles was estimated. MNP–DMSA concentration was determined by drying and weighting a known volume of particles. The concentration of particles subsequently modified with PLLib and Nav–bPEG was estimated by correction of MNP–DMSA concentration value with the respective dilution factor (resulting from the coating reactions and dialyses). The iron content in MNP samples was determined by Inductively Coupled Plasma-Atomic Emission Spectroscopy (ICP-AES) (Horiba Jobin-Yvon, Ultima).Examination of pH dependent Nav–bPEG release
The pH dependence of Nav–bPEG release was investigated by dialyzing MNP–DMSA–PLLib–Nav–bPEG inside 300 kDa Float-a-Lyzers (Spectrum Laboratories) to PBS at pH 5, pH 6.5, pH 7.4 or pH 8.5 during approximately 20 h to ensure that an equilibrium between the nanoparticles dispersion and the PBS was reached and all the released Nav–bPEG diffused to the PBS. After the 20 h of dialysis, the total protein content of the dialysates was quantified using the bicinchoninic acid test39 (QuantiPro BCA assay kit) and normalized to the mass of MNP and to the mass of Nav–bPEG used for the coating reaction (mass(Nav–bPEG)in), quantified in the same assay. To correct the total protein concentration values regarding PLLib that is also released during dialysis, control dialyses of MNP–DMSA–PLLib were performed and PLLib released to the dialysates was quantified using the same test. PLLib released per mg of MNP at each pH was then subtracted from the total protein released per mg of MNP to obtain the mass of Nav–bPEG released per mg of MNP at each pH condition. Normalization to mass(Nav–bPEG)in gives the percentage of Nav–bPEG released per mg of MNP (% Nav–bPEGin per mg MNP).Characterization of in vitro cell–MNP interactions
For magnetic cell labelling, cells were seeded in 24-well plates (at 1 × 105 cells per well) with either regular (pH 7.4) or acidic (pH 6.5) culture medium (0.4 ml) and incubated for approximately 24 h to allow cell adhesion. Then, the culture medium was replaced by fresh medium (either regular or acidic) containing the magnetic nanoparticles (MNP–DMSA–PLLib or MNP–DMSA–PLLib–Nav–bPEG) at 10 μg Fe per ml. After 5 h of incubation,11,13,20 the cell–MNP interaction assays detailed hereafter were carried out. DMEM medium was acidified to pH 6.5 by adding some drops of HCl (between 0.1 M and 5 M) and filtered under sterile conditions with a 0.2 μm syringe filter prior to cell seeding. MNP–DMSA–PLLib–Nav–bPEG were subjected to pre-treatments at pH 7.4 and pH 6.5 for 20 h before being dispersed in culture medium and added to the cells.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (v/v in PBS) or 1 drop of ProLong Gold Antifade Mountant with DAPI (Life Technologies) for fluorescent staining of cell nuclei. The slides were observed under bright-field and fluorescent illumination using an Olympus BX51 microscope equipped with an Olympus DP50 camera and the AnalySIS Soft Imaging software.
3 (v/v in PBS) or 1 drop of ProLong Gold Antifade Mountant with DAPI (Life Technologies) for fluorescent staining of cell nuclei. The slides were observed under bright-field and fluorescent illumination using an Olympus BX51 microscope equipped with an Olympus DP50 camera and the AnalySIS Soft Imaging software.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v)) for 30 min at 90 °C, diluted to 1 ml with MilliQ water and analysed separately for iron by ICP-AES. The iron in the cellular fraction (sum of fraction 1 and fraction 2) was normalized to the number of cells and to the total mass of iron (sum of the three fractions). A control sample containing only cells was also quantified to provide a calibration for the native iron content of cells.
1 (v/v)) for 30 min at 90 °C, diluted to 1 ml with MilliQ water and analysed separately for iron by ICP-AES. The iron in the cellular fraction (sum of fraction 1 and fraction 2) was normalized to the number of cells and to the total mass of iron (sum of the three fractions). A control sample containing only cells was also quantified to provide a calibration for the native iron content of cells.T2-Weighted MR images were obtained in a magnetic field of 7 T, at 25 °C, using a Bruker Advance III Spectrometer (160 G cm−1 imaging gradient) and a fast low angle shot gradient (FLASH) imaging sequence with repetition time (TR) = 110 ms, echo time (TE) = 1.7 ms, excitation angle of 20° and number of excitations (NEX) = 32. Due to space restriction inside the spectrometer's sample holder, the NMR tubes with the samples were imaged separately.
T2 relaxation times were also determined for the final cell samples with particles at pH 6.5 and pH 7.4. Briefly, the average signal was measured as the MR image intensity in a circular region of interest (1.2 × 106 μm2) placed in the centre of each cell phantom, for different echo times. The signal intensities, measured separately for each sample, were then plotted against the respective echo times, and the signal intensity (SI) function was fitted to the data according to the exponential decay equation SI = A + Ce(−t/T2), where SI is the signal intensity, t is the echo time, A is an off-set constant and C is a pre-factor constant. Resulting from these fittings, the transverse relaxation times T2 were obtained.
Statistical analysis
All data in figures and text is given as mean ± standard deviation. Statistical analysis was performed with GraphPad Prism 6.0 software. One-way or two-way ANOVA complemented with Tukey's test or Bonferroni's test for multiple comparisons were used when applicable.The threshold for significance was P = 0.1 and P-values < 0.1 (*), <0.05 (**), <0.005 (***) and <0.0001 (****) were considered significant.
Acknowledgements
This work was supported by the Unidade de Ciências Biomoleculares Aplicadas-UCIBIO, which is financed by national funds from FCT/MEC (UID/Multi/04378/2013) and co-financed by the ERDF under the PT2020 Partnership Agreement (POCI-01-0145-FEDER-007728). The authors thank Fundação para a Ciência e Tecnologia (FCT) for SFRH/BD/51112/2010 doctoral grant (Susana Palma) and contract PTDC/EBB-BIO/118317/2010, PTDC/BBB-NAN/1812/2012. The authors acknowledge Pedro V. Baptista and Pedro Martins, from UCIBIO-Departamento de Ciências da Vida, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, for scientific and technical contribution on cellular assays, Alexandra Carvalho, from CENIMAT – I3N, Departamento de Ciência dos Materiais, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, for the MRI analyses and Carla Rodrigues from Laboratório de Análises, REQUIMTE for the ICP analyses.Notes and references
- M. Colombo, S. Carregal-Romero, M. F. Casula, L. Gutiérrez, M. P. Morales, I. B. Böhm, J. T. Heverhagen, D. Prosperi and W. J. Parak, Chem. Soc. Rev., 2012, 41, 4306–4334 RSC.
- S. Sharifi, H. Seyednejad, S. Laurent, F. Atyabi, A. A. Saei and M. Mahmoudi, Contrast Media Mol. Imaging, 2015, 10(5), 329–335 CrossRef CAS PubMed.
- T.-H. Shin, Y. Choi, S. Kim and J. Cheon, Chem. Soc. Rev., 2015, 44, 4501–4516 RSC.
- R. Vinhas, M. Cordeiro, F. Carlos, S. Mendo, A. R. Fernandes, S. Figueiredo and P. V. Baptista, Nanobiosensors in Disease Diagnosis, 2015, 4, 11–23 Search PubMed.
- F. Danhier, O. Feron and V. Préat, J. Controlled Release, 2010, 148, 135–146 CrossRef CAS PubMed.
- J.-Z. Du, C.-Q. Mao, Y.-Y. Yuan, X.-Z. Yang and J. Wang, Biotechnol. Adv., 2014, 32, 789–803 CrossRef CAS PubMed.
- S. F. Medeiros, A. M. Santos, H. Fessi and A. Elaissari, Int. J. Pharm., 2011, 403, 139–161 CrossRef CAS PubMed.
- L. Tian and Y. H. Bae, Colloids Surf., B, 2012, 99, 116–126 CrossRef CAS PubMed.
- V. Estrella, T. Chen, M. Lloyd, J. Wojtkowiak, H. H. Cornnell, A. Ibrahim-Hashim, K. Bailey, Y. Balagurunathan, J. M. Rothberg, B. F. Sloane, J. Johnson, R. A. Gatenby and R. J. Gillies, Cancer Res., 2013, 73, 1524–1535 CrossRef CAS PubMed.
- S. H. Crayton and A. Tsourkas, ACS Nano, 2011, 5, 9592–9601 CrossRef CAS PubMed.
- H. Mok, O. Veiseh, C. Fang, F. M. Kievit, F. Y. Wang, J. O. Park and M. Zhang, Mol. Pharm., 2010, 7, 1930–1939 CrossRef CAS PubMed.
- I. Saha, K. E. Chaffee, C. Duanmu, B. M. Woods, A. M. Stokes, L. E. Buck, L. L. Walkup, N. Sattenapally, J. Huggenvik, Y. Gao and B. M. Goodson, J. Phys. Chem. C, 2013, 117, 1893–1903 CAS.
- D. Ling, W. Park, S.-J. Park, Y. Lu, K. S. Kim, M. J. Hackett, B. H. Kim, H. Yim, Y. S. Jeon, K. Na and T. Hyeon, J. Am. Chem. Soc., 2014, 136, 5647–5655 CrossRef CAS PubMed.
- Avidin-Biotin Technology, ed. M. Wilchek and E. A. Bayer, Elsevier, 1990, vol. 184 Search PubMed.
- K. Hofmann, G. Titus, J. A. Montibeller and F. M. Finn, Biochemistry, 1982, 21, 978–984 CrossRef CAS PubMed.
- G. Orr, J. Biol. Chem., 1981, 256, 761–766 CAS.
- F. Garret-Flaudy and R. Freitag, Biotechnol. Bioeng., 2000, 71, 223–234 CrossRef CAS PubMed.
- S. Sun, M. Ma, N. Qiu, X. Huang, Z. Cai, Q. Huang and X. Hu, Colloids Surf., B, 2011, 88, 246–253 CrossRef CAS PubMed.
- H. Inoue, K. Sato and J. Anzai, Biomacromolecules, 2004, 6, 27–29 CrossRef PubMed.
- Z. Poon, D. Chang, X. Zhao and P. T. Hammond, ACS Nano, 2011, 5, 4284–4292 CrossRef CAS PubMed.
- S. I. C. J. Palma, M. Marciello, A. Carvalho, S. Veintemillas-Verdaguer, M. d. P. Morales and A. C. A. Roque, J. Colloid Interface Sci., 2015, 437, 147–155 CrossRef CAS PubMed.
- M. K. Yu, J. Park and S. Jon, Theranostics, 2012, 2, 3–44 CrossRef CAS PubMed.
- J. Xie, C. Xu, N. Kohler, Y. Hou and S. Sun, Adv. Mater., 2007, 19, 3163–3166 CrossRef CAS.
- A. Ruiz, Y. Hernández, C. Cabal, E. González, S. Veintemillas-Verdaguer, E. Martínez and M. P. Morales, Nanoscale, 2013, 5, 11400–11408 RSC.
- A. Ruiz, G. Salas, M. Calero, Y. Hernández, A. Villanueva, F. Herranz, S. Veintemillas-Verdaguer, E. Martínez, D. F. Barber and M. P. Morales, Acta Biomater., 2013, 9, 6421–6430 CrossRef CAS PubMed.
- M. L. Mojica Pisciotti, E. Lima, M. Vasquez Mansilla, V. E. Tognoli, H. E. Troiani, A. A. Pasa, T. B. Creczynski-Pasa, A. H. Silva, P. Gurman, L. Colombo, G. F. Goya, A. Lamagna and R. D. Zysler, J. Biomed. Mater. Res., Part B, 2014, 102, 860–868 CrossRef CAS PubMed.
- Z. Poon, J. B. Lee, S. W. Morton and P. T. Hammond, Nano Lett., 2011, 11(5), 2096–2103 CrossRef CAS PubMed.
- M. Calero, M. Chiappi, A. Lazaro-Carrillo, M. J. Rodríguez, F. J. Chichón, K. Crosbie-Staunton, A. Prina-Mello, Y. Volkov, A. Villanueva and J. L. Carrascosa, J. Nanobiotechnol., 2015, 13, 16 CrossRef PubMed.
- M. M. Yallapu, N. Chauhan, S. F. Othman, V. Khalilzad-Sharghi, M. C. Ebeling, S. Khan, M. Jaggi and S. C. Chauhan, Biomaterials, 2015, 46, 1–12 CrossRef CAS PubMed.
- M. Mahmoudi, I. Lynch, M. R. Ejtehadi, M. P. Monopoli, F. B. Bombelli and S. Laurent, Chem. Rev., 2011, 111, 5610–5637 CrossRef CAS PubMed.
- L. H. Reddy, J. L. Arias, J. Nicolas and P. Couvreur, Chem. Rev., 2012, 112, 5818–5878 CrossRef CAS PubMed.
- A. Theumer, C. Gräfe, F. Bähring, C. Bergemann, A. Hochhaus and J. H. Clement, J. Magn. Magn. Mater., 2015, 380, 27–33 CrossRef CAS.
- J. Chen, J. A. Hessler, K. Putchakayala, B. K. Panama, D. P. Khan, S. Hong, D. G. Mullen, S. C. Dimaggio, A. Som, G. N. Tew, A. N. Lopatin, J. R. Baker, M. M. B. Holl and B. G. Orr, J. Phys. Chem. B, 2009, 113, 11179–11185 CrossRef CAS PubMed.
- S. I. C. J. Palma, C. A. Rodrigues, A. Carvalho, M. P. Morales, F. Freitas, A. R. Fernandes, J. S. Cabral and A. C. A. Roque, Nanoscale, 2015, 7, 14272–14283 RSC.
- K. M. Krishnan, IEEE Trans. Magn., 2010, 46, 2523–2558 CrossRef CAS PubMed.
- S. Sun, H. Zeng, D. B. Robinson, S. Raoux, P. M. Rice, S. X. Wang and G. Li, J. Am. Chem. Soc., 2004, 126, 273–279 CrossRef CAS PubMed.
- M. Lattuada and T. A. Hatton, Langmuir, 2007, 23, 2158–2168 CrossRef CAS PubMed.
- I. L. Batalha, A. Hussain and A. C. A. Roque, J. Mol. Recognit., 2010, 23, 462–471 CrossRef CAS PubMed.
- K. J. Wiechelman, R. D. Braun and J. D. Fitzpatrick, Anal. Biochem., 1988, 175, 231–237 CrossRef CAS PubMed.
- S. I. C. J. Palma, A. Carvalho, J. Silva, P. Martins, M. Marciello, A. R. Fernandes, M. P. Morales and A. C. A. Roque, Contrast Media Mol. Imaging, 2015, 10, 320–328 CrossRef CAS PubMed.
- C. Riggio, M. P. Calatayud, C. Hoskins, J. Pinkernelle, B. Sanz, T. E. Torres, M. R. Ibarra, L. Wang, G. Keilhoff, G. F. Goya, V. Raffa and A. Cuschieri, Int. J. Nanomed., 2012, 7, 3155–3166 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1 to S5. See DOI: 10.1039/c6ra17217b |
| This journal is © The Royal Society of Chemistry 2016 |