Poly(vinyl alcohol): review of its promising applications and insights into biodegradation
Nihed Ben Halima
*
National Engineering School of Sfax (ENIS), Street Soukra Km 3.5 B.P. 1173, 3038-Sfax, University of Sfax & Street El Aïn Km 6, 3042-Sfax, Tunisia. E-mail: nihedbenhalima@mail.com; Tel: +21626061987
First published on 14th April 2016
Abstract
Poly(vinyl alcohol) (PVA) is a water soluble synthetic polymer, with a backbone composed only of carbon atoms and is biodegradable under both aerobic and anaerobic conditions. This polymer can be prepared by the hydrolysis of polyvinylacetate and is one of the most important synthetic polymers used in commercial, industrial, medical and nutraceutical applications. The environmental issues caused by PVA industrial practice have increased globally. Several methods have been used to treat PVA industrial discharge including in particular physicochemical methods such as electrocoagulation. Nowadays, use of bioremediation for PVA release, which has caused serious pollution problems in the natural environment, has attracted much interest. The bioremediation ability of microorganisms and their PVA degrading enzymes, especially PVA oxidases/hydrolases, has long been perceived. These enzymes as well as symbiotic microorganisms could be an effective means for biodegradation of PVA.
1. Introduction
Poly(vinyl alcohol) (PVA) is a water soluble synthetic polymer. It has a backbone composed only of carbon atoms, and is biodegradable under both aerobic and anaerobic conditions.1,2 It has been prepared by the hydrolysis of polyvinyl acetate. The discovery of PVA dates back to 1924, when a solution of poly(vinyl alcohol) was obtained by saponifying poly(vinyl ester) with caustic soda solution.3 Indeed, the physical characteristics of PVA are deeply related to its method of preparation from either the complete or partial hydrolysis of polyvinyl acetate (Fig. 1). Therefore, PVA can be classified into two groups namely: fully hydrolyzed (A) and partially hydrolyzed (B); and the partially hydrolyzed PVA is known to be used in the foods.4 Varying the initial length of vinyl acetate polymer as well as the hydrolysis conditions yielded PVA products of different properties i.e. molecular weights (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–400
000–400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), flexibility, solubility and adhesiveness.5 General physicochemical properties of PVA are given in Table 1. PVA exhibits many versatile physicochemical properties such as viscosity, film forming, emulsifying, dispersing power, tensile strength, and flexibility; it is thermostable; it is adhesive and displays tolerance towards solvents. Therefore, PVA is used in a wide range of industries especially in fabric and paper sizing, fiber coating, adhesives, emulsion polymerization, films for packing and farming, and the production of poly(vinyl butyral).6,7
000), flexibility, solubility and adhesiveness.5 General physicochemical properties of PVA are given in Table 1. PVA exhibits many versatile physicochemical properties such as viscosity, film forming, emulsifying, dispersing power, tensile strength, and flexibility; it is thermostable; it is adhesive and displays tolerance towards solvents. Therefore, PVA is used in a wide range of industries especially in fabric and paper sizing, fiber coating, adhesives, emulsion polymerization, films for packing and farming, and the production of poly(vinyl butyral).6,7
| Chemical identity and physical properties | References | |
|---|---|---|
| a Variable based on PVA grade. | ||
| CAS no. | 9002-89-5 | 92 |
| Molecular weighta | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–200 000–200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
93 |
| Structural formulaa | (–CH2CHOH–)–n–(–CH2CHOCOCH3–)–m | 94 |
| Empirical formulaa | (C2H4O)n(C4H6O2)m | 94 |
| Physical appearance | Odorless, white to cream-colored granular powder | 93 |
| Specific gravity | 1.19–1.31 | 93 |
| Solubility | Insoluble in aliphatic and aromatic hydrocarbons, esters, ketones, and oils; water soluble | 93 |
Over the last decade, much attention has been recorded in PVA as biocompatible, low cytotoxic and degradable polymer for biomedical and biomaterial research fields.8–10
PVA has also been used as an immobilization carrier and it has been considered as a type of polymer widely used in biocatalysts.11,12 Moreover, PVA complexed with other materials can be efficient for medical application such as PVA-nanogold composite membranes which have been used for antibacterial application.13
The most enjoyable applications of PVA were the formation of PVA hydrogels for biomedical uses. Such hydrogels were prepared by crosslinking of PVA solutions with chemicals (e.g., boric acid, glutaraldehyde, formaldehyde, and sodium nitrate), physicals (alternate freezing and thawing with or without organic solvents), or radiations (e.g., UV light, gamma radiation, and electron beam).14 The PVA hydrogels were found in drug delivery (hollow microparticles with or without gas), cell encapsulation, wound dressings, artificial meniscus, vascular grafts, and as biosensors and nucleus pulposus implants.8,15–18 PVA hydrogels were used also as disposable contact lenses thanks to its high moisture retention, high oxygen permeability, optical clarity, compliance and softness.19,20
However, hydrogels made using chemicals could be cytotoxic because of leaching of the residual crosslinking agents from the gels.21 Whereas, hydrogels made by physicals or radiations have shown very low toxicity.10
PVA as a cross-linked gel or linear polymer is being involved in cancer therapy.22 Moreover, PVA's applications were extended to the field of ophthalmology in treating dry eyes using artificial tear drops and in solutions for contact lens. In addition, PVA was used in electrospinning for biomedical applications.10 It is worth noting that PVA orally or intravenously administered is quickly removed from the body, with very little bioaccumulation and limited adverse effects.5
On the other hand, PVA was a useful tool as a polymer matrix for enzymes entrapment. In this context, recent studies have proven the importance of lipase entrapment in PVA and chitosan23 and entrapped alpha-amylase in pectin-PVA blend was reported to improve starch digestion.24
As far as, PVA was investigated as a surfactant system by the mean of nanoparticle formulation in inner aqueous phase to prevent aggregation and to stabilize hexameric insulin in poly(lactide-co-glycolide) based this nanoparticle formulation.25 To this end, differential scanning calorimetry suggested prevention by PVA in insulin aggregation, and the in silico study (molecular docking) justified maximum stability of the release PVA-insulin. Furthermore, in vivo studies showed that nanoparticle formulation of loaded insulin with PVA was bioactive and proved effective reduction of blood glucose level with rapid action's onset in diabetic rats. Otherwise, PVA nanoparticle formulation was found to be a good protectant of C-terminal proline residue, charged residues, and residues of antiparallel β-sheet of hexameric form of released insulin from the PVA-nanoparticle as supported by in silico and various spectroscopic and calorimetric studies.25
As the utilization of PVA is expected to increase globally,26 the environmental threat and the industrial menace posed by PVA usage will discernibly increase. From industrial effluents, the important amount of discharged PVA, a well-known recalcitrant pollutant,27 is harmful to the ecosystems and to human health. Thus, there is a significant pollution problem caused by PVA.5,28 In fact, PVA can be removed either by biological treatments under anaerobic or aerobic conditions with separated or mixed culture of different microorganisms6,29–32 or by non-biological treatments e.g. photochemically initiated degradation processes,33 ultrasonic techniques,34 radiation induced degradation,35 adsorption by various materials,36,37 and electrocoagulation using monopolar and bipolar electrodes.38 Biodegradability of PVA could be enhanced by Fenton's reagent as suggested by Xiao et al.39 and Larking et al.40 In many cases, microbial symbiotic as well as supplemental compounds such as exogenous pyrroloquinoline quinone (PQQ) that is presumed to be necessary as a cofactor for some PVA degrading enzyme especially PQQ-dependent PVA dehydrogenase (PVADH), are required for PVA degradation.41,42 PVA biodegradation would be due to a random chain cleavage in which two enzymes catalyzed oxidation process and brook the carbon backbone of the vinyl alcohol polymer.43 The enzymes responsible for the polymer's cleavage have been shown to be oxidases19,43–45 and/or hydrolases.46 Thus, many organisms that utilized these enzyme systems were studied to degrade PVA in diverse environments.7,46 Kawai and Hu7 have established a two-step process for the biodegradation of PVA. Firstly the oxidation of hydroxyl groups to form diketone or monoketone structures involving PVA oxidases (secondary alcohol oxidases (SAO) and PVA dehydrogenase (PVADH)) and on the other hand the hydrolysis of carbonyl structures that involved especially oxidized PVA hydrolase (OPH).
Although PVA is a biodegradable polymer, it has become among the major pollutants of industrial wastewater, e.g. in the textile industry.2 Consequently, it is very interesting to cover PVA bio-degraders.
In this review, biological mechanisms by which diverse categories of microorganisms and their enzymes were involved in the degradation and metabolism of PVA.
2. Importance of biological removal of PVA
The promising particularity of such synthetic polymer (PVA) is its potential as a biodegradable polymer which is considered as an unusual trait among the other synthetic carbon-chain polymers. Indeed, PVA has been found to be the only vinyl polymer which can be utilized by some microorganisms as a source of carbon and energy.47–49Due to the high mass production and utilization, considerable amounts of PVA were expected to be released into the environment. It was very difficult to remove PVA from an effluents contained many toxic chemicals and a lot of hardly degradable materials by simple treatment facilities such as non-biological treatment i.e. adsorption, chemical coagulation, membrane filtration, ultrasonic degradation, catalytic oxidation or Fenton's methods. Furthermore, the afore-mentioned processes generated excessive sludge as well as the operation cost was expensive. Thus, PVA degradation by biological treatments would solve these problems; although, there was some limitations of biological methods especially slow biodegradability which could be enhanced by potential specific microorganisms degrading PVA.50–52 In fact, PVA-biodegradation dates back to 1936 when the degradation by Fusarium lini B was been the first report53 followed by Pseudomonas O-3 in 1973.47 Therefore, PVA biodegradability is attached in some particular taxa of bacteria and fungi. Among these bacterial PVA-degraders, Pseudomonas (Sphingomonads) were the well-studied genus microorganisms.43,47,54,55 Sphingomonas species have well-known to be linked to the degradation of synthetic polymers42,55 and have also been isolated as a PVA degrader.28,49 It seems that this particular genus has some adaptations permitting the utilization of macromolecular substrates. Beside, recent reports were focused on the isolation and characterization of novel PVA-degrading bacteria related either to Sphingopyxis genus41,56 or to other genera from different biotopes such as a novel genus (Povalibacter) of PVA-degrading bacterium isolated from grapes57 and a novel species of the genus Thalassospira of a marine bacterium for degrading PVA.58 However, fungal PVA-degraders included the genera of Aspergilus,59,60 Fomitopsis,31 and Penicillium.61 In general, PVA is ideally biodegraded under aerobic aquatic environment with diverse and active organisms; while under some soils or some composting conditions, PVA was insufficiently been degraded.62,63 Whereas, Tsujiyama et al.64 have proven that biodegradation of PVA was carried out under an unsubmerged culture of Flammulina velutipes probably because its mycelium was sufficiently developed to produce PVA-degrading enzymes. Perhaps, certain acclimation phase of microbial communities was necessary to accomplish effectively PVA degradation63 as well as certain addition of various biomolecules such as fatty acids and sugars to the hydroxyl group of PVA would make it an excellent backbone for exploring novel biodegradable plastics with enhanced properties.65
However, there are some attempts to remove PVA by anaerobic processes in anaerobic baffled reactor29 or by partial anaerobic removal especially when PVA was blended with other easily biodegradable constituents66 and when nitrates was added in a sequential anaerobic-aerobic bioreactor to enhance PVA removal.67 Several other reports suggested also partial anaerobic removal of relatively low molecular weight PVA.68,69 Recently, Marusincová et al.1 have proven biodegradation of PVA under denitrifying conditions. Nevertheless, some other authors have focused on the isolation of a single bacterium with PVA as the sole carbon source in the absence of supplemental compounds,41 and mixed culture of PVA-degrading strains would be the most probable and efficient weapon to decompose recalcitrant synthetic polymers such as PVA.63,70,71 Table 2 summarize some most important PVA bio-degraders with biodegradation efficiency, microbial species/community, PVA-degrading enzymes and environmental conditions.
| Microbial species/community | PVA-degrading enzymes | Environmental conditions | Biodegradation efficiency | References |
|---|---|---|---|---|
| Sphingopyxis sp. PVA3 | The initial oxidation of PVA is mediated via a PVA oxidase, and not a PQQ-dependent dehydrogenase | Isolated from an activated sludge sample (dyeing factory) | Over 90% of PVA, at an initial concentration of 0.1%, was degraded within a 6 day cultivation | 41 |
| Degradation of PVA in the absence of symbionts or cofactors such as pyrroloquinoline quinone (PQQ) | ||||
| Sphingomonas sp./Rhodococcus erythropolis strain | PVA oxidase activity and alcohol dehydrogenase activity for various low-molecular-weight secondary alcohols | Isolated from activated sludge sampled during a waste water treatment process | Under optimal conditions, the isolate was able to degrade 500 mg of PVA per litre in 2 weeks | 42 |
| The strain required pyrroloquinoline quinone (PQQ) and another growth factor, the later could be supplied by a co-isolated Rhodococcus erythropolis strain | ||||
| Sphingopyxis sp. 113P3 (NBRC 111507) | PVA is depolymerized by PVA dehydrogenase (PVA-DH), linked with cytochrome c and oxidized-PVA hydrolase (OPH) | Isolated from activated sludge | Only three genes are directly relevant to the metabolism of PVA and comprise the pva operon, which was deposited as accession no. AB190228 | 56 |
| Identified as a polyvinyl alcohol (PVA)-degrading Pseudomonas species; it was later reidentified as Sphingopyxis species | ||||
| Flammulina velutipes | The enzymes contributing to PVA degradation could be characteristically produced in the solid and unsubmerged cultures similar to the ligninolytic enzymes, such as laccase and Mn-dependent peroxidase | Unsubmerged and solid cultures containing woody waste (quartz sand cultures) | F. velutipes initially oxidizes hydroxyl groups mainly in the syndiotactic portion and then depolymerizes the hydroxyl groups. It is still unknown whether depolymerization is the result of hydrolysis or oxidation | 64 |
| A mixture of two strains Microbacterium barkeri KCCM 10507 and Paenibacillus amylolyticus KCCM 10508 | PVA-oxidases and PVA-hydrolase or aldolase. (In fact, extensive published studies have established a two-step process for the biodegradation of PVA. The first step is either (1) oxidation of two adjacent hydroxyl groups leading to β-diketone structures or (2) oxidation of one hydroxyl group yielding monoketone structures. According to the products produced by the first step, there were two possible pathways for the second step; either hydrolysis of β-diketone structures of oxidized PVA by a β-diketone hydrolase or/and the aldolase reaction involving the monoketone structures of oxidized PVA) | Activated sludge sampled from the waste water treatment facilities in textile and dyeing factories consumed a lot of PVA, and it was used as a bacterial source | The degradation rate by a mixed culture of the two isolated strains was higher than that by single strain only. Also, the effect of polymerization degree on biodegradation was negligible, but initial PVA concentration was very sensitive to biodegradation. Forty-two per cent of PVA and 55% of chemical oxygen demand in textile waste water were removed by a mixed culture of the two isolated strains after 5 days | 70 |
2.1. Symbiotic behaviors
Several studies have identified symbiotic or mixed culture of microorganisms with the capacity to degrade PVA.28,72,73 In many cases, a cooperative function of two strains is required for degrading PVA. Shimao et al.74 and Sakazawa et al.75 have first described such a symbiosis of two bacterial strains during PVA degradation. They identified that pyrroloquinoline quinone (PQQ) plays a key growth factor for PVA degradation with the strain Pseudomonas sp. VM15C and is necessary as co-factor of its PVA dehydrogenase. Similar bacteria were isolated by other authors49,51 that required a PQQ supply either from their local environment or from a symbiotic strain.Moreover, Hashimoto and Fujita52 isolated the strain Pseudomonas vesicularis var. povalolyticus PH, that found to require not PQQ but thiamine as a growth factor and cystine, isoleucine and tyrosine for the expression of PVA decomposition activity; and these factors were supplied by co-isolated bacteria Flavobacterium sp.
In addition, Mori et al.76 described the efficiency of symbiotic microorganisms using Bacillus megaterium BX1 and its Gram-positive counterpart to degrade PVA without the need of PQQ.
Choi et al.30 also described mixed culture of two strains, M. barkeri KCCM 10507 and P. amylolyticus KCCM 10508 from dyeing wastewater which could remove PVA efficiently.
Novel aspects of symbiotic PVA biodegradation were well-documented by Vaclavkova et al.42 which demonstrated that the PVA-degrading strain required the addition of both PQQ and catalase on the agar plates and also in liquid media, suggesting a unique and more complex dependency on the symbiotic partner(s) in the natural environment.
Chung et al.70 have isolated and identified a mixture of two strains Microbacterium barkeri KCCM 10507 and Paenibacillus amylolyticus KCCM 10508 for the degradation of PVA contained in textile waste water. They demonstrated that degradation rate by the mixed strains was higher than that by single strain only. To this end, forty-two per cent of PVA and 55% of chemical oxygen demand in textile waste water were removed by a mixed culture of the two above mentioned isolated strains after 5 days.
2.2. Enzymatic degradation
The PVA chemical structure comprises mainly repeated 1,3-diol units which can be assimilated by microorganisms in two steps: oxidation and hydrolysis (cleavage).Since the discovery of the first PVA-degrading bacteria,47 many studies on biodegradation of PVA have increased significantly and have been focused on new microbial isolates, PVA-biodegradation mechanisms and PVA-degrading enzyme (PVAase) purification and characterization.2,46,73,77–79 Many authors were interested to improve the productivity and activity of PVAase by developing optimal conditions such as pH control strategy on the batch fermentation with mixed culture2 and by using efficient compounds such as 1,4-butanediol and controlling designed fermentation to enhance PVAase production by mixed microbial culture.77
Sakai et al.80 have reported the purification and characterization of a specific esterase involved in hydrolyzing the acetyl groups of PVA from the cytoplasmic fraction of Pseudomonas vesicularis PD, which can assimilate PVA by PVA oxidase.
However, the principal enzyme systems involved in PVA-degradation relied on two key enzymes: PVA-oxidizing enzymes and oxidized PVA(oxi-PVA)-hydrolysing enzymes. The first type one may be a secondary alcohol oxidase (SAO) with oxygen as an electron acceptor, as well as a periplasmic PVA dehydrogenase (PVADH) whose pathway degradation would be begun with the action of PQQ and from which electrons can be conducted to soluble cytochrome c and oxygen can be further a terminal acceptor.54 These degradation pathways are most likely done under aerobic conditions; but they should be feasible under anaerobic environment with an alternative electron acceptor such as nitrogen.1,81,82 Subsequently, oxi-PVA undergone to PVA hydrolase (β-diketone hydrolase (BDH) also called OPH) or related hydrolase is present as an aldolase-type cleavage reaction in which β-hydroxyl ketone products of PVADH (monoketone) would be hydrolyzed,7,83 and probably the resulting shorter fragments of PVA can be then assimilated.
Yet, several bacteria have been able to utilize PVA as a carbon and energy source, including, for example, Pseudomonas sp. strain VM15C. Indeed, the PVA-biodegradation in such strain involved the combination of PVA dehydrogenase (PVADH) and oxidized PVA hydrolase (OPH).84 Firstly, PVA is oxidized by PVADH or secondary alcohol oxidase to form β-diketone structure. Then, this latter structure along the polymer chain is cleaved (hydrolyzed) by OPH with formation of methyl ketone, carboxylic acid and acetic acid, etc.7,48,85
Kawai and Hu7 mentioned that the basic metabolic pathway is common to all PVA-utilizing microorganisms, centering on two-step degradation either based on extracellular depolymerization (Fig. 2) or just in the periplasm (Fig. 3), by oxidation of hydroxyl groups to form diketone or monoketone structures involving PVA oxidases (secondary alcohol oxidases (SAO) or PVA dehydrogenase (PVADH)) and hydrolytic cleavage of a C–C bond at diketone structures that involved especially oxidized PVA hydrolase (OPH or BDH).
 | ||
| Fig. 2 Proposed metabolic pathway for PVA by SAO and BDH, based on extracellular depolymerization.7 | ||
 | ||
| Fig. 3 Proposed metabolic pathway for PVA by PVA-DH and BDH (OPH) in the periplasm.7 | ||
Similar simplified proposed model for PVA degradation were illustrated by several other authors,84,86 which consists of the two parallel biodegradation pathway by oxidation and hydrolysis (Fig. 4 and 5).
 | ||
| Fig. 4 PVA degradation. Two parallel pathways of PVA oxidation are catalyzed by SAO and PDH, and the resulting OPA (oxi-PVA) can be cleaved by aldolase or OPH.86 | ||
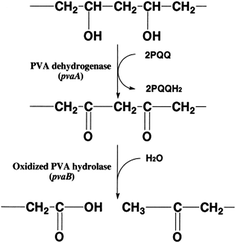 | ||
| Fig. 5 Proposed mechanism for cleavage of the main chain of PVA molecules by the pvaA and B products.84 | ||
Shimao et al.84 have also proposed a mechanism for hydrolyzing the main chain of PVA by the gene pvaA and pvaB products (PVA-DH and OPH, respectively) in Pseudomonas sp. strain VM15C (Fig. 5). They reported that the pvaB (that encodes oxidized PVA hydrolase) forms an operon with the PVA dehydrogenase gene pvaA.
Several PVA-oxidizing enzymes (SAO or PVA-DH) have already been purified and characterized. To this end, Sakai et al.87 and Kawagoshi and Fujita88 have focused on the purification and properties of oxidases with an acidic isoelectric point, and with broad substrate range from Pseudomonas vesicularis var. povalolyticus PH, respectively. Whereas, Shimao et al.89 have studied the existence of a novel enzyme, pyroloquinoline quinonedependent polyvinyl alcohol dehydrogenase, in a bacterial symbiont, Pseudomonas sp. strain VM15C.
Recently, Jia et al.90 have investigated in PVA-dehydrogenase (PVADH, EC 1.1.99.23) and reported high efficient preparation and characterization of bioactive and full-length mature PVADH from Sphingopyxis sp.113P3 in Escherichia coli by inclusion bodies renaturation. Thus, their approach would provide a useful platform to further characterization of intact PVA-oxidases.
The OPH is encoded by the bdh and oph gene in Pseudomonas and in Sphingopyxis, respectively; and the corresponding enzymes (pOPH and sOPH) which belong to the α/β-hydrolase superfamily, share 65% amino-acid sequence identity. Yang et al.86 have reported novel α/β-hydrolase structures of OPH from the two known PVA-degrading microorganisms, Pseudomonas sp. VM15C and Sphingopyxis sp. 113P3 (Fig. 6).
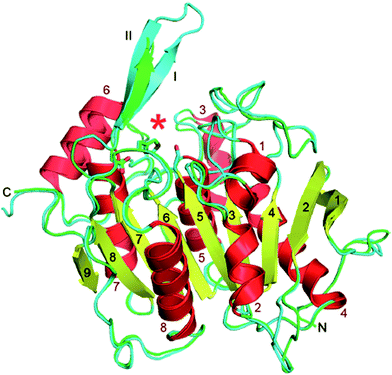 | ||
| Fig. 6 OPH structure. The ribbons drawings of pOPH and sOPH are colored red for α-helices and yellow for β-strands, except for the two strands (I and II) that forms the lid (colored as for the loops: cyan for pOPH and green for sOPH). The active site (red asterisk), is adjacent to the lid. The side chains of the catalytic triad are shown as stick models.86 | ||
The complexes of pOPH with butyric acid, PEG and DMSO were illustrated by Yang et al.91 who highlighted the roles of tryptophan residue and disulfide bond in the variable lid region of OPH (Fig. 7 and 8).
 | ||
| Fig. 7 The complexes of pOPH with butyric acid, PEG and DMSO. (A) In the S172A/pNPB crystal, only butyric acid was observed. The enzyme is shown as a ribbon diagram with red α-helices and yellow β-strands except for the lid, which is colored cyan as the loops. The ligand is bound with two different orientations, shown as green sticks. The catalytic triad is depicted as cyan sticks. (B) The butyric acid is hydrogen bonded to the protein side chains in either orientation. Here the amino acid residues are also shown as stick models. (C) The bound PEG in the WT crystal (cyan) and DMSO in the S172C mutant (pink) occupy the same place as the substrate. The hydrogen bonds in (B) and (C) are shown as dashed lines (for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).91 | ||
 | ||
| Fig. 8 Structural difference of the lid region. (A) On the left side the two lid residues Cys257/267 formed a disulfide bond in the pOPH crystal (green). On the right the equivalent Cys241/248 in sOPH (cyan) did not show a similar bond. (B) By using pNPA as the substrate, little effect of DTT on the sOPH activity was observed. The double mutant C241A/C248A of sOPH remained active. In contrast, DTT reduced the activity of pOPH (red bar at the right). (C) The surface of pOPH is viewed in two opposite directions. (D) That of sOPH is shown in two equivalent orientations as in (C). The surfaces are colored according to electrostatic potential, with blue and red representing positive and negative charges. The lid region is shown by using red circle (for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).91 | ||
3. Conclusion
PVA constitute the promising class of synthetic polymer used in a variety of industries including biomedical, biomaterial, textile, adhesive, pharmaceutical and food industries and may cause environmental menaces e.g. it can represent major component in wastewater from these industrial applications. Fortunately, PVA is biodegradable by different organisms under a two-step metabolism consisting of an oxidation and hydrolysis.PVA-degrading microorganisms would include fungi, as well as Gram-negative and Gram-positive bacteria. Both aerobic and anaerobic process can be involved in PVA-biodegradation but the well-established one was occurred in aerobic conditions. Certainly, PVA-degradation would be affected by chemical structural characteristics, such as stereotacticity and 1,2-diol units. For example, degradation scheme of PVA by Flammulina velutipes (Fig. 9) is initiated by oxidizing hydroxyl groups mainly in the syndiotactic portion and then depolymerization of the hydroxyl groups is been taken into account.64
 | ||
| Fig. 9 Possible degradation scheme of PVA by Flammulina velutipes.64 | ||
Symbiotic behaviors in mixed culture of microorganisms are efficient to remove significant amount of PVA, and the principal enzymes involved in its biodegradation are PVA-oxidases (SAO and PVADH) as well as PVA-hydrolases (OPH). However, with regards to limited studies on PVA degradation, further works are needed to elucidate the metabolism of PVA.
References
- H. Marusincová, L. Husárová, J. Ruzicka, M. Ingr, V. Navrátil, L. Bunková and M. Koutny, Int. Biodeter. Biodegr., 2013, 84, 21–28 CrossRef.
- M. Li, D. Zhang, G. Du and J. Chen, J. Microbiol. Biotechnol., 2012, 22, 220–225 CrossRef CAS PubMed.
- W. Haehnel and W. O. Herrmann, German Patent, 450, 286, 1924.
- FAO, Polyvinyl alcohol (PVA) Chemical and Technical Assessment (CTA) First draft prepared by S.K., Saxena. 61st JECFA, 2004.
- C. C. DeMerlis and D. R. Schoneker, Food Chem. Toxicol., 2003, 41, 319–326 CrossRef CAS PubMed.
- M. Shimao, Curr. Opin. Biotechnol., 2001, 12, 242–247 CrossRef CAS PubMed.
- F. Kawai and X. Hu, Appl. Microbiol. Biotechnol., 2009, 84, 227–237 CrossRef CAS PubMed.
- M. Chaouat, C. L. Visage, W. E. Baille, B. Escoubett, F. Chaubet, M. M. Alexandru and D. Letourneur, Adv. Funct. Mater., 2008, 18, 2855–2861 CrossRef CAS.
- G. Paradossi, F. Cavalieri, E. Chiessi, C. Spagnoli and M. K. Cowman, J. Mater. Sci.: Mater. Med., 2003, 14, 687–691 CrossRef CAS PubMed.
- S. G. Pathan, L. M. Fitzgerald, S. M. Ali, S. M. Damrauer, M. J. Bide, D. W. Nelson, C. Ferran, T. M. Phaneuf and M. D. Phaneuf, J. Biomed. Mater. Res., Part B, 2015, 103, 1652–1662 CrossRef CAS PubMed.
- M. H. El-Naas, A. H. I. Mourad and R. Surkatti, Int. Biodeter. Biodegr., 2013, 85, 413–420 CrossRef CAS.
- R. Surkatti and M. H. El-Naas, Journal of Water Process Engineering, 2014, 1, 84–90 CrossRef.
- R. K. Balasubramanian, Colloids Surf., A, 2014, 455, 174–178 CrossRef.
- N. A. Peppast and S. R. Schauna, Polymer, 1992, 18, 3932–3936 Search PubMed.
- M. Kobayashi, Y. S. Chang and M. Oka, Bio-Med. Mater. Eng., 2004, 14, 505–515 Search PubMed.
- M. Wu, B. Bo, F. Yoshii and K. Makuuchi, J. Radioanal. Nucl. Chem., 2001, 250, 391–395 CrossRef CAS.
- A. F. Spivak, Y. A. Dzenis and D. H. Reneker, Mech. Res. Commun., 2000, 27, 37–42 CrossRef.
- J. H. Finley, Anal. Chem., 1961, 33, 1925–1927 CrossRef CAS.
- C. A. Finch, Polyvinyl alcohol developments. John Wiley and Sons Ltd., New York, N.Y, 1992 Search PubMed.
- C. A. Finch, Polyvinyl Alcohol—Properties and Applications, Wiley, London, 1973 Search PubMed.
- G. B. Mckenna and F. Horkay, Polymer, 1994, 35, 5737–5742 CrossRef CAS.
- B. Cerroni, E. Chiessi, S. Margheritelli, L. Oddo and G. Paradossi, Biomacromolecules, 2011, 12, 593–601 CrossRef CAS PubMed.
- K. A. Batista, F. M. Lopes, F. Yamashita and K. F. Fernandes, Mater. Sci. Eng., C, 2013, 33, 1696–1701 CrossRef CAS PubMed.
- M. Cruz, K. Fernandes, C. Cysneiros, R. Nassar and S. Caramori, BioMed Res. Int., 2015, 2015, 145903 Search PubMed.
- S. Rawat, P. Gupta, A. Kumar, P. Garg, C. R. Suri and D. K. Sahoo, Mol. Pharmaceutics, 2015, 12, 1018–1030 CrossRef CAS PubMed.
- HIS, Chemical Economics Handbook, (Polyvinyl Alcohol), IHS, Englewood, USA, 2007 Search PubMed.
- J. A. Giroto, R. Guardani, A. C. S. C. Teixeira and C. A. O. Nascimento, Chem. Eng. Process., 2006, 45, 523–532 CrossRef CAS.
- Y. Tokiwa, G. Kawabata and A. Jarerat, Biotechnol. Lett., 2001, 23, 1937–1941 CrossRef CAS.
- J. L. Xu, T. L. Huang and Z. Y. Wang, Biodegradation of PVA in an anaerobic baffled reactor. Future of Urban Wastewater Systems Decentralisation and Reuse, China Architecture & Building Press, Beijing, 2005, pp. 429–436 Search PubMed.
- K. K. Choi, C. W. Park, S. Y. Kim, W. S. Lyoo, S. H. Lee and J. W. Lee, J. Microbiol. Biotechnol., 2004, 14, 1009–1013 CAS.
- S. Tsujiyama and A. Okada, Biotechnol. Lett., 2013, 35, 1907–1911 CrossRef CAS PubMed.
- F. Kawai, S. Kitajima, K. Oda, T. Higasa, J. Charoenpanich, X. Hu and R. Mamoto, Arch. Microbiol., 2013, 195, 131–140 CrossRef CAS PubMed.
- Y. X. Chen, Z. S. Sun, Y. Yang and Q. Ke, J. Photochem. Photobiol., A, 2001, 142, 85–89 CrossRef CAS.
- A. Gronroos, P. Pirkonen, J. Heikkinen, J. Ihalainen, H. Mursunen and H. Sekki, Ultrason. Sonochem., 2001, 8, 259–264 CrossRef CAS PubMed.
- S. J. Zhang and H. Q. Yu, Water Res., 2004, 38, 309–316 CrossRef CAS PubMed.
- S. G. de Bussetti and E. A. Ferreiro, Clays Clay Miner., 2004, 52, 334–340 CrossRef CAS.
- S. K. Behera, J. H. Kim, X. J. Guo and H. S. Park, J. Hazard. Mater., 2008, 153, 1207–1214 CrossRef CAS PubMed.
- S. Altin, Int. J. Chem. React. Eng., 2011, 9, A33 Search PubMed.
- Y. T. Xiao, S. S. Xu and Z. H. Li, J. Cent. South Univ. Technol., 2011, 18, 96–100 CrossRef CAS.
- D. M. Larking, R. J. Crawford, G. B. Y. Christie and G. T. Lonergan, Appl. Environ. Microbiol., 1999, 65, 1798–1800 CAS.
- A. Yamatsu, R. Matsumi, H. Atomi and T. Imanaka, Appl. Microbiol. Biotechnol., 2006, 72, 804–811 CrossRef CAS PubMed.
- T. Vaclavkova, J. Ruzicka, M. Julinova, R. Vicha and M. Koutny, Appl. Microbiol. Biotechnol., 2007, 76, 911–917 CrossRef CAS PubMed.
- Y. Watanabe, M. Morita, M. Hamada and Y. Tsujisaka, Arch. Biochem. Biophys., 1976, 174, 573–579 CrossRef CAS.
- M. Shimao and N. Kato, Mixed continuous cultures of polyvinyl alcohol-utilizing symbionts Pseudomonas putida VM15A and Pseudomonas sp. strain VM15C, inInternational Symposium on Biodegradable Polymers. Program and abstracts. Biodegradable Plastics Society, Tokyo, Japan, 1990, p. 56 Search PubMed.
- T. Suzuki and A. Tsuchii, Process Biochem., 1983, 18, 13–16 CAS.
- Y. Kawagoshi and M. Fujita, World J. Microbiol. Biotechnol., 1998, 14, 95–100 CrossRef CAS.
- T. Suzuki, Y. Ichihara and M. Yamada, Agric. Biol. Chem., 1973, 37, 747–756 CrossRef CAS.
- K. Sakai, N. Hamada and Y. Watanabe, Agric. Biol. Chem., 1986, 50, 989–996 Search PubMed.
- B. C. Kim, C. K. Shon, S. K. Lim, J. W. Lee and W. Park, J. Ind. Microbiol. Biotechnol., 2003, 30, 70–74 CrossRef CAS PubMed.
- R. Fukae, T. Fujii, M. Takeo and T. Yamamoto, J. Polym., 1994, 26, 1381–1386 CAS.
- J. A. Lee and M. N. Kim, Polym. Degrad. Stab., 2003, 81, 303–308 CrossRef CAS.
- S. Hashimoto and M. Fujita, J. Ferment. Technol., 1985, 63, 471–474 CAS.
- F. F. Nord, Naturwissenschaften, 1936, 24, 763 CrossRef CAS.
- M. Shimao, T. Tamogami, K. Nishi and S. Harayama, Biosci., Biotechnol., Biochem., 1996, 60, 1056–1062 CrossRef CAS PubMed.
- F. Kawai, J. Ind. Microbiol. Biotechnol., 1999, 23, 400–407 CrossRef CAS PubMed.
- Y. Ohtsubo, Y. Nagata, M. Numata, K. Tsuchikane, A. Hosoyama, A. Yamazoe, M. Tsuda, N. Fujita and F. Kawai, Genome Announcements, 2015, 3(5), e01169, DOI:10.1128/genomeA.01169-15.
- Y. Nogi, M. Yoshizumi, K. Hamana, M. Miyazaki and K. Horikoshi, Int. J. Syst. Evol. Microbiol., 2014a, 64, 2712–2717 Search PubMed.
- Y. Nogi, M. Yoshizumi and M. Miyazaki, Int. J. Syst. Evol. Microbiol., 2014b, 64, 1149–1153 Search PubMed.
- L. Jecu, A. Gheorghe, A. Rosu, I. Raut, E. Grosu and M. Ghiurea, J. Polym. Environ., 2010, 18, 284–290 CrossRef CAS.
- A. Stoica-Guzun, L. Jecu, A. G. I. Raut, M. Stroescu, M. G. M. Danila, I. Jipa and V. Fruth, J. Polym. Environ., 2011, 19, 69–79 CrossRef CAS.
- D. Qian, G. Du and J. Chen, World J. Microbiol. Biotechnol, 2004, 20, 587–591 CrossRef CAS.
- M. Kimura, K. Toyota, M. Iwatsuki and H. Sawada, Effects of soil conditions on the biodegradation of plastics and responsible microorganisms, in Biodegradable plastics and polymers, ed. Y. Doi and K. Fukuda, Elsevier, Amsterdam, 1994, pp. 92–106 Search PubMed.
- E. Chiellini, A. Corti and R. Solaro, Polym. Degrad. Stab., 1999, 64, 305–312 CrossRef CAS.
- S. Tsujiyama, T. Nitta and T. Maoka, J. Biosci. Bioeng., 2011, 112, 58–62 CrossRef CAS PubMed.
- Y. Tokiwa, H. Fan, Y. Hiraguri, R. Kurane, M. Kitagawa, S. Shibatani and Y. Maekawa, Macromolecules, 2000, 33, 1636–1639 CrossRef CAS.
- J. Hrncirík, J. Pseja, J. Kupec and S. Bernkopfová, J. Polym. Environ., 2010, 18, 98–103 CrossRef.
- H. Yu, G. Gu and L. Song, Environ. Technol., 1996, 17, 1261–1267 CrossRef CAS.
- S. Matsumura, H. Kurita and H. Shimokobe, Biotechnol. Lett., 1993, 15, 749–754 CrossRef CAS.
- S. Gartiser, M. Wallrabenstein and G. Stiene, J. Environ. Polym. Degrad., 1998, 6, 159–173 CrossRef CAS.
- J. Chung, S. Kim, K. Choi and J. O. Kim, Environ. Technol., 2015, 17, 1–7 Search PubMed.
- C. W. Lee, C. K. Kim, Y. J. Choi, Y. T. Rim, J. K. Ryu and Y. S. Suh, Journal of Korean Society on Water Quality, 1994, 10, 112–119 Search PubMed.
- A. Corti, R. Solaro and E. Chiellini, Polym. Degrad. Stab., 2002, 75, 447–458 CrossRef CAS.
- J. Chen, Y. Zhang, G. C. Du, Z. Z. Hua and Y. Zhu, Enzyme Microb. Technol., 2007, 40, 1686–1691 CrossRef CAS.
- M. Shimao, H. Yamamoto, K. Ninomiya, N. Kato, O. Adachi, M. Ameyama and C. Sakazawa, Agric. Biol. Chem., 1984, 48, 2873–2876 CAS.
- C. Sakazawa, M. Shimao, Y. Taniguchi and N. Kato, Appl. Environ. Microbiol., 1981, 4, 1261–1267 Search PubMed.
- T. Mori, M. Sakimoto, T. Kagi and T. Sakai, Biosci., Biotechnol., Biochem., 1996, 60, 330–332 CrossRef CAS.
- B. Tang, X. Liao, D. Zhang, M. Li, R. Li, K. Yan and G. Du, Polym. Degrad. Stab., 2010, 95, 557–563 CrossRef CAS.
- S. Matsumura, N. Tomizawa and A. Toki, Macromolecules, 1999, 32, 7753–7761 CrossRef CAS.
- Y. Zhang, Y. Li and W. Shen, World J. Microbiol. Biotechnol, 2006, 22, 625–628 CrossRef CAS.
- K. Sakai, M. Fukuba, Y. Hasui, K. Moriyoshi, T. Ohmoto, T. Fujita and T. Ohe, Biosci., Biotechnol., Biochem., 1998, 62, 2000–2007 CrossRef CAS PubMed.
- W. G. Zumft, Microbiol. Mol. Biol. Rev., 1997, 61, 533–616 CAS.
- J. Reimann, U. Flock, H. Lepp, A. Honigmann and P. Adelroth, Biochim. Biophys. Acta, 2007, 1767, 362–373 CrossRef CAS PubMed.
- R. Hirota-Mamoto, R. Nagai, S. Tachibana, M. Yasuda, A. Tani, K. Kimbara and F. Kakai, Microbiology, 2006, 152, 1941–1949 CrossRef CAS PubMed.
- M. Shimao, T. Tamogami and S. Kishida, Microbiology, 2000, 146, 649–657 CrossRef CAS PubMed.
- R. Solaro, A. Corti and E. Chiellini, Polym. Adv. Technol., 2000, 11, 873–878 CrossRef CAS.
- Y. Yang, T. P. Ko, L. Liu, J. Li, C. H. Huang, H. C. Chan, F. Ren, D. Jia, A. H. J. Wang, R. T. Guo, J. Chen and G. Du, ChemBioChem, 2014a, 15, 1882–1886 Search PubMed.
- K. Sakai, N. Hamada and Y. Watanabe, Agric. Biol. Chem., 1984, 49, 817–825 Search PubMed.
- Y. Kawagoshi and M. Fujita, World J. Microbiol. Biotechnol., 1997, 13, 273–277 CrossRef CAS.
- M. Shimao, K. Ninomiya, O. Kuno, N. Kato and C. Sakazawa, Appl. Environ. Microbiol., 1986, 51, 268–275 CAS.
- D. Jia, Y. Yang, Z. Peng, D. Zhang, J. Li, L. Liu, G. Du and J. Chen, Appl. Biochem. Biotechnol., 2014, 172, 2540–2551 CrossRef CAS PubMed.
- Y. Yang, T. P. Ko, L. Liu, J. Li, C. H. Huang, J. Chen, R. T. Guo and G. Du, Biochem. Biophys. Res. Commun., 2014b, 452, 509–514 Search PubMed.
- USP/NF, United States Pharmacopoeia (24) and National Formulary (19), U.S. Pharmacopeial Convention, Rockville, MD, 2000, pp. 1352–1353 Search PubMed.
- Handbook of Pharmaceutical Excipients, ed. A. Wade and P. J. Weller, American Pharmaceutical Association, Washington, DC, 2nd edn, 1994, pp. 383–384 Search PubMed.
- The Japanese Pharmaceutical Excipients Directory, Monograph on Polyvinyl Alcohol, 1996, p. 355 Search PubMed.
| This journal is © The Royal Society of Chemistry 2016 |


