Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Extrinsic surface-enhanced Raman scattering detection of influenza A virus enhanced by two-dimensional gold@silver core–shell nanoparticle arrays†
Kullavadee
Karn-orachai
ab,
Kenji
Sakamoto
a,
Rawiwan
Laocharoensuk
c,
Suwussa
Bamrungsap
c,
Sirirurg
Songsivilai
c,
Tararaj
Dharakul
cd and
Kazushi
Miki
*ab
aNational Institute for Materials Science (NIMS), 1-1 Namiki, Tsukuba, Ibaraki 305-0044, Japan. E-mail: MIKI.Kazushi@nims.go.jp; Fax: +81-29-860-4718; Tel: +81-29-860-4718
bFaculty of Pure and Applied Sciences, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8571, Japan
cNational Nanotechnology Center (NANOTEC), National Science and Technology Development Agency (NSTDA), Pathumthani 12120, Thailand
dDepartment of Immunology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok 10700, Thailand
First published on 5th October 2016
Abstract
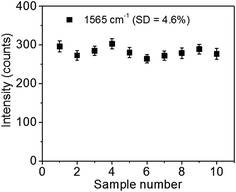
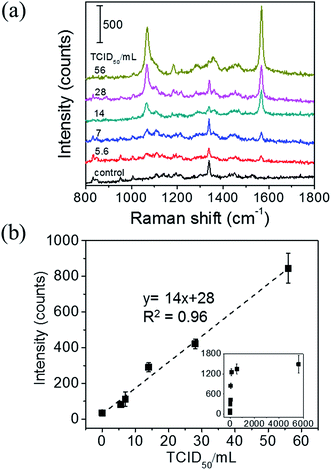
A surface-enhanced Raman scattering (SERS) based biosensor using a direct immunoassay platform is demonstrated for influenza A detection. The nucleoprotein of influenza A virus, which is one of the most conserved and abundant structural proteins on the virion, was used as a target. In this study, highly sensitive biosensors were realized by combining specific recognition of antibody–antigen interactions and high signal enhancement of the SERS effect. SERS probes were fabricated by decorating PEGylated, 4,4′-thiobisbenzenethiol (TBBT)-labeled gold nanoparticles (NPs) with influenza A antibodies. To improve the sensitivity, a SERS immunoassay was performed on two-dimensional (2D) arrays of gold@silver core–shell (Au@Ag) NPs, which work as SERS substrates. The SERS signal of TBBT was utilized to detect the selective nucleoprotein–antibody recognition. The SERS signal was enhanced ∼4 times by using the SERS substrates instead of a flat Au film. These results indicate that using a well-tuned Au@Ag 2D array as a SERS substrate is an effective way of improving sensitivity of SERS-based biosensors. Our SERS immunoassay system revealed high selectivity and good reproducibility with a sample-to-sample variation of 4.6% (relative standard deviation). To demonstrate the applicability of our SERS immunoassay system to real biological samples, the detection of influenza A using infected allantoic fluid was also performed. The linear relation between the concentration of infected allantoic fluid and the SERS signal was obtained in the range of 5 to 56 TCID50 per mL (R2 = 0.96 for the TBBT Raman bands at 1565 cm−1) with the lowest detection limit of 6 TCID50 per mL. These findings demonstrated the potential of this SERS immunosensor platform for the highly sensitive and specific detection of target molecules in a complex matrix commonly found in clinical specimens.
1. Introduction
Surface enhanced Raman scattering (SERS) is regarded as a powerful technique for detecting trace amounts of biological and chemical substances1,2 adsorbed on metal nanostructures,3 because of its inherent advantages: e.g. nondestructive, no photobleaching, sensitive and fast detection.4,5 Highly sensitive biosensors for clinical and early diagnosis of diseases can be realized by combining specific recognition of antibody–antigen interactions and high signal enhancement of the SERS effect.6,7 Since proteins have common basic chemical compositions, it is difficult to make a distinction between different proteins from Raman spectra. Thus, most SERS-based biosensors adopt the extrinsic detection method, in which a target biomolecule is labeled with a Raman reporter molecule that has unique vibrational fingerprints. By properly selecting a Raman reporter molecule, the presence of biomolecules can be detected without interference from undesired background Raman signals.For extrinsic detection methods, target biomolecules are first immobilized on a substrate, and then SERS probes containing Raman reporter molecules are selectively labeled on the target biomolecules. The SERS probes should have a very large effective Raman cross-section with selective binding ability to a target biomolecule and sufficient stability in air and aqueous solution. Typical SERS probes are metallic nanoparticles (NPs) that are labeled with Raman reporter molecules, stabilized with a protecting shell, and have specific targeting moieties on the outer surface.8,9 By designing metallic NPs properly, the scattering efficiency of Raman reporter molecules can be strongly enhanced via localized surface plasmon resonance (LSPR) of the metallic NPs.
Typically SERS-based immunosensors rely on the use of flat substrates, e.g. Au or glass substrates.10,11 In those cases, the enhancement of Raman signals solely comes from the LSPR of isolated SERS probe and/or plasmon coupling among SERS probes. This is because no significant electric field enhancement can be expected from such a flat substrate. Thus, the structural design of SERS probes is crucial for achieving sufficient detection sensitivity. For instance a higher electromagnetic field enhancement can be obtained by using Au nanoflowers,12 Au nanostars,13 or aggregates of AuNPs14 as the metallic core of SERS probes, instead of spherical AuNPs. In addition, the use of a nanostructured metallic substrate (SERS substrate) instead of a flat substrate is useful for improving the sensitivity. By properly designing SERS substrates, an additional Raman signal enhancement can be expected through strong plasmon coupling between the SERS substrate and SERS probes. According to these strategies, some groups reported good achievements in SERS biosensors on the nanostructure-based substrate, for example, two-dimensional (2D) array of Au nanotriangle,15 Au–Ag coated GaN substrate,12 and 2D array of Au nanostar,13 with high sensitivity and/or low detection limit. Therefore, the development of SERS substrates that can be electromagnetically coupled with SERS probes is essential to realize highly sensitive biosensors.
Recently, we reported promising plasmonic substrates for SERS-based sensors, which are 2D arrays of gold@silver core–shell (Au@Ag) NPs formed on indium-tin-oxide (ITO) substrates.16 These arrays were obtained by our original hybrid method,17 which is a bottom-up deposition method based on self-assembly of NPs. Large-area, uniform, high-coverage 2D arrays with a constant interparticle gap distance can be obtained, and they possess strong mechanical stability. The LSPR of the 2D arrays is easily tuned by changing the Au@Ag NP size and the Au/Ag molar ratio. Therefore, we can design the 2D arrays optimized to an excitation laser wavelength of Raman measurement. Indeed, a high SERS enhancement factor of 107 at an excitation wavelength of 633 nm (a He–Ne laser line) was observed for rhodamine 6G molecules directly deposited on the 2D Au@Ag NP array. Thus, this SERS substrate is expected to be a key component for successful fabrication of high performance SERS-based sensors.
In this article, we report a proof of principle study of a SERS-based immunosensor for detecting the nucleoprotein of influenza A virus, and its performance. In this sensor, the immunoreaction occurs on 2D arrays of Au@Ag NPs on an ITO substrate, served as a SERS substrate. The SERS probes were fabricated by decorating PEGylated, 4,4′-thiobisbenzenethiol (TBBT)-labeled AuNPs with antibodies specific to the target nucleoprotein. TBBT was chosen as a Raman reporter molecule in this SERS immunoassay because of its outstanding characteristic Raman spectra. The strong Raman bands assigned to vibrations of νCS and νCC(ring) are observed at 1065 and 1565 cm−1, respectively. Although both the bands can be used for detection of the nucleoprotein of influenza A virus, the 1565 cm−1 band was used in this study. This is because both the bands provide us the same information and also because the baseline around the 1565 cm−1 band was more structureless than that around the 1065 cm−1 band. The ∼4 times sensitivity enhancement was achieved by replacing a flat Au film with a 2D array of Au@Ag NPs, whose LSPR was tuned to the excitation laser wavelength of 633 nm. Finally, we demonstrated the applicability of our SERS immunoassay system to complex biological samples using an infected allantoic fluid containing 5.6 × 103 TCID50 per mL (50% tissue culture infectious dose) of pandemic influenza A H1N1 2009 virus.
2. Material and method
2.1 Chemical and biomaterials
Chloroauric acid (HAuCl4), TBBT, 1-ethyl-3[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), and mercaptohexadecanoic acid (MHDA) were obtained from Sigma Aldrich. Phosphate buffered saline (PBS) solution was prepared by mixing disodium hydrogen phosphate (Na2HPO4), potassium dihydrogen phosphate (KH2PO4), and sodium chloride (NaCl) provided by Sigma Aldrich: one liter of 10 mM PBS solution includes 2.17 g of Na2HPO4, 0.26 g of KH2PO4, 8.71 g of NaCl. Silver nitrate (AgNO3), sodium citrate, ascorbic acid, dodecanethiol, and octadecanethiol were purchased from Nacalai tesque. Polyethylene glycol methyl ether thiol (PEG-SH, MW 5000 Da) and polyethylene glycol 2-mercaptoethyl ether acetic acid (COOH–PEG-SH, MW 3400 Da) were obtained from Nanocs, Inc. All the commercially available reagents were used as received without further purification. Distilled water (18.2 MΩ cm) was used in all experiments.Monoclonal antibody specific to influenza A nucleoprotein was purchased from Innova Biotechnology and used without further purification. The recombinant influenza A nucleoprotein used in this study was generated and characterized as described previously.18 The allantoic fluid infected with influenza A/Thailand/104/2009 (H1N1) (Prof. Pilaipan Puthavathana, Mahidol University, Bangkok, Thailand) was prepared and inactivated as described.19 For dilution of the infected allantoic fluid, an extraction buffer solution (150 mM NaCl, 5 mM EDTA, 16 mM Triton X-100, 12 mM sodium deoxycholate, and 3.5 mM SDS in 10 mM phosphate buffer pH 7.4) was used as solvent.
2.2 Preparation of SERS probes
SERS probes were prepared in three steps: (i) synthesis of 25 nm citrate-capped AuNPs, (ii) modification of AuNPs with TBBT (Raman reporter) and PEG molecules, and (iii) antibody immobilization. In brief, 25 nm AuNPs were synthesized by reducing HAuCl4 with sodium citrate.20 Then, the citrate-capped AuNPs were simultaneously labeled with TBBT and thiolated PEG acid (COOH–PEG-SH) by adding their mixed aqueous solution to a colloidal solution of AuNPs. The particle aggregation and precipitation during TBBT-labeling treatment is prevented by this simultaneous modification. To acquire long-term stability of particles, the AuNPs were further treated with an aqueous solution of thiolated PEG (PEG-SH). After removal of excess TBBT, COOH–PEG-SH and PEG-SH molecules from the colloidal solution by two cycles of centrifugation and re-dispersion in water, the resultant colloidal solution can be stored at 4 °C for more than 6 months. Finally, the carboxyl groups of COOH–PEG-SH molecules on AuNPs were activated with EDC/NHS, and the antibody was covalently immobilized on the TBBT-labeled AuNPs. The final antibody immobilization was conducted in a 10 mM PBS solution (pH 7.4), and the particles were washed by two cycles of centrifugation and re-dispersion in PBS solution. Hereafter, these particles are called “SERS probes”. The details of the SERS probe preparation are described in ESI.†2.3 Preparation of hydrophilic SERS substrates
The SERS substrate used in this study is a 2D array of Au@Ag NPs formed on a 0.6 mm-thick quartz substrate with a 10 nm-thick ITO film. The structure of Au@Ag NPs was designed so that the LSPR energy of 2D Au@Ag NP arrays matches the excitation wavelength (633 nm) of the Raman measurement. Additionally considering the feasibility of narrow size distribution and shape uniformity of Au@Ag NPs, the Au core diameter and the Ag shell thickness were determined to be 40 nm and 5 nm, respectively.16 Alkanethiolate-capped Au@Ag NPs were synthesized according to the procedure reported previously,16 except for using a mixed solution of octadecanethiol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dodecanethiol = 1
dodecanethiol = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 (molar ratio) in acetone21 for the mixed self-assembled monolayer (SAM) formation on the Au@Ag NP surface. The details of Au@Ag NP synthesis were described in ESI.†
6 (molar ratio) in acetone21 for the mixed self-assembled monolayer (SAM) formation on the Au@Ag NP surface. The details of Au@Ag NP synthesis were described in ESI.†
The alkanethiolate-capped Au@Ag NPs were arrayed on an ITO substrate (10 × 10 mm2) by a hybrid method, in which uniform and high-coverage 2D arrays of metallic NPs are formed through solvent-evaporation-induced self-assembly under the presence of electric field.17,22 Before arraying Au@Ag NPs, the ITO substrate was successively functionalized with 3-mercaptopropyl trimethoxysilane (MPTMS) and 1,6-hexanedithiol. (See ESI.†) The arraying of Au@Ag NPs was performed according to the procedure reported previously.16 After arraying, the 2D array samples were annealed at 60 °C overnight to enable chemisorptions of Au@Ag NPs on the functionalized ITO substrate, and then washed by sonication in hexane for 30 s to remove excess of Au@Ag NPs.
Since the 2D array substrate obtained by this method is hydrophobic, it is not suitable for immunoassay for influenza A detection. Thus, the 2D array substrate was hydrophilized by exchanging the capping molecules with MHDA. First, the 2D array substrate was annealed at 90 °C for 1 h. Then the substrate was immersed in an ethanolic solution of MHDA (15 mM, 1 mL) for 12 h at 50 °C. The substrates were rinsed with ethanol three times and dried under nitrogen atmosphere. By performing the annealing process, the MHDA treatment time was significantly reduced from 7 days to 12 h.
2.4 Immunoassay protocol
The SERS immunosensor fabricated in this research is based on a direct immunoassay (Scheme 1). After the SERS substrate (10 × 10 mm2) was divided in quarters, a 5 × 5 mm2 SERS substrate was used as the solid support for immunoassay. First, the freshly prepared hydrophilic SERS substrate was activated by successively dropping 10 μL of a 15 mM EDC aqueous solution and 10 μL of a 15 mM NHS solution. The mixed droplet was kept on the substrate for 30 min. Then, the substrate was washed with distilled water three times and dried by nitrogen gas. Next, 1 μL of a recombinant nucleoprotein solution with different concentrations was directly dropped onto the SERS substrate and incubated for 30 min for immobilizing recombinant nucleoprotein. The incubation time was optimized (see Section 3.3.1). After that, the SERS substrate was washed for 5 min each in 10 mM PBS solution followed by DI water under stirring condition. Subsequently 5 μL of a SERS probe aqueous solution (optical density = 0.5) was dropped onto the substrate to react with the target nucleoprotein. The incubation time for immobilizing SERS probes on recombinant nucleoproteins was 1 h, which was also optimized (see Section 3.3.1). Finally, the substrate was washed twice in stirred PBS solution for 5 min and then twice in stirred water for 5 min.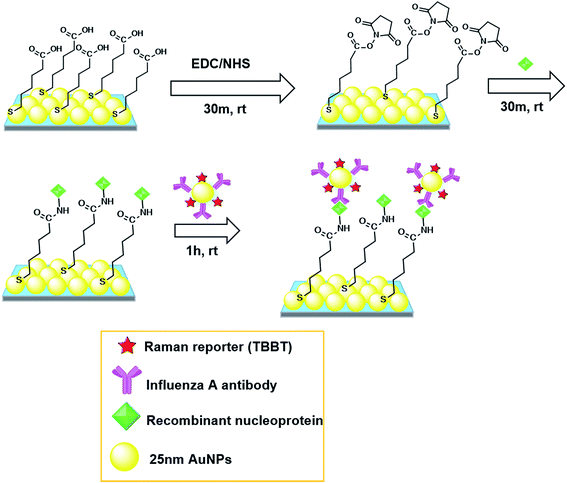 | ||
| Scheme 1 Schematic illustration of direct immunoassay on 2D Au@Ag NP array substrate. rt: room temperature. | ||
2.5 Instrumental
UV-Vis-NIR spectroscopy was carried out with a V-670 spectrometer (JASCO, Japan) to examine the optical property of nanoparticles and SERS substrates. SEM images were obtained with FE-SEM S-4800 (Hitachi, Japan) to evaluate the morphology and coverage of particles on the substrate. Raman spectra were measured with a micro-Raman spectrometer system with a He–Ne laser and a charge-coupled device (CCD) detector (NT-MDT NTEGRA Spectra or Lambda Vision MicroRAM-300). For the NT-MDT NTEGRA spectra system, the excitation laser light was focused on the sample surface using a 40× objective lens with a numerical aperture of 0.65, and the Raman scattered light was collected with the same objective lens. The Raman scattered light was dispersed with a single spectrograph of a focal length of 520 nm with a grating of 1200 grooves per mm. For the MicroRAM-300 system, a 50× objective lens with a numerical aperture of 0.75 was used for the excitation and collection of Raman scattered light. A single spectrograph of a focal length of 300 nm with a grating of 600 grooves per mm was used to record Raman spectra. For both Raman spectrometer systems, a 632.8 nm line of the He–Ne laser was used for excitation, and the laser power was set to 100 μW at the sample surface. The laser spot size was estimated about to be 4 μm in diameter. Stokes Raman shift spectra were obtained with an exposure time of 50 s. To improve the signal-to-noise ratio, typically five to ten spectra were averaged.3. Results and discussion
3.1 Characterization of SERS probes
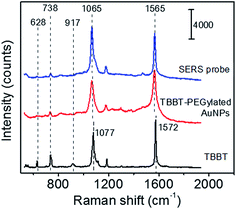
As mentioned above, the SERS probes for influenza A nucleoprotein detection were prepared in three steps; (i) synthesis of 25 nm citrate-capped AuNPs, (ii) modification of AuNPs with TBBT (Raman reporter) and PEG molecules, and (iii) immobilization of influenza A antibody. The high uniformity of citrate-capped AuNPs in shape (spherical) and in size (average diameter of 25 ± 3 nm) was confirmed by SEM (Fig. S1 in ESI†). Fig. 1 shows the Raman spectra of solid TBBT, TBBT-PEGylated AuNPs, and SERS probes. The characteristic Raman bands of solid TBBT were observed at 1077 and 1572 cm−1, which were assigned to the vibrations of νCS and νCC(ring), respectively.23,24 The corresponding bands were observed at 1065 and 1565 cm−1 for TBBT-PEGylated AuNPs and SERS probes. These vibrational frequency shifts were reported for TBBT adsorbed on AuNPs,24 and AgNPs,25 indicating chemisorptions of TBBT. The δCSH band of solid TBBT at 917 cm−1 completely disappeared on the TBBT-PEGylated AuNPs and SERS probes, indicating that TBBT molecules were chemisorbed on the colloidal AuNP surface by losing two H atoms of S–H bonds. Based on TBBT molecular structure, there are two possible double-end adsorption structures; one is bidentate adsorption on a single AuNP, and the other is bridging adsorption between adjacent AuNPs, which leads to aggregation of AuNPs. The extinction spectra of colloidal solutions of citrate-capped AuNPs, TBBT-PEGylated AuNPs, and SERS probes showed a single narrow LSPR peak at around 523 nm (see Fig. S3 in ESI†), indicating that colloidal particles were well dispersed and no aggregation occurred even after labeling AuNPs with TBBT. Thus the plausible adsorption structure is bidentate adsorption on a single AuNP. This adsorption structure was also reported on AgNPs.25 After conjugation of influenza A antibody, the characteristic peaks of TBBT were still dominant; hence, both peaks at 1065 and 1565 cm−1 can be used to detect the completion of specific antigen–antibody recognition. Since the background signal originating from the SERS substrate is more structureless around the 1565 cm−1 band than that around the 1065 cm−1 band, in this study the 1565 cm−1 band was used for detection of the nucleoprotein of influenza A virus.3.2 Characterization of hydrophilic SERS substrates
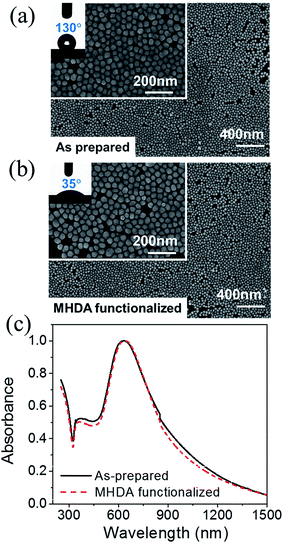
Fig. 2(a) shows high (100k) and low (25k) magnification SEM images of a 2D array prepared by the hybrid method, indicating the high uniformity and surface coverage of the 2D array. The solid curve of Fig. 2(c) shows the extinction spectrum of the as-prepared 2D array. The extinction peak assigned to LSPR of the 2D array was observed around 627 nm. This result showed successful tuning of LSPR energy to the excitation (633 nm) of our Raman measurement. Hence, the highest enhancement effect was expected from the 2D array SERS substrate.Since the constituent Au@Ag NPs of the as-prepared 2D array are stabilized with mixed alkanethiol molecules, this substrate surface is hydrophobic. As shown in the inset of Fig. 2(a), the contact angle of a 1 μL water droplet was 130°, indicating a strong water repellent characteristic. Thus, the 2D array must be hydrophilized before utilizing it as a substrate in an immunoassay using aqueous solutions. The hydrophilization was performed by annealing the hydrophobic 2D array at 90 °C for 1 h and then immersing it in an ethanolic solution of MHDA at 50 °C for 12 h. Fig. 2(b) shows the SEM images of the MHDA-functionalized 2D array and the contact angle of a water droplet. The MHDA-functionalization significantly reduced the water contact angle from 130° to 35°, indicating successful hydrophilization. This hydrophilization was also confirmed by attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy (see ESI†). The arrangement of Au@Ag NPs was not influenced by MHDA-functionalization as confirmed by the SEM images. In addition, the extinction spectrum shown by the dashed curve in Fig. 2(c) can be used to confirm that optical property of the SERS substrate remains unchanged. Since the LSPR wavelength of the resultant hydrophilic SERS substrate is matched with the Raman excitation wavelength (633 nm), this 2D array SERS substrate is suitable for an immunoassay.
3.3 SERS immunoassay
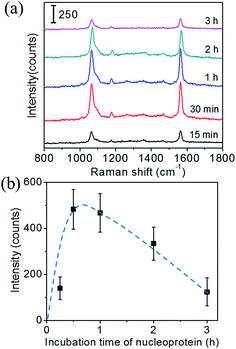
Next, the incubation time of the SERS probes was optimized. Fig. 4(a) shows the Raman spectra of the immunoassay performed with different incubation time of SERS probes, under a fixed immobilization condition of the recombinant nucleoprotein: 1.34 μg mL−1 and 30 min. The characteristic peak intensity related to incubation time of SERS probes is shown in Fig. 4(b). These results suggest that the incubation time of 1 h is required for sufficient nucleoprotein–antibody recognition reaction. For incubation time longer than 1 h, ∼40% decrease in intensity was observed. It could be also due to the denaturation of protein structure induced by a long incubation of SERS probes. Thus, the suitable incubation time for SERS probes is 1 h.
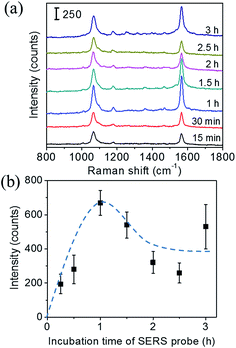
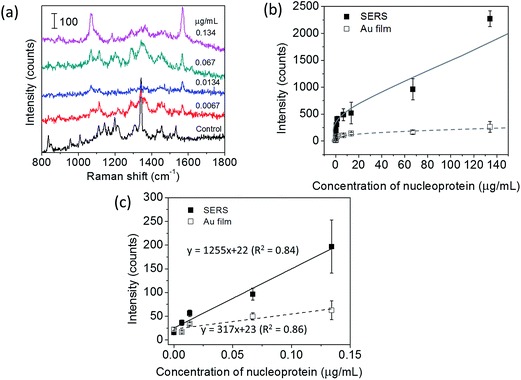
Fig. 5(a) shows the Raman spectra of the immunoassay conducted on the SERS substrates. For the control, PBS solution without recombinant nucleoprotein was dropped onto the substrates. The result demonstrated that several Raman bands were observed for the control sample on a SERS substrate in the range of 600 to 1600 cm−1, which can be tentatively assigned to the vibrational modes of MPTMS and activated MHDA. The assignment was presented in ESI (Table S1†). These bands are found to be well separated from the characteristic Raman band of TBBT at 1565 cm−1. As expected, the 1565 cm−1 band was negligibly weak for the control samples, and its intensity increased with increasing concentration of recombinant nucleoprotein.
Fig. 5(b) show the sensitivity curves of the immunoassay conducted on the SERS substrate and the Au film substrate in the entire measurement range. The sensitivity curves were obtained by plotting the intensity of the 1565 cm−1 band as a function of the concentration of recombinant nucleoprotein. The stronger Raman signal was observed for the SERS substrate than that of the Au film substrate with the same concentration of the target. Both sensitivity curves show a similar pattern of dependence; i.e. initially the Raman intensity increases almost linearly with increasing concentration of recombinant nucleoprotein, and then the increasing rate gradually decreases. The linear relation was observed in the concentration range of 0.0067 to 0.134 μg mL−1 for both substrates.
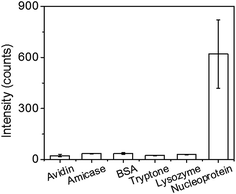
The linear sensitivity curves are shown in Fig. 5(c). Comparing the slopes, we found that the sensitivity of immunoassay improved about 4 times by using the 2D Au@Ag NP array substrate, instead of the flat Au film substrate. The limit of detection (LOD) of the immunoassay was estimated using the linear sensitivity curves. The definition of LOD used in this study is described in ESI.† The LOD of the immunoassay using the SERS substrate was 8 ng mL−1, while that using the Au film substrate was 59 ng mL−1. Since the increase of the surface area of the 2D-NP array against the flat surface is at the outside twice, the improvement in the sensitivity and detection limit can be mainly attributed to plasmon coupling between the SERS probe and the SERS substrate. Finally, comparison of the LOD with those of other techniques previously reported for detecting the same analyte. The LOD of our SERS immunoassay system (8 ng mL−1) is much lower than those of fluorescence-based LFIA (250 ng mL−1),26 an immune-interferometric sensor (1 μg mL−1),27 and comparable to that of electrochemical immunoassay (e-ELISA) (10 ng mL−1).28
4. Conclusion
A SERS immunoassay with high sensitivity and selectivity for influenza A was demonstrated utilizing both PEGylated TBBT-labeled AuNPs covered with influenza A antibody as the SERS probes and hydrophilic Au@Ag 2D array as the SERS substrates. The antibody–antigen recognition was detected by observing characteristic Raman signal of TBBT (Raman reporter). Immunoassay taken place on SERS substrates has about 4 times higher sensitivity than immunoassay on Au film substrates, because the Raman signal from the SERS probes is amplified by the electromagnetic field enhancement effect of the SERS substrate. This immunoassay shows the good performance to detect the target nucleoprotein in a complex biological matrix that contains various kinds of interferences with the detection limit of 6 TCID50 per mL. These results indicate that using a well-tuned Au@Ag 2D array as a SERS substrate is an effective way of improving sensitivity of SERS-based biosensors.Acknowledgements
We thank Dr S. Nishiyama and Dr K. Nittoh for their technical supports. We thank both the Japan Science and Technology Agency (JST) and the National Science and Technology Development Agency (NSTDA Thailand) for financial support of the East Asia Science and Innovation Area Joint Research Program (e-ASIA JRP). A part of this work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 24656040, by the Yazaki Memorial Foundation for Science and Technology, by NIMS and by NANOTEC. R. L., S. B. and T. D. would acknowledge funding from NANOTEC, NSTDA. K. K. would acknowledge the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) for the Japanese Government Scholarship for Research Students.References
- T. Vo-Dinh, Y. Liu, A. M. Fales, H. Ngo, H.-N. Wang, J. K. Register, H. Yuan, S. J. Norton and G. D. Griffin, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7(1), 17–33 CrossRef CAS PubMed.
- J. Kneipp, H. Kneipp and K. Kneipp, Chem. Soc. Rev., 2008, 37, 1052–1060 RSC.
- C. J. Orendorff, A. Gole, T. K. Sau and C. J. Murphy, Anal. Chem., 2005, 77(10), 3261–3266 CrossRef CAS PubMed.
- C. Wang and C. Yu, Nanotechnology, 2015, 26, 092001 CrossRef CAS PubMed.
- S. McAughtrie, K. Lau, K. Faulds and D. Graham, Chem. Sci., 2013, 4, 3566–3572 RSC.
- Y.-J. Lin, C.-Y. Wu, T. Li, P.-W. Hsiao and D.-K. Chang, J. Biosens. Bioelectron., 2014, 5, 1000150 Search PubMed.
- A. M. Paul, Z. Fan, S. S. Sinha, Y. Shi, L. Le, F. Bai and P. C. Ray, J. Phys. Chem. C, 2015, 119, 23669–23675 CAS.
- A. J. Driscoll, M. H. Harpster and P. A. Johnson, Phys. Chem. Chem. Phys., 2013, 15, 20415–20433 RSC.
- C. David, N. Guillot, H. Shen, T. Toury and M. L. de la Chapelle, Nanotechnology, 2010, 21, 475501 CrossRef PubMed.
- C.-C. Lin, Y.-M. Yang, Y.-F. Chen, T.-S. Yang and H.-C. Chang, Biosens. Bioelectron., 2008, 24, 178–183 CrossRef CAS PubMed.
- J. D. Driskell, K. M. Kwarta, R. J. Lipert, M. D. Porter, J. D. Neill and J. F. Ridpath, Anal. Chem., 2005, 77, 6147–6154 CrossRef CAS PubMed.
- A. Kamińska, E. Witkowska, K. Winkler, I. Dzięcielewski, J. L. Weyher and J. Waluk, Biosens. Bioelectron., 2015, 66, 461–467 CrossRef PubMed.
- Y. Pei, Z. Wang, S. Zong and Y. Cui, J. Mater. Chem. B, 2013, 1, 3992–3998 RSC.
- T. Büchner, D. Drescher, H. Traub, P. Schrade, S. Bachmann, N. Jakubowski and J. Kneipp, Anal. Bioanal. Chem., 2014, 406, 7003–7014 CrossRef PubMed.
- M. Li, S. K. Cushing, J. Zhang, S. Suri, R. Evans, W. P. Petros, L. F. Gibson, D. Ma, Y. Liu and N. Wu, ACS Nano, 2013, 7, 4967–4976 CrossRef CAS PubMed.
- F. Pincella, Y. Song, T. Ochiai, K. Isozaki, K. Sakamoto and K. Miki, Chem. Phys. Lett., 2014, 605–606, 115–120 CrossRef CAS.
- K. Isozaki, T. Ochiai, T. Taguchi, K. Nittoh and K. Miki, Appl. Phys. Lett., 2010, 97, 221101 CrossRef.
- K. Watcharatanyatip, S. Boonmoh, K. Chaichoun, T. Songserm, M. Woratanti and T. Dharakul, J. Virol. Methods, 2010, 163, 238–243 CrossRef CAS PubMed.
- D. J. King, Avian Dis., 1991, 35, 505–514 CrossRef CAS PubMed.
- S. Link, Z. L. Wang and M. A. El-Sayed, J. Phys. Chem. B, 1999, 103, 3529–3533 CrossRef CAS.
- U. T. D. Thuy, K. Sakamoto, S. Nishiyama, S. Yanagida, N. Q. Liem and K. Miki, Langmuir, 2015, 31, 13494–13500 CrossRef CAS PubMed.
- T. Ochiai, K. Isozaki, S. Nishiyama and K. Miki, Appl. Phys. Express, 2014, 7, 0065001 CrossRef.
- T.-T. You, P.-G. Yin, L. Jiang, X.-F. Lang, L. Gou and S.-H. Yang, Phys. Chem. Chem. Phys., 2012, 14, 6817–6825 RSC.
- Y. Wang, L. Gan, H. Chen, S. Dong and J. Wang, J. Phys. Chem. B, 2006, 110, 20418–20425 CrossRef CAS PubMed.
- Y. Wang, H. Chen, S. Dong and E. Wang, J. Raman Spectrosc., 2008, 39, 389–394 CrossRef CAS.
- S. Bamrungsap, C. Apiwat, W. Chantima, T. Dharakul and N. Wiriyachaiporn, Microchim. Acta, 2014, 181, 223–230 CrossRef CAS.
- L. R. Farris, N. Wu, W. Wang, L.-J. A. Clarizia, X. Wang and M. J. McDonald, Anal. Bioanal. Chem., 2010, 396, 667–674 CrossRef CAS PubMed.
- K. Ohtsuka, H. Endo, K. Morimoto, B. N. Vuong, H. Ogawa, K. Imai and S. Takenaka, Anal. Sci., 2008, 24, 1619–1622 CrossRef CAS PubMed.
- N. Shah and D. C. Naseby, Biosens. Bioelectron., 2015, 68, 447–453 CrossRef CAS PubMed.
- N. Wiriyachaiporn, W. Maneeprakorn, C. Apiwat and T. Dharakul, Microchim. Acta, 2015, 182, 85–93 CrossRef CAS.
- C. Apiwat, N. Wiriyachaiporn, W. Maneeprakorn, T. Dharakul, C. Thepthai, P. Puthavathana, S. Siritantikorn and N. Horthongkham, Arch. Virol., 2014, 159, 1603–1611 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra17143e |
| This journal is © The Royal Society of Chemistry 2016 |