Open Access Article
Open Access ArticleNon-invasive monitoring of the osteogenic differentiation of human mesenchymal stem cells on a polycaprolactone scaffold using Raman imaging†
Yu
Gao
 a,
Chenjie
Xu
*bc and
Lianhui
Wang
*a
a,
Chenjie
Xu
*bc and
Lianhui
Wang
*a
aKey Laboratory for Organic Electronics and Information Displays & Institute of Advanced Materials (IAM), Jiangsu National Synergistic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts & Telecommunications, 9 Wenyuan Road, Nanjing 210023, China. E-mail: iamlhwang@njupt.edu.cn
bSchool of Chemical and Biomedical Engineering, Nanyang Technological University, 70 Nanyang Drive, Singapore 637457. E-mail: cjxu@ntu.edu.sg
cNTU-Northwestern Institute for Nanomedicine, Nanyang Technological University, 50 Nanyang Avenue, Singapore 639798
First published on 23rd June 2016
Abstract
Scaffold-based bone tissue engineering often involves the use of human mesenchymal stem cells (hMSCs) that are seeded into three-dimensional (3D) scaffolds and induced to generate new bone by osteoinductive cues. In order to obtain an efficacious reconstruction of bone tissue, hMSCs must grow and differentiate in osteogenic conditions on biomaterial scaffolds to be subsequently implanted in vivo. Traditional evaluation of the osteogenic differentiation of hMSCs on scaffolds depends on time-consuming and cell-destroying end-point assays. This work explores the use of Raman spectroscopy as a non-invasive and real-time imaging method for continuous monitoring of the osteogenic differentiation of hMSCs on a polycaprolactone scaffold. In a period of 28 days, Raman spectroscopic imaging with a single cell resolution provided fingerprint chemical information and structural information on the differentiating hMSCs. Delayed mineralization was observed for hMSC osteogenic differentiation on PCL scaffolds in comparison to that on tissue culture plates.
Introduction
In the past decades, bone tissue-engineering with the goal of regenerating the bone tissue on a porous permanent or temporary scaffold has been developed to represent a cost-effective and patient-friendly approach.1,2 As bone is the second most transplanted tissue in the world, it has been reported that over two million replacement procedures are carried out annually globally.3 The successful bone tissue engineering processes in the clinic always involve loading, culturing and osteogenic induction of human mesenchymal stem cells (hMSCs) on biocompatible scaffolds in vitro and subsequent implantation in vivo. The ideal scaffold should allow the regeneration of bones that can withstand stress or forces without breaking, which requires the successful adhesion and subsequent induction of the osteogenic differentiation of hMSCs on the scaffold before transplantation.4–6The traditional approaches for monitoring the osteogenic differentiation of MSCs on scaffold include quantitative alkaline phosphatase (ALP) activity measurement, gene expression of osteogenic markers like ALP, type 1 collagen, osteocalcin and osteopontin, histological (e.g. von Kossa and Alizarin red staining) and immunochemical staining of osteogenic markers.7 Most of these methods are poorly quantified by pixel measurements and supply limited distinguishable information of the volume, mechanical properties and chemical composition of newly formed bone deposition.8 More importantly, they are invasive that disable the following usage of bone scaffolds for transplantation. While the primary method for assessing fracture healing (i.e. computed tomography) could allow better assessment of the efficacy and performance of grafts, they are lack of resolution for distinguishing the subtle structures and information of chemical composition during the bone healing.9 Fluorescence based nanosensors were developed to monitor the osteogenesis of hMSCs.10,11 However, the influences on cellular viability and bone functionality generated from the fluorescent dye are still not well understood. Therefore, it is highly desired to have a label-free imaging methodology which can non-invasively detect the osteogenic differentiation of MSCs, visualize the fine structure of mineralized nodules, and acquire the chemical information during the osteogenic differentiation on the scaffolds.
Raman imaging is an attractive analytical tool because of its high specificity (finger-print chemical information), low sensitivity to water (advantageous to biological samples), and minimal sample preparation (label-free).12 It has been used in non-invasive identification of the osteogenic differentiation of hMSCs13–17 and mapping of bone tissue sections previously.18In situ Raman spectrum was acquired and analyzed during the osteogenic differentiation of stem cells on cover slides or Raman substrates. The development of hydroxyapatite was revealed to be associated with the differentiation of hMSCs into osteoblasts.13–18 By analyzing the mineralized nodules formed in vitro, Raman spectroscopy can further reveal cell-source-dependent differences in multiple bone-like mineral environments.19 However, all of these works were performed in 2D tissue culture plates or glass slides, where hMSCs behave differently from polymeric scaffolds.20 Thus the previous observation might not be helpful in identifying the suitable time for transplanting the engineered scaffold.
In this report, we explore the usage of Raman imaging to monitor the osteogenic differentiation of MSCs on the polymeric scaffold for the first time. Specifically, hMSCs were seeded on polycaprolactone (PCL) film scaffold, and their osteogenic differentiation were monitored by Raman imaging for 28 days. Delayed mineralization was observed for hMSCs osteogenic differentiation on PCL scaffolds in comparison to that on tissue culture plates. Based on chemical and structural information provided by Raman imaging, two stages of mineralization on PCL scaffolds were found after 3 and 4 weeks post osteogenic induction respectively.
Results and discussions
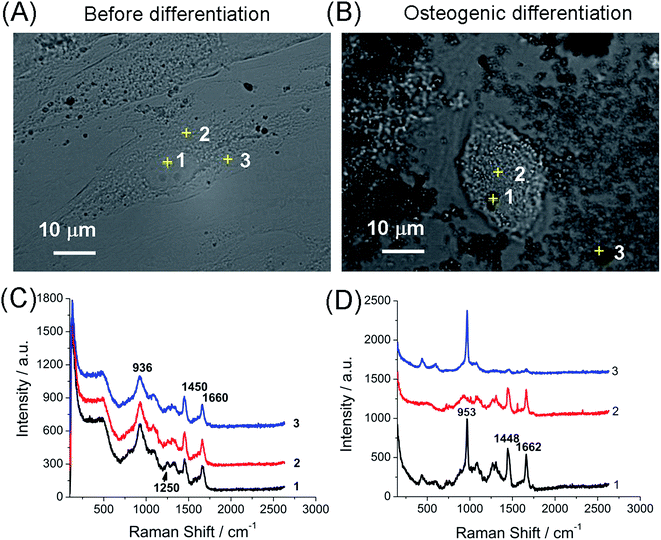
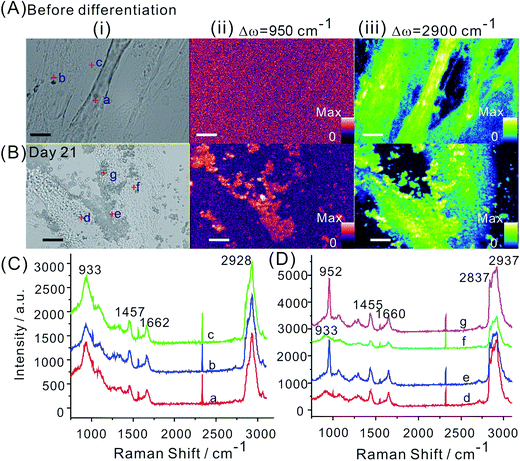
The Raman spectrum of hMSCs before and after osteogenic induction for 21 days on tissue culture plates were first acquired. Undifferentiated hMSCs exhibited a spindle shape (Fig. 1A). Their Raman spectrum of nucleus (1 in Fig. 1C) and cytoplasm regions (2 and 3 in Fig. 1C) were marked by major cellular components features including protein (skeletal C–C vibrations at 936 cm−1, amide I band at 1660 cm−1 and amide III vibrations at 1250 cm−1) and carbohydrates (C–H deformation at 1450 cm−1).13,19 After induction of osteogenic differentiation for 21 days, hMSCs differentiated and mineralized that exhibited morphology change (from spindle shaped into cuboidal shaped, Fig. 1B) with hydroxyapatite deposition on top of and around the cells. Hydroxyapatite Ca10(PO4)6(OH)2 is the mineral component of bone and is usually deposited on an extracellular matrix during the osteogenic differentiation of hMSCs. The specific Raman peak near 953 cm−1 indicated the PO4−3ν1 symmetric stretch from the hydroxyapatite (1 and 3 in Fig. 1D), while similar Raman spectrum to that of undifferentiated hMSCs was obtained at the cellular region without mineralization (2 in Fig. 1B and D). Therefore, the Raman intensity near 950 cm−1 was assigned to identify the mineralized nodules from undifferentiated cells.By line illumination and beam scanning the interest areas, Raman imaging were performed for hMSCs before and after osteogenic differentiation on tissue culture plates. Integration of the intensities at 950 cm−1 of each pixel generates the image of mineralization while integration at 2900 cm−1 (C–H vibration) depicts the profile of cells. The mineralization was absent for undifferentiated hMSCs as expected (Fig. 2A and C), where only background signal was shown in the Raman mapping image at 950 cm−1. After the osteogenic induction for 21 days, clear mineralized nodules presented in the Raman mapping image (Δω = 950 cm−1, Fig. 2B and D, S1 in ESI†). The distribution of mineralized nodules was mainly associated with and adjacent to the cells. The chemical information obtained from the Raman spectrum during the scanning allowed multiple components imaging simultaneously. For example, adipogenic differentiation of hMSCs which do not directly contribute to bone formation can be clearly identified by integration the intensity at 2900 cm−1 generated from the abundant lipid droplets in adipocytes that is much stronger than that from other cellular components (Fig. S2 in ESI†).
After establishing the imaging method on tissue culture plates, we next set to monitor the osteogenic differentiation of hMSCs on cell loaded scaffolds. PCL has been widely used in biodegradable scaffolds and drug containing implants for tissue engineering and cancer therapy.21–24 For example, the Osteopore products (PCL based scaffolds) has been obtained FDA approval and a CE mark, and been implanted in more than 1500 patients.6 In this work, a PCL film scaffold fabricated by cryomilling and subsequent heat pressing presented a smooth and continuous surface (Fig. S3 in ESI†). The mechanical properties were also obtained from a tensile test with 10.8 ± 1.9 MPa of yield strength, 192.9 ± 38.4 MPa of Young's modulus and 5.7 ± 0.8 MJ m−3 of toughness, which are in good agreements with reported studies.22,24 The Raman spectroscopy of the PCL scaffold was marked by νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching (1734 cm−1), νC–H stretching (2900 cm−1) and other crystalline domains (800 to 1500 cm−1).25 Although the strong Raman peak of PCL at 2900 cm−1 might influence the visualization of cells, the Raman peak near 900 cm−1 (927 cm−1) coincidentally avoid overlapping with the signal of phosphate at 950 cm−1 (Fig. S3 in ESI†). As a result, the Raman peak at 950 cm−1 generated from the mineral deposition during the osteogenic differentiation of hMSCs still can be distinguished from the background signal of PCL scaffolds theoretically.
O stretching (1734 cm−1), νC–H stretching (2900 cm−1) and other crystalline domains (800 to 1500 cm−1).25 Although the strong Raman peak of PCL at 2900 cm−1 might influence the visualization of cells, the Raman peak near 900 cm−1 (927 cm−1) coincidentally avoid overlapping with the signal of phosphate at 950 cm−1 (Fig. S3 in ESI†). As a result, the Raman peak at 950 cm−1 generated from the mineral deposition during the osteogenic differentiation of hMSCs still can be distinguished from the background signal of PCL scaffolds theoretically.
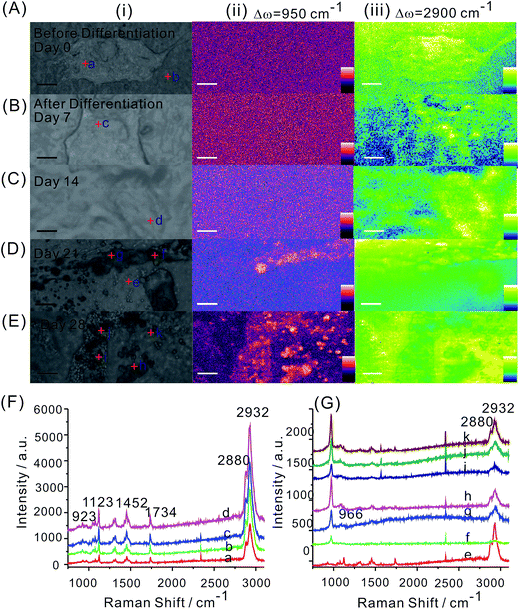
hMSCs were seeded on the PCL scaffolds and imaged at day 0. Then they were induced osteogenic differentiation and cultured for 28 days. At predetermined time points, the hMSCs loaded PCL scaffolds were scanned by Raman imaging for evaluation of the osteogenic differentiation. Both images at 950 and 2900 cm−1 and the Raman spectrum random selected from the bright field images were presented (Fig. 3). Before differentiation (i.e. day 0), there was no signal of mineralized nodules showing in the Raman image (Fig. 3A). The Raman peak at 950 cm−1 was also absent for hMSCs on PCL scaffold cultured in non-differentiation inducing medium (i.e. growth medium) for 28 days (Fig. S4 in ESI†). Clear mineralization started at day 21, that was 3 weeks after osteogenic induction. At this moment, mineral nodules were random distributed on the scaffolds suggesting the mineralization stage of bone evolution (Fig. 3D).8 The time point that mineralization happened on tissue culture plates was reported around 1 to 2 weeks after osteogenic induction.10 Our observation on PCL scaffolds suggested the differences of mineralization of osteogenic induced hMSCs in different culture environment. The delayed mineralization exhibited by hMSCs on scaffolds might be caused by the differences between materials that resulting in different cell adhesion and proliferation. If culture were kept for a longer period, the nucleation and rapid growth of mineral nodules occurred. This was confirmed by the Raman image at day 28 where significant increase of mineral nodules presented (Fig. 3E). The subtle structure of mineral nodules exhibited disjointed fine plate-like networks of two connecting fibres randomly oriented, revealing the woven bone was formed. This finding was supported by previous studies that the woven bone stage happened at 3 to 6 weeks.8 In terms of the Raman mapping image at 2900 cm−1, the visualization of cell profile was significantly compromised by the background signal of PCL scaffolds, only undifferentiated hMSCs spread on the scaffolds was identified.
Conclusions
In summary, a non-invasive and label-free imaging methodology for evaluation the osteogenic differentiation of MSCs on stem cell loaded polymeric scaffolds was developed. This method was based on Raman imaging that can provide chemical information and subtle structural information of differentiated MSCs on the scaffolds before transplantation. As a proof of concept, the evolution of osteogenic induced hMSCs on PCL scaffolds were monitored for 28 days. This method holds great promise in assessment osteogenic differentiation of hMSCs in tissue-engineered bone scaffolds before transplantation non-invasively.Experimental
Cell culture and hMSC differentiation assay
hMSCs were purchased from Lonza (USA), and were harvested from normal human bone marrow. All the experiments of hMSCs culture and differentiation assay were performed according to the protocol provided by the vendor.hMSCs were cultured in hMSC growth medium (Lonza) supplemented with 10% Fetal Bovine Serum (FBS, Gibco) and 1% penicillin–streptomycin (Gibco), at 37 °C with 5% CO2 and 90% humidity. hMSCs (passage 2) were seeded at a density of 6000 cells per cm2, fed with hMSC growth medium every 3 days, and sub-cultured when they were approximately 90% confluent (6 or 7 days). hMSCs in all assays were used by passage 5.
The osteogenic differentiation was induced by replacing the growth medium with osteogenesis induction medium (Lonza). Feed the induced hMSCs every 3–4 days. Adipogenic differentiation was induced with 3 cycles of induction/maintenance. In each cycle, hMSCs were fed with adipogenic induction medium (Lonza) for 3 days and then replaced with adipogenic maintenance medium (Lonza) for another 2 days. After 3 complete cycles, hMSCs were cultured in adipogenic maintenance medium for 7 days. The non-induced control group was fed with hMSC growth medium on the same schedule.
Fabrication of PCL scaffolds
PCL (Osteopore International, Singapore) was used as received. PCL films were fabricated according to our previous protocol.23,24 They were fabricated by a cryomilling (Retsch®, Germany) and thermally pressed process. PCL powder were loaded into the cryogenic vial. The cryomilling protocol was 6–8 min of pre-cooling in liquid nitrogen and 20 min of continuous milling for one cycle. Then the formed PCL powders were thermally pressed into films of thickness approximately 30–60 μm. Briefly, a known mass of PCL was placed between two stainless steel sheets on the Carver system (Carver Inc, USA). Temperature was elevated to 80 °C and pressure added for 30 min. The film was then stored in a dry cabinet (Digi-Cabi AD-100, Singapore) before use.Cell seeding on PCL scaffolds
The pressed film scaffolds were cut into round pieces (12 mm in diameter) and then glued onto 12 mm coverslips. The whole compartment were sterilized by immersing into 100% ethanol for 24 hours. After washing with PBS for more than 3 times, the scaffolds were placed into a 24 well plate with 0.5 mL cell medium for 1 hour before seeding. All cells were seeded at the density of 2 × 104 per well and incubated at 37 °C with 5% CO2.High speed slit-scanning confocal Raman spectroscopy measurements
Raman spectra and Raman mapping were obtained with the sample mounted on the Ramantouch microspectrometer (Nanophoton Inc, Osaka, Japan). A 532 nm laser was used as an excitation laser. The excitation laser light was focused into a line on a sample through a cylindrical lens26 and an air objective lens (LU Plan Fluor 100× NA 0.9). The back-scattered Raman signal from the line illuminated site was collected with the same objective lens, and a one-dimensional Raman image (1D space and Raman spectra) was obtained with a two-dimensional image sensor (Princeton Instrument, PIXIS 400 BR, −70 °C, 1340 × 400 pixels) at once. At a single acquisition, line-shaped illumination is shone on the sample, where 400 Raman scattering points are then collected simultaneously in the x-direction. Two-dimensional (2D) Raman spectral images were obtained by scanning the line-shaped laser focus in a single direction. The line illumination drastically reduces the acquisition time for x–y axis Raman mapping to less than half an hour for a 6400 μm2 area, as compared to the few hours required when using conventional Raman system. The excitation laser power was 0.09 mW on the sample plane. The exposure time for each line and slit width of the spectrometer were 3 s and 50 μm for 2D Raman imaging. The line scan mode with the resolution of y direction around 300 nm was used for x–y imaging.For single point spectra collection, at least three spectrum was repeated for each sample. For the Raman imaging, the spectra was extracted from the obtained Raman mapping image. Each pixel corresponded to one spectra. The software will export the Raman spectra for selected pixels. The data treatment of raw spectra was done by the software provided by Nanophoton.
Conflict of interest
The authors declare no conflict of interest.Acknowledgements
The work was supported by Ministry of Education Tier 1 Academic Research Fund (RG131/15) and NTU-Northwestern Institute for Nanomedicine (M4081502.F40 to C. J. X). L. H. Wang acknowledges the National Basic Research Program of China (2012CB933301), the Jiangsu National Synergistic Innovation Center for Advanced Materials (SICAM), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R37), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, YX03001) for support.Notes and references
- S. Bose, M. Roy and A. Bandyopadhyay, Trends Biotechnol., 2012, 30, 546–554 CrossRef CAS PubMed.
- A. R. Amini, C. T. Laurencin and S. P. Nukavarapu, Crit. Rev. Bioeng., 2012, 40, 363–408 Search PubMed.
- K.-U. Lewandrowski, J. D. Gresser, D. L. Wise and D. J. Trantolo, Biomaterials, 2000, 21, 757–764 CrossRef CAS PubMed.
- Z.-Y. Zhang, S. H. Teoh, W.-S. Chong, T.-T. Foo, Y.-C. Chng, M. Choolani and J. Chan, Biomaterials, 2009, 30, 2694–2704 CrossRef CAS PubMed.
- Z.-Y. Zhang, S.-H. Teoh, M. S. K. Chong, E. S. M. Lee, L.-G. Tan, C. N. Mattar, N. M. Fisk, M. Choolani and J. Chan, Biomaterials, 2010, 31, 608–620 CrossRef CAS PubMed.
- Y. Liu, J. Lim and S.-H. Teoh, Biotechnol. Adv., 2013, 31, 688–705 CrossRef CAS PubMed.
- Z.-Y. Zhang, S.-H. Teoh, J. H. P. Hui, N. M. Fisk, M. Choolani and J. K. Y. Chan, Biomaterials, 2012, 33, 2656–2672 CrossRef CAS PubMed.
- Y. Liu, J. K. Y. Chan and S.-H. Teoh, J. Tissue Eng. Regener. Med., 2015, 9, 85–105 CrossRef CAS PubMed.
- B. B. Kalpakcioglu, K. Engelke and H. K. Genant, Bone, 2011, 48, 1221–1231 CrossRef PubMed.
- C. Wiraja, D. C. Yeo, M. S. K. Chong and C. Xu, Small, 2016, 12, 1342–1350 CrossRef CAS PubMed.
- M. Wang, X. Hou, C. Wiraja, L. Sun, Z. J. Xu and C. Xu, ACS Appl. Mater. Interfaces, 2016, 8, 5877–5886 Search PubMed.
- S. Stewart, R. J. Priore, M. P. Nelson and P. J. Treado, Annu. Rev. Anal. Chem., 2012, 5, 337–360 CrossRef CAS PubMed.
- H. K. Chiang, F.-Y. Peng, S.-C. Hung and Y.-C. Feng, J. Raman Spectrosc., 2009, 40, 546–549 CrossRef CAS.
- L. L. McManus, G. A. Burke, M. M. McCafferty, P. O'Hare, M. Modreanu, A. R. Boyd and B. J. Meenan, Analyst, 2011, 136, 2471–2481 RSC.
- A. Ghita, F. C. Pascut, V. Sottile and I. Notingher, Analyst, 2014, 139, 55–58 RSC.
- P.-S. Hung, Y.-C. Kuo, H.-G. Chen, H.-H. K. Chiang and O. K.-S. Lee, PLoS One, 2013, 8, e65438 CrossRef CAS PubMed.
- V. V. Pully, A. Lenferink, H.-J. van Manen, V. Subramaniam, C. A. van Blitterswijk and C. Otto, Anal. Chem., 2010, 82, 1844–1850 CrossRef CAS PubMed.
- M. Kazanci, H. D. Wagner, N. I. Manjubala, H. S. Gupta, E. Paschalis, P. Roschger and P. Fratzl, Bone, 2007, 41, 456–461 CrossRef CAS PubMed.
- E. Gentleman, R. J. Swain, N. D. Evans, S. Boonrungsiman, G. Jell, M. D. Ball, T. A. V. Shean, M. L. Oyen, A. Porter and M. M. Stevens, Nat. Mater., 2009, 8, 763–770 CrossRef CAS PubMed.
- C. Wiraja, D. C. Yeo, S. Y. Chew and C. Xu, J. Mater. Chem. B, 2015, 3, 6148–6156 RSC.
- M. A. Woodruff and D. W. Hutmacher, Prog. Polym. Sci., 2010, 35, 1217–1256 CrossRef CAS.
- J. Lim, M. S. K. Chong, J. K. Y. Chan and S.-H. Teoh, Small, 2014, 10, 2495–2502 CrossRef CAS PubMed.
- Y. Gao, J. Lim, Y. Han, L. Wang, M. S. K. Chong, S.-H. Teoh and C. Xu, Nanoscale, 2016, 8, 2568–2574 RSC.
- Y. Gao, J. Lim, D. C. L. Yeo, S. Liao, M. Lans, Y. Wang, S.-H. Teoh, B. T. Goh and C. Xu, ACS Appl. Mater. Interfaces, 2016, 8, 6336–6343 Search PubMed.
- O. Hartman, C. Zhang, E. L. Adams, M. C. Farach-Carson, N. J. Petrelli, B. D. Chase and J. E. Rabolt, Biomaterials, 2010, 31, 5700–5718 CrossRef CAS PubMed.
- H. Yamakoshi, K. Dodo, A. Palonpon, J. Ando, K. Fujita, S. Kawata and M. Sodeoka, J. Am. Chem. Soc., 2012, 134, 20681–20689 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra11636a |
| This journal is © The Royal Society of Chemistry 2016 |