Penicillanthone and penicillidic acids A–C from the soil-derived fungus Penicillium aculeatum PSU-RSPG105†
Charuwan Daengrota,
Vatcharin Rukachaisirikul*a,
Kwanruthai Tadpetcha,
Souwalak Phongpaichitb,
Kawitsara Bowornwiriyapanb,
Jariya Sakayarojc and
Xu Shend
aDepartment of Chemistry and Center of Excellence for Innovation in Chemistry, Faculty of Science, Prince of Songkla University, Hat Yai, Songkhla 90112, Thailand. E-mail: vatcharin.r@psu.ac.th
bNatural Products Research Center of Excellence and Department of Microbiology, Faculty of Science, Prince of Songkla University, Hat Yai, Songkhla 90112, Thailand
cNational Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand Science Park, Khlong Luang, Pathum Thani 12120, Thailand
dShanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China
First published on 14th April 2016
Abstract
A new xanthone (penicillanthone, 1) and three new diphenyl ether derivatives (penicillidic acids A–C, 2–4) together with 14 known compounds (5–18) were isolated from the soil-derived fungus Penicillium aculeatum PSU-RSPG105. The structures were elucidated by spectroscopic analyses. The absolute configuration of compound 1 was determined using the Snatzke's method whereas those of compounds 2–4 was established by comparison of their optical rotations with those of structurally related compounds. Compound 5 exhibited mild antimycobacterial activity against Mycobacterium tuberculosis with an MIC value of 25 μg mL−1 and was noncytotoxic to noncancerous Vero cells. In addition, altenusin (12) displayed moderate antibacterial activity against methicillin-resistant Staphylococcus aureus with an MIC value of 32 μg mL−1 and mild activity towards Vero cells with an IC50 value of 19.46 μM.
Introduction
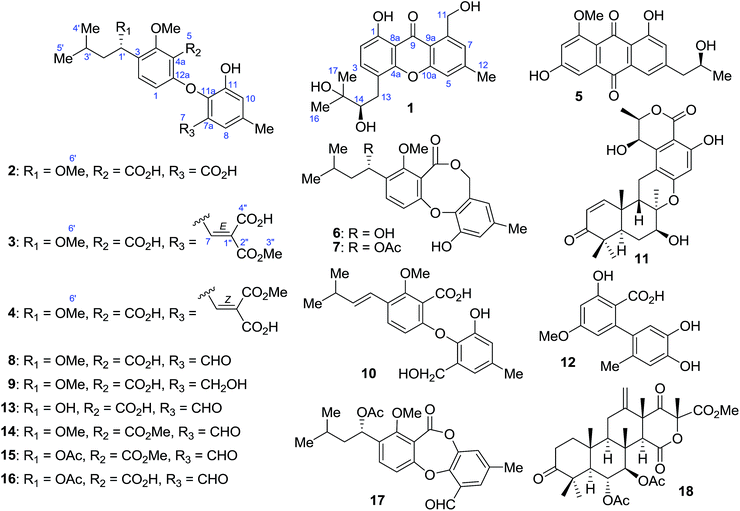
Fungi in the genus Penicillium are known as potential sources for the production of bioactive compounds.1,2 Our recent investigation of soil-derived fungi in the genus Penicillium resulted in the isolation of antioxidant herqueinone from P. herquei PSU-RSPG93,3 cytotoxic (7R,8R)-α-diversonolic ester and decarboxydihydrocitrinin from P. citrinum PSU-RSPG95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and antimycobacterial and cytotoxic GKK1032B from Penicillium sp. PSU-RSPG99.5 Accordingly, we continued to search for bioactive metabolites from the soil-derived fungus P. aculeatum PSU-RSPG105 collected from the Plant Genetic Conservation Project under the Royal Initiation of Her Royal Highness Princess Maha Chakri Sirindhorn at Ratchaprapa Dam in Suratthani Province, Thailand. The broth ethyl acetate extract of P. aculeatum PSU-RSPG105 exhibited antibacterial activity against Staphylococcus aureus ATCC25923 with an MIC value of 200 μg mL−1. The mycelial ethyl acetate extract displayed antibacterial and antifungal activities against S. aureus and Cryptococcus neoformans ATCC90113 flucytosine-resistant with MIC values of 128 and 200 μg mL−1, respectively, as well as cytotoxic activity against oral cavity cancer (KB) cell lines with an IC50 value of 27.56 μg mL−1. In addition, the mycelial hexane extract showed antifungal and antimycobacterial activities against C. neoformans and Mycobacterium tuberculosis, H37Ra strain, with the MIC values of 200 and 50 μg mL−1, respectively, together with antimalarial (Plasmodium falciparum, K1 strain), and cytotoxic [KB and human breast cancer (MCF-7) cell lines] activities with respective IC50 values of 3.75, 21.14 and 35.74 μg mL−1. All of the crude extracts exhibited cytotoxic activity against African green monkey kidney fibroblast (Vero) cell lines with IC50 values in the range of 17.74–28.51 μg mL−1. Chemical investigation of the broth extract led to the isolation of nine secondary metabolites including one new xanthone, penicillanthone (1), along with penipurdin A (5),6 1′S-penicillide (6),7,8 1′S-purpactin A (7),7 tenellic acid A (8),9 1′S-6-[2-hydroxy-6-(hydroxymethyl)-4-methylphenoxy]-2-methoxy-3-(1-methoxy-3-methylbutyl)benzoic acid (9),10 penikellide A (10),11 5R,7S,8R,9S,10S,7′R,8′R-chrodrimanin A (11)12,13 and altenusin (12).14 In addition, three new diphenyl ether derivatives, penicillidic acids A–C (2–4), and two known diphenyl ether derivatives, tenellic acid B (13)9 and tenellic acid A methyl ester (14),9 were obtained from the mycelial ethyl acetate extract while four known compounds, 1′S-secopenicilide B (15),7 tenellic acid C (16),9 purpactin C′ (17)15 and 5R,6R,7R,8R,9R,10R,13R,14S,16R-penisimplicin A (18),16 were isolated from the corresponding hexane extract. Tenellic acid A methyl ester (14) was isolated as a natural product for the first time. Moreover, this investigation established the absolute configuration at C-1′ of compounds 8, 13, 14, 16 and 17. Some isolated compounds were evaluated for antimicrobial (S. aureus, methicillin-resistant S. aureus, and C. neoformans), antimycobacterial (M. tuberculosis, H37Ra strain), antimalarial (P. falciparum, K1 strain), and cytotoxic (KB, MCF-7 and Vero cell lines) activities.
4 and antimycobacterial and cytotoxic GKK1032B from Penicillium sp. PSU-RSPG99.5 Accordingly, we continued to search for bioactive metabolites from the soil-derived fungus P. aculeatum PSU-RSPG105 collected from the Plant Genetic Conservation Project under the Royal Initiation of Her Royal Highness Princess Maha Chakri Sirindhorn at Ratchaprapa Dam in Suratthani Province, Thailand. The broth ethyl acetate extract of P. aculeatum PSU-RSPG105 exhibited antibacterial activity against Staphylococcus aureus ATCC25923 with an MIC value of 200 μg mL−1. The mycelial ethyl acetate extract displayed antibacterial and antifungal activities against S. aureus and Cryptococcus neoformans ATCC90113 flucytosine-resistant with MIC values of 128 and 200 μg mL−1, respectively, as well as cytotoxic activity against oral cavity cancer (KB) cell lines with an IC50 value of 27.56 μg mL−1. In addition, the mycelial hexane extract showed antifungal and antimycobacterial activities against C. neoformans and Mycobacterium tuberculosis, H37Ra strain, with the MIC values of 200 and 50 μg mL−1, respectively, together with antimalarial (Plasmodium falciparum, K1 strain), and cytotoxic [KB and human breast cancer (MCF-7) cell lines] activities with respective IC50 values of 3.75, 21.14 and 35.74 μg mL−1. All of the crude extracts exhibited cytotoxic activity against African green monkey kidney fibroblast (Vero) cell lines with IC50 values in the range of 17.74–28.51 μg mL−1. Chemical investigation of the broth extract led to the isolation of nine secondary metabolites including one new xanthone, penicillanthone (1), along with penipurdin A (5),6 1′S-penicillide (6),7,8 1′S-purpactin A (7),7 tenellic acid A (8),9 1′S-6-[2-hydroxy-6-(hydroxymethyl)-4-methylphenoxy]-2-methoxy-3-(1-methoxy-3-methylbutyl)benzoic acid (9),10 penikellide A (10),11 5R,7S,8R,9S,10S,7′R,8′R-chrodrimanin A (11)12,13 and altenusin (12).14 In addition, three new diphenyl ether derivatives, penicillidic acids A–C (2–4), and two known diphenyl ether derivatives, tenellic acid B (13)9 and tenellic acid A methyl ester (14),9 were obtained from the mycelial ethyl acetate extract while four known compounds, 1′S-secopenicilide B (15),7 tenellic acid C (16),9 purpactin C′ (17)15 and 5R,6R,7R,8R,9R,10R,13R,14S,16R-penisimplicin A (18),16 were isolated from the corresponding hexane extract. Tenellic acid A methyl ester (14) was isolated as a natural product for the first time. Moreover, this investigation established the absolute configuration at C-1′ of compounds 8, 13, 14, 16 and 17. Some isolated compounds were evaluated for antimicrobial (S. aureus, methicillin-resistant S. aureus, and C. neoformans), antimycobacterial (M. tuberculosis, H37Ra strain), antimalarial (P. falciparum, K1 strain), and cytotoxic (KB, MCF-7 and Vero cell lines) activities.
Results and discussion
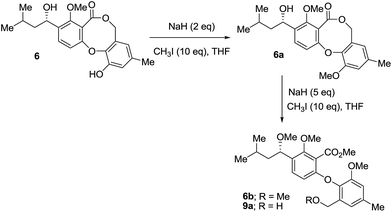
Compounds (1–18) (Fig. 1) were obtained using various chromatographic techniques. Their structures were determined by spectroscopic analyses. The relative configuration was assigned according to the NOEDIFF data. The absolute configuration of the isolated compounds was established by comparison of their optical rotations with those of known or structurally related compounds (Table S1†). For compound 1, the absolute configuration was assigned by Snatzke's method.17 The absolute configuration at C-1′ in 6 was established to be S, identical to previous assignment,7,8 by Mosher's method18 (Fig. 2).Methylation of 6 with sodium hydride and methyl iodide yielded known penicillide monomethyl ether (6a)8 as a major product (Fig. 3). Surprisingly, when 6a was further reacted with an excess of sodium hydride, compounds 6b and 9a with lactone ring opening were obtained (Fig. 3).
Methylation of (1′S)-9 afforded 9a, indicating S configuration at C-1′ of 9a. Based on the S configuration at C-1′ of the starting precursor 6 and the product 9a, 6b which was the methylated product of 9a would have identical configuration. The negative optical rotation of 8, [α]24D −4.8 (c 0.18, CH3OH), the same sign as those of 6b, [α]24D −10.0 (c 0.18, CH3OH), and 9a, [α]24D −9.3 (c 0.18, CH3OH), established S configuration at C-1′ for 8. Furthermore, comparison of optical rotations of 2–4 and 14 and 13 with those of 8 and hydroxytenellic acid B,10 respectively (Table S1†), indicated that all of them had identical absolute configuration. For compounds 16 and 17, the absolute configuration at C-1′ was determined to be the same as that of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 and 15 by comparison of their optical rotations (Table S1†) and the circular dichroism (Table S2†).
19 and 15 by comparison of their optical rotations (Table S1†) and the circular dichroism (Table S2†).
Penicillanthone (1) was obtained as a yellow solid and had the molecular formula C20H22O6 determined by HRESIMS peak at m/z 359.1495 [M + H]+. It showed UV absorption bands of a xanthone chromophore at 233, 259, 304 and 369 nm20 while IR absorption bands were found at 3116 cm−1 for a hydroxy group and 1647 cm−1 for a xanthone carbonyl group.20 The 1H NMR data (Table 1) consisted of signals for one hydrogen-bonded hydroxy proton (δH 12.70, s, 1H), three free hydroxy protons [δH 4.56 (t, J = 6.0 Hz, 1H), 3.74 (d, J = 5.5 Hz, 1H) and 3.54 (s, 1H)], two ortho-coupled aromatic protons (δH 7.63 and 6.70, each d, J = 8.0 Hz, 1H), two meta-coupled aromatic protons (δH 7.58 and 7.35, each brs, 1H), a 1-substituted-2,3-dihydroxy-3-methylbutyl group [δH 3.68 (ddd, J = 2.0, 5.5 and 10.0 Hz, 1H), 3.31 (dd, J = 2.0 and 14.0 Hz, 1H), 2.68 (dd, J = 10.0 and 14.0 Hz, 1H), 1.33 (s, 3H) and 1.30 (s, 3H)], one hydroxymethyl group (δH 5.21, d, J = 6.0 Hz, 2H) and one methyl group (δH 2.53, s, 3H). The 13C NMR spectrum (Table 1) displayed signals for a xanthone carbonyl (δC 185.5), nine quaternary, five methine, two methylene and three methyl carbons. A chelated hydroxy proton (δH 12.70) was placed at C-1 (δC 161.3), a peri position to the xanthone carbonyl group, and displayed HMBC correlations with C-1, C-2 (δC 110.5) and C-9a (δC 110.1) (Table 1). The ortho-coupled aromatic proton resonating at δH 6.70 was identified as H-2 due to its HMQC correlation to C-2. Therefore, the other ortho-coupled aromatic proton (δH 7.63) was assigned as H-3 and showed the HMBC correlations with C-1 and C-4a (δC 154.3). 1H–1H COSY correlations of Hab-13 (δH 3.31 and 2.68)/H-14 (δH 3.68) and H-14/14-OH (δH 3.74), and HMBC correlations of H3-16 (δH 1.30) and H3-17 (δH 1.33) with C-14 (δC 79.0) and C-15 (δC 73.1) as well as those of 15-OH (δH 3.54) with C-14, C-15, C-16 (δC 25.6) and C-17 (δC 26.2) established the 1-substituted-2,3-dihydroxy-3-methylbutyl unit. HMBC correlations from Hab-13 to C-3 (δC 139.8), C-4 (δC 119.4) and C-4a as well as that of H-3 with C-13 (δC 32.5) attached the 1-substituted-2,3-dihydroxy-3-methylbutyl unit at C-4. The hydroxymethyl group was placed at C-8 (δC 146.4) on the basis of the appearance of the hydroxymethyl protons (δH 5.21) at much lower field due to the anisotropic effect of the carbonyl group and HMBC correlations with C-7 (δC 124.8), C-8 and C-8a (δC 116.2). The meta-coupled aromatic proton resonating at δH 7.58 was then attributed to H-7 on the basis of its HMQC correlation with C-7. Thus, the other meta-coupled aromatic proton (δH 7.35) was attributed to H-5. The HMBC correlations from H3-12 (δH 2.53) with C-5 (δC 117.5), C-6 (δC 148.2) and C-7 indicated the attachment of a methyl group at C-6. The absolute configuration of C-14 was assigned by Snatzke's method.17 The negative Cotton effect observed at 303 nm (Δε −3.8) permitted the assignment of the R configuration at C-14. Thus, 1 was a dihydroxy derivative of paeciloxanthone.20
| Position | 1 | ||
|---|---|---|---|
| δC, type | δH, mult. (J in Hz) | HMBC | |
| a Recorded in acetone-d6 (300 and 75 MHz). | |||
| 1 | 161.3, C | ||
| 1-OH | 12.70, s | C-1, C-2, C-9a | |
| 2 | 110.5, CH | 6.70, d (8.0) | C-1, C-4, C-9a |
| 3 | 139.8, CH | 7.63, d (8.0) | C-1, C-4a, C-13 |
| 4 | 119.4, C | ||
| 4a | 154.3, C | ||
| 5 | 117.5, CH | 7.35, brs | C-7, C-8a, C-9, C-10a, C-12 |
| 6 | 148.2, C | ||
| 7 | 124.8, CH | 7.58, brs | C-5, C-8a, C-11, C-12 |
| 8 | 146.4, C | ||
| 8a | 116.2, C | ||
| 9 | 185.5, C | ||
| 9a | 110.1, C | ||
| 10a | 158.7, C | ||
| 11 | 64.2, CH2 | 5.21, d (6.0) | C-7, C-8, C-8a, C-9, C-10a |
| 11-OH | 4.56, t (6.0) | C-8, C-11 | |
| 12 | 22.2, CH3 | 2.53, s | C-5, C-6, C-7 |
| 13 | 32.5, CH2 | a: 3.31, dd (2.0, 14.0) | C-3, C-4, C-4a, C-14, C-15 |
| b: 2.68, dd (10.0, 14.0) | C-3, C-4, C-4a, C-14, C-15 | ||
| 14 | 79.0, CH | 3.68, ddd (2.0, 5.5, 10.0) | C-4, C-15, C-16, C-17 |
| 14-OH | 3.74, d (5.5) | C-13, C-14, C-15 | |
| 15 | 73.1, C | ||
| 15-OH | 3.54, s | C-14, C-15, C-16, C-17 | |
| 16 | 25.6, CH3 | 1.30, s | C-14, C-15, C-17 |
| 17 | 26.2, CH3 | 1.33, s | C-14, C-15, C-16 |
Penicillidic acid A (2) was obtained as a colorless gum. The molecular formula was C22H26O8 on the basis of the HRESIMS peak at m/z 441.1516 [M + Na]+. The UV spectrum showed absorption bands of benzene chromophore at 207 and 286 nm.10 The IR spectrum displayed absorption bands at 3376 cm−1 for a hydroxy group and 1736 and 1718 cm−1 for two carboxyl functional groups.7 The 1H NMR spectroscopic data (Table 2) contained signals for two ortho-coupled aromatic protons (δH 6.98 and 6.69, each d, J = 8.5 Hz, 1H), two meta-coupled aromatic protons (δH 6.76 and 6.57, each d, J = 2.0 Hz, 1H), one oxymethine proton (δH 4.54, dd, J = 4.5 and 9.0, 1H), one methine proton (δH 1.66, m, 1H), one set of nonequivalent methylene protons [δH 1.53 (ddd, J = 5.5, 9.0 and 14.0 Hz) and 1.28 (ddd, J = 4.5, 8.5 and 14.0 Hz), each 1H], two methoxy groups (δH 3.84 and 3.08, each s, 3H) and three methyl groups [δH 2.19 (s), 0.87 (d, J = 7.0 Hz) and 0.85 (d, J = 6.6 Hz), each 3H]. The 13C NMR spectrum (Table 2) displayed signals for two carbonyl (δC 174.0 and 173.0), eight quaternary, six methine, one methylene, two methoxy and three methyl carbons. Detailed comparison of the 1H and 13C NMR (Table 2) spectral data of 2 with those of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 revealed the similarity of their structures. The difference was the replacement of the aldehyde signal (δH 10.35, s, δC 189.6) in 8 with a carboxyl group (δC 174.0) in 2. The attachment of the carboxyl group at C-7a (δC 135.9) was confirmed using an HMBC correlation of H-8 (δH 6.76) with C-7 (δC 174.0) (Fig. 4). Compound 2 gave a similar optical rotation, [α]24D −4.1 (c 0.18, CH3OH), to that of the co-metabolite 8, [α]24D −4.8 (c 0.18, CH3OH).9 Thus, the absolute configuration at C-1′ in 2 was proposed to be S configuration.
9 revealed the similarity of their structures. The difference was the replacement of the aldehyde signal (δH 10.35, s, δC 189.6) in 8 with a carboxyl group (δC 174.0) in 2. The attachment of the carboxyl group at C-7a (δC 135.9) was confirmed using an HMBC correlation of H-8 (δH 6.76) with C-7 (δC 174.0) (Fig. 4). Compound 2 gave a similar optical rotation, [α]24D −4.1 (c 0.18, CH3OH), to that of the co-metabolite 8, [α]24D −4.8 (c 0.18, CH3OH).9 Thus, the absolute configuration at C-1′ in 2 was proposed to be S configuration.
| Position | 2 | 3 | 4 | |||
|---|---|---|---|---|---|---|
| δC, typea | δH, mult.b (J in Hz) | δC, typec | δH, mult.d (J in Hz) | δC, typea | δH, mult.b (J in Hz) | |
| a Recorded in CD3OD (125 MHz).b Recorded in CD3OD (500 MHz).c Recorded in CD3OD (75 MHz).d Recorded in CD3OD (300 MHz). | ||||||
| 1 | 112.2, CH | 6.69, d (8.5) | 113.4, CH | 6.54, d (8.4) | 112.4, CH | 6.54, d (8.5) |
| 2 | 125.3, CH | 6.98, d (8.5) | 127.2, CH | 7.10, d (8.4) | 125.6, CH | 7.06, d (8.5) |
| 3 | 129.4, C | 131.3, C | 129.9, C | |||
| 4 | 154.5, C | 156.0, C | 154.6, C | |||
| 4-OMe | 60.9, CH3 | 3.84, s | 62.2, CH3 | 3.93, s | 60.8, CH3 | 3.91, s |
| 4a | 125.3, C | 126.7, C | 125.4, C | |||
| 5 | 173.0, C | 174.4, C | 172.6, C | |||
| 7 | 174.0, C | 130.2, CH | 7.65, s | 132.5, CH | 7.71, s | |
| 7a | 135.9, C | 134.6, C | 135.3, C | |||
| 8 | 119.2, CH | 6.76, d (2.0) | 120.6, CH | 7.44, d (1.8) | 118.6, CH | 6.66, brs |
| 9 | 135.1, C | 136.8, C | 135.3, C | |||
| 9-Me | 19.7, CH3 | 2.19, s | 21.2, CH3 | 2.26, s | 19.8, CH3 | 2.23, s |
| 10 | 117.6, CH | 6.57, d (2.0) | 120.9, CH | 6.70, d (1.8) | 118.9, CH | 6.67, brs |
| 11 | 150.0, C | 151.3, C | 150.1, C | |||
| 11a | 138.4, C | 142.5, C | 140.8, C | |||
| 12a | 154.5, C | 155.7, C | 154.2, C | |||
| 1′ | 75.7, CH | 4.54, dd (4.5, 9.0) | 77.1, CH | 4.62, dd (4.5, 9.0) | 75.7, CH | 4.61, dd (4.5, 9.0) |
| 2′ | 46.7, CH2 | 1.53, ddd (5.5, 9.0, 14.0) | 48.1, CH2 | 1.61, ddd (5.4, 8.7, 13.8) | 46.7, CH2 | 1.60, ddd (5.5, 9.0, 14.0) |
| 1.28, ddd (4.5, 8.5, 14.0) | 1.36, ddd (4.2, 8.1, 13.8) | 1.35, ddd (4.5, 8.5, 14.0) | ||||
| 3′ | 24.7, CH | 1.66, m | 26.1, CH | 1.75, m | 24.7, CH | 1.73, m |
| 4′ | 22.4, CH3 | 0.85, d (6.6) | 23.8, CH3 | 0.92, d (6.9) | 22.3, CH3 | 0.91, d (6.5) |
| 5′ | 21.2, CH3 | 0.87, d (7.0) | 22.5, CH3 | 0.94, d (6.9) | 21.1, CH3 | 0.94, d (6.5) |
| 6′ | 55.5, CH3 | 3.08, s | 56.9, CH3 | 3.16, s | 55.4, CH3 | 3.15, s |
| 1′′ | 130.0, C | 129.8, C | ||||
| 2′′ | 168.4, C | 170.4, C | ||||
| 3′′ | 52.5, CH3 | 3.74, s | 51.6, CH3 | 3.77, s | ||
| 4′′ | 174.6, C | 169.6, C | ||||
Penicillidic acid B (3) with the molecular formula C26H30O10 deduced from HRESIMS peak at m/z 525.1736 [M + Na]+ was obtained as a colorless gum. The UV spectrum showed absorption bands of conjugated carbonyl and benzene chromophores at 206, 225 and 294 nm.9 The IR spectrum showed absorption bands at 3406 cm−1 for a hydroxy group, 1742 cm−1 for an ester carbonyl functional group and 1725 and 1710 cm−1 for two carboxyl functional groups.7 Comparison of the 1H and 13C (Table 2) NMR spectra of 3 with those of 2 indicated that the carboxyl signal (δC 174.0) in 2 was replaced by a 2-(methoxycarbonyl)acrylic acid unit (δH 7.65, s, 1H and 3.74, s, 3H, δC 174.6, 168.4, 130.2, 130.0 and 52.5) in 3. This assignment was confirmed by HMBC correlations from H-7 (δH 7.65) with C-7a (δC 134.6), C-8 (δC 120.6), C-11a (δC 142.5), C-1′′ (δC 130.0), C-2′′ (δC 168.4) and C-4′′ (δC 170.6) and from H3-3′′ (δH 3.74) to C-2′′ (Fig. 5). The configuration of a trisubstituted double bond was assigned as E on the basis of signal enhancement of H3-3′′ upon irradiation of H-7 in an NOEDIFF experiment (Fig. 5). Comparison of the optical rotation of 3, [α]24D −5.6 (c 0.18, CH3OH), with that of the co-metabolite 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 indicated that they possessed S configuration at C-1′ (δC 77.08).
9 indicated that they possessed S configuration at C-1′ (δC 77.08).
Penicillidic acid C (4) was obtained as a colorless gum. The molecular formula C26H30O10 determined by HRESIMS peak at m/z 525.1738 [M + Na]+ was identical to that of 3. Detail analysis of the UV, IR, 1H and 13C NMR spectra data (Table 2) as well as 1H–1H COSY and HMBC correlations of 4 revealed that its chemical structure was almost identical to that of 3. However, the configuration of the trisubstituted double bond at C-7 (δC 132.5) and C-1′′ (δC 129.8) was assigned as Z according to signal enhancement of H-1 (δH 6.54) and H-8 (δH 6.66), but not H3-3′′ (δH 3.77), after irradiation of H-7 (δH 7.71) in an NOEDIFF experiment. Finally, 4 was a Z isomer of 3. Compound 4 displayed the observed optical rotation, [α]24D −5.5 (c 0.18, CH3OH), similar to those of 3 and 8,9 indicating their identical absolute configuration at C-1′ (δC 75.68).
The biosynthetic pathway of depsidones, penicillide analogues, tenellic acid derivatives and xanthones has been proposed to proceed from a benzophenone precursor.15,21–24 Purpactin A (7) has been reported to be derived from the benzophenone A1 via the spirofuran-3-one B1 by oxidative cyclization and subsequent prenylation with mevalonic acid to obtain a prenyl side chain, reduction and benzylic oxidation of the side chain to form C2. Acetylation of C2 would yield purpactin B (C1) (Fig. 6). Hydrolysis of C1 and then lactone formation would afford 7.15 Similarly, 6 and 9 would be derived from B1 via C2 and C3, the methylated product of C2, respectively. Tenellic acid C (16) and purpactin C′ (17) would be synthesized from the spirofuran-3-one B2 via purpactin C (C4) using the similar biosynthetic pathway15,23 which would also give 8, 13 and 16 from C6, C5 and C4, respectively. Oxidation of the aldehyde to a carboxylic acid in 8 would afford 2 whereas 3 and 4 would be derived by condensation of the aldehyde with malonic acid dimethyl ester and subsequent partial hydrolysis. Esterification of 8 and 16 would result in the formation of 14 and 15, respectively, while cyclization of 16 would yield 17. The xanthone 1 would be obtained from A1 upon cyclization to produce a xanthone which would undergo prenylation with mevalonic acid, epoxidation of the prenyl side chain and subsequent opening of epoxide ring with water.
The isolated compounds 6–9, 12–13, 15–16 and 18 with sufficient amount were tested for antimicrobial activity against S. aureus ATCC25923, methicillin-resistant S. aureus and C. neoformans ATCC90113 flucytosine-resistant (Table 3). Compound 12 showed moderate antibacterial activity against methicillin-resistant S. aureus with an MIC value of 32 μg mL−1 and was four fold less active against S. aureus. Only compound 6 displayed antifungal activity against C. neoformans with an MIC value of 128 μg mL−1. Others were inactive against all tested microorganisms at the concentration of 200 μg mL−1. Additionally, compounds 3, 5, 6–9, 12–13, 15–16 and 18 were evaluated for antimycobacterial (M. tuberculosis, H37Ra strain), antimalarial (P. falciparum, K1 strain), and cytotoxic (Vero, KB and MCF-7 cells) activities (Table 3). Compounds 3, 13 and 18 showed no activities. For antimycobacterial activity, compound 5 displayed mild activity with an MIC value of 25 μg mL−1 while 6, 8–9 and 16 were twice less active. Compounds 7 and 15 were approximately three fold more active against P. falciparum than 6 with IC50 values of 5.69 and 5.11 μM, respectively. These data indicated that the acetoxy group at C-1′ and the methyl ester functional group at C-5 significantly increased antimalarial activity. For cytotoxic activity toward KB cell lines, 6–7, 9, 12 and 15 showed weak activity with IC50 values in the range of 24.97–154.24 μM. Only compounds 7, 9 and 15 displayed mild activity against MCF-7 cell lines with IC50 values ranging from 45.35–75.28 μM. However, 6–9, 12 and 15 exhibited weak cytotoxic activity to Vero cells. Previously, penicillide (6) was active against the human laryngeal carcinoma (Hep-2) and human rhabdomyosarcoma (RD) cells with IC50 values of 6.7 and 7.8 μM.25 Interestingly, purpactin A (7) has been previously reported to be moderately active against several cancer cell lines including MCF-7 and MCF-7/ADR cells with IC50 values of 11.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 26 and 16.4 μM,27 respectively, whereas tenellic acid A methyl ester (14), the ester derivative of 8, exhibited moderate activity against MCF-7/ADR cells with an IC50 value of 8.2 μM.27 In addition, compound 9 showed moderate antibacterial activity against Escherichia coli with an MIC value of 32 μg mL−1
26 and 16.4 μM,27 respectively, whereas tenellic acid A methyl ester (14), the ester derivative of 8, exhibited moderate activity against MCF-7/ADR cells with an IC50 value of 8.2 μM.27 In addition, compound 9 showed moderate antibacterial activity against Escherichia coli with an MIC value of 32 μg mL−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 Up to the present, there are a few reports on the biological activities of penicillide and tenellic acid analogues. Therefore, this investigation provided additional biological data for compounds of these types.
10 Up to the present, there are a few reports on the biological activities of penicillide and tenellic acid analogues. Therefore, this investigation provided additional biological data for compounds of these types.
| Compound | Antimicrobial (MIC, μg mL−1) | Antimycobacterial (MIC, μg mL−1) | Antimalarial (IC50, μM) | Cytotoxic (IC50, μM) | ||||
|---|---|---|---|---|---|---|---|---|
| Sa | MRSA | Cn | M. tuberculosis, H37Ra strain | P. falciparum | KB | MCF-7 | Vero | |
| a Sa = Staphylococcus aureus ATCC25923, MRSA = methicillin-resistant Staphylococcus aureus SK1, Cn = Cryptococcus neoformans ATCC90113 flucytosine-resistant, IN = inactive.b Vancomycin.c Amphotericin B.d Rifampicin.e Streptomycin.f Isoniazid.g Ofloxacin.h Ethambutol.i Dihydroartemisinine (nM).j Mefloquine.k Ellipticine.l Doxorubicin.m Tamoxifen. | ||||||||
| 5 | IN | IN | IN | 25.00 | IN | IN | IN | IN |
| 6 | IN | IN | 128 | 50.00 | 16.41 | 43.77 | IN | 53.73 |
| 7 | IN | IN | IN | IN | 5.69 | 52.50 | 75.28 | 32.57 |
| 8 | IN | IN | IN | 50.00 | IN | IN | IN | 45.99 |
| 9 | IN | IN | IN | 50.00 | IN | 24.97 | 45.35 | 25.02 |
| 12 | 128 | 32 | IN | IN | IN | 154.24 | IN | 19.46 |
| 15 | IN | IN | IN | IN | 5.11 | 32.71 | 56.65 | 25.96 |
| 16 | IN | IN | IN | 50.00 | IN | IN | IN | IN |
| Control | 0.2b | 0.5b | 1c | 0.0020d | 2.74i | 12.75k | 19.08l | 4.55k |
| 0.6250e | 0.0326j | 2.08l | 22.31m | |||||
| 0.0469f | ||||||||
| 0.3910g | ||||||||
| 0.9380h | ||||||||
Experimental
General experimental procedures
The melting points were determined on an Electrothermal 9100 melting point apparatus and reported without correction. Optical rotations were recorded on a JASCO P-1020 polarimeter. The ultraviolet (UV) absorption spectra were measured in CH3OH on a SHIMADZU UV-VIS spectrophotometer UV-2600 Series. The CD spectra were obtained on a Jasco J-815 spectrometer. The infrared (IR) spectra were recorded neat using a Perkin-Elmer 783 FTS165 FT-IR spectrometer. Mass spectra were obtained from a MAT 95 XL mass spectrometer (Thermo Finnigan), Bruker MicrOTOF mass spectrometer. 1H and 13C NMR spectra were recorded on a 300 or 500 MHz Bruker FTNMR Ultra Shield spectrometer. Chemical shifts are expressed in δ (parts per million, ppm) referring to the tetramethylsilane peak. Thin-layer chromatography (TLC) and preparative TLC (PTLC) were performed on silica gel 60 GF254 (Merck). Column chromatography (CC) was carried out on Sephadex LH-20 with CH3OH, silica gel (Merck) type 60 (230–400 mesh ASTM) or type 100 (70–230 mesh ASTM), or reversed phase C18 silica gel.Fungal material
The soil fungus PSU-RSPG105 (BCC56846, GenBank accession no. KC478547) was isolated from soil collected from Rajjaprabha Dam, Surat Thani Province, Thailand in 2010. Its colony grown on potato dextrose agar (PDA) was deep green with white edge and powdery. Microscopic morphology showed 3-stage branched conidiophores bearing dense brush-like penicillin with chains of smooth conidia. ITS rDNA sequence analysis revealed that PSU-RSPG105 was closely related to several strains of Penicillium aculeatum (JN542525, EU076930, EU781668, HQ392495, GU565131, HQ392496, HQ392497, JN899378 and AF033397) with 100% robust bootstrap values and 99.0–99.5% nucleotide identity. Therefore, PSU-RSPG105 was identified as Penicillium aculeatum.Extraction and isolation
The extracts obtained from the culture broth and mycelia of the soil fungus PSU-RSPG105 were prepared using the same procedure as described for P. sclerotiorum PSU-A13.28 The broth extract (2.65 g) was separated by CC over Sephadex LH-20 using CH3OH/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford four fractions (A–D). Fraction B (920.9 mg) was purified by CC over silica gel using a gradient of acetone/petroleum ether (1
1) to afford four fractions (A–D). Fraction B (920.9 mg) was purified by CC over silica gel using a gradient of acetone/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 to 1
5 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) to provide nine fractions (B1–B9). Fraction B2 (37.9 mg) was purified by CC over silica gel using EtOAc/petroleum ether (1
0) to provide nine fractions (B1–B9). Fraction B2 (37.9 mg) was purified by CC over silica gel using EtOAc/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) followed by CC over silica gel using EtOAc/hexane (3
3) followed by CC over silica gel using EtOAc/hexane (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7) to afford 7 (30.1 mg). Fraction B4 (212.2 mg) was purified by CC over silica gel using EtOAc/petroleum ether (1
7) to afford 7 (30.1 mg). Fraction B4 (212.2 mg) was purified by CC over silica gel using EtOAc/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give 6 (165.4 mg). Fraction B5 (28.1 mg) was further separated by CC over silica gel using EtOAc/petroleum ether (3
1) to give 6 (165.4 mg). Fraction B5 (28.1 mg) was further separated by CC over silica gel using EtOAc/petroleum ether (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7) followed by dissolving with CH3OH to give compound 11 (2.7 mg) from the insoluble fraction. Fraction B6 (25.2 mg) was subjected to CC over silica gel using a gradient of EtOAc/petroleum ether (1
7) followed by dissolving with CH3OH to give compound 11 (2.7 mg) from the insoluble fraction. Fraction B6 (25.2 mg) was subjected to CC over silica gel using a gradient of EtOAc/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 to 1
3 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) follow by PTLC using EtOAc/CH2Cl2 (1
0) follow by PTLC using EtOAc/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) as a mobile phase (3 runs) to provide three subfractions. The third subfraction (3.8 mg) was separated by dissolving with hexane to give compound 1 (1.5 mg) from the insoluble fraction. Fraction B8 (255.8 mg) was subjected to CC over silica gel using CH3OH/CH2Cl2 (1
5) as a mobile phase (3 runs) to provide three subfractions. The third subfraction (3.8 mg) was separated by dissolving with hexane to give compound 1 (1.5 mg) from the insoluble fraction. Fraction B8 (255.8 mg) was subjected to CC over silica gel using CH3OH/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19 to 1
19 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) to give four subfractions (B8A–B8D). Subfraction B8B (27.9 mg) was rechromatographed on CC over reversed phase C18 silica gel using CH3OH/H2O (3
0) to give four subfractions (B8A–B8D). Subfraction B8B (27.9 mg) was rechromatographed on CC over reversed phase C18 silica gel using CH3OH/H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) followed by PTLC using CH3OH/CH2Cl2 (3
2) followed by PTLC using CH3OH/CH2Cl2 (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17) as a mobile phase (3 runs) to provide 8 (6.8 mg). Subfraction B8C (84.5 mg) was purified by CC over reversed phase C18 silica gel using CH3OH/H2O (3
17) as a mobile phase (3 runs) to provide 8 (6.8 mg). Subfraction B8C (84.5 mg) was purified by CC over reversed phase C18 silica gel using CH3OH/H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to give four subfractions. The second subfraction (28.1 mg) was rechromatographed on CC over Sephadex LH-20 using CH3OH/CH2Cl2 (1
2) to give four subfractions. The second subfraction (28.1 mg) was rechromatographed on CC over Sephadex LH-20 using CH3OH/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) followed by PTLC using CH3OH/CH2Cl2 (1
1) followed by PTLC using CH3OH/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19) as a mobile phase (2 runs) to provide 9 (7.4 mg). The third subfraction (9.7 mg) was purified by PTLC using CH3OH/CH2Cl2 (1
19) as a mobile phase (2 runs) to provide 9 (7.4 mg). The third subfraction (9.7 mg) was purified by PTLC using CH3OH/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19) as a mobile phase (6 runs) to afford 10 (3.6 mg). Fraction C (245.7 mg) was subjected to the same procedure as fraction B8 to provide seven fractions (C1–C7). Compound 5 (3.0 mg) was obtained from fractions C2. Fraction C6 (114.7 mg) was separated by CC over reversed phase C18 silica gel using CH3OH/H2O (2
19) as a mobile phase (6 runs) to afford 10 (3.6 mg). Fraction C (245.7 mg) was subjected to the same procedure as fraction B8 to provide seven fractions (C1–C7). Compound 5 (3.0 mg) was obtained from fractions C2. Fraction C6 (114.7 mg) was separated by CC over reversed phase C18 silica gel using CH3OH/H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) to afford 12 (70.7 mg). The mycelial ethyl acetate extract (6.63 g) was separated using the same procedure as the broth extract to afford four fractions (CE1–CE4). Fraction CE2 (4.47 g) was purified using the same procedure as the mycelial ethyl acetate extract and subsequent purification by CC over silica gel using a gradient of CH3OH/CH2Cl2 (0
3) to afford 12 (70.7 mg). The mycelial ethyl acetate extract (6.63 g) was separated using the same procedure as the broth extract to afford four fractions (CE1–CE4). Fraction CE2 (4.47 g) was purified using the same procedure as the mycelial ethyl acetate extract and subsequent purification by CC over silica gel using a gradient of CH3OH/CH2Cl2 (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) to give five subfractions (CE2A–CE2E). Subfraction CE2B (85.2 mg) was rechromatographed on CC over silica gel using EtOAc/petroleum ether (1
0) to give five subfractions (CE2A–CE2E). Subfraction CE2B (85.2 mg) was rechromatographed on CC over silica gel using EtOAc/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) followed by PTLC using EtOAc/petroleum ether (1
4) followed by PTLC using EtOAc/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) as a mobile phase (9 runs) to afford 14 (1.8 mg). Subfraction CE2D (457.4 mg) was purified by CC over reversed phase C18 silica gel using a gradient of CH3OH/H2O (1
9) as a mobile phase (9 runs) to afford 14 (1.8 mg). Subfraction CE2D (457.4 mg) was purified by CC over reversed phase C18 silica gel using a gradient of CH3OH/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) subsequent purification by CC over reversed phase C18 silica gel using CH3OH/H2O (2
0) subsequent purification by CC over reversed phase C18 silica gel using CH3OH/H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) followed by CC over Sephadex LH-20 using CH3OH/CHCl3 (1
3) followed by CC over Sephadex LH-20 using CH3OH/CHCl3 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give 3 (3.4 mg). Fraction CE3 (709.0 mg) was purified using the same procedure as the broth extract to provide four subfractions. The second subfraction (69.5 mg) was separated by CC over reversed phase C18 silica gel using a gradient of CH3OH/H2O (3
1) to give 3 (3.4 mg). Fraction CE3 (709.0 mg) was purified using the same procedure as the broth extract to provide four subfractions. The second subfraction (69.5 mg) was separated by CC over reversed phase C18 silica gel using a gradient of CH3OH/H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 to 1
2 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) followed by PTLC using CH3OH/CH2Cl2 (1
0) followed by PTLC using CH3OH/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17) as a mobile phase (8 runs) to provide 2 (1.6 mg) and 4 (1.4 mg). The third subfraction (66.7 mg) was purified by CC over reversed phase C18 silica gel using CH3OH/H2O (3
17) as a mobile phase (8 runs) to provide 2 (1.6 mg) and 4 (1.4 mg). The third subfraction (66.7 mg) was purified by CC over reversed phase C18 silica gel using CH3OH/H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) subsequent purification by CC over reversed phase C18 silica gel using CH3OH/H2O (1
2) subsequent purification by CC over reversed phase C18 silica gel using CH3OH/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) followed by CC over Sephadex LH-20 using CH3OH/CHCl3 (1
1) followed by CC over Sephadex LH-20 using CH3OH/CHCl3 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 13 (6.8 mg). The mycelial hexane extract (675.4 mg) was separated by CC over Sephadex LH-20 using CH3OH/CH2Cl2 (1
1) to afford 13 (6.8 mg). The mycelial hexane extract (675.4 mg) was separated by CC over Sephadex LH-20 using CH3OH/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) followed by CC over silica gel using a gradient of CH3OH/CH2Cl2 (0
1) followed by CC over silica gel using a gradient of CH3OH/CH2Cl2 (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) to provide seven fractions (CH2A–CH2G). Compound 17 (2.6 mg) was obtained from fraction CH2B. Fraction CH2D (255.1 mg) was subjected to CC over silica gel using a gradient of acetone/petroleum ether (1
0) to provide seven fractions (CH2A–CH2G). Compound 17 (2.6 mg) was obtained from fraction CH2B. Fraction CH2D (255.1 mg) was subjected to CC over silica gel using a gradient of acetone/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 to 1
9 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) to give five subfractions. The second subfraction (37.8 mg) was purified on CC over silica gel using EtOAc/CH2Cl2 (1
0) to give five subfractions. The second subfraction (37.8 mg) was purified on CC over silica gel using EtOAc/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19) followed by CC over silica gel using acetone/petroleum ether (1
19) followed by CC over silica gel using acetone/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17) to provide 15 (7.3 mg). The fourth subfraction (33.0 mg) was separated by dissolving with EtOAc/petroleum ether (1
17) to provide 15 (7.3 mg). The fourth subfraction (33.0 mg) was separated by dissolving with EtOAc/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) to give compound 18 (23.4 mg) from the insoluble fraction. Fraction CH2F (70.7 mg) was separated by CC over reversed phase C18 silica gel using CH3OH/H2O (3
4) to give compound 18 (23.4 mg) from the insoluble fraction. Fraction CH2F (70.7 mg) was separated by CC over reversed phase C18 silica gel using CH3OH/H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to afford 16 (14.7 mg).
2) to afford 16 (14.7 mg).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 233 (3.52), 259 (3.45), 304 (3.04), 369 (2.71) nm; IR (neat) νmax 3116, 1647 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z: [M + H]+ calcd for C20H23O6, 359.1495; found, 359.1495; CD (CH3OH, c 1.0 × 10−3 mol L−1) Δε (nm): −6.4 (211), +2.6 (223), −4.8 (235), +2.7 (243), −5.0 (265), +0.5 (310).
ε): 233 (3.52), 259 (3.45), 304 (3.04), 369 (2.71) nm; IR (neat) νmax 3116, 1647 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z: [M + H]+ calcd for C20H23O6, 359.1495; found, 359.1495; CD (CH3OH, c 1.0 × 10−3 mol L−1) Δε (nm): −6.4 (211), +2.6 (223), −4.8 (235), +2.7 (243), −5.0 (265), +0.5 (310).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 207 (3.49), 286 (2.46) nm; IR (neat) νmax 3376, 1736, 1718 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS m/z: [M + Na]+ calcd for C22H26O8Na, 441.1525; found, 441.1516.
ε) 207 (3.49), 286 (2.46) nm; IR (neat) νmax 3376, 1736, 1718 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS m/z: [M + Na]+ calcd for C22H26O8Na, 441.1525; found, 441.1516.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 206 (3.83), 225 (3.83), 294 (3.40) nm; IR (neat) νmax 3406, 1742, 1725, 1710 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS m/z: [M + Na]+ calcd for C26H30O10Na, 525.1737; found, 525.1736.
ε) 206 (3.83), 225 (3.83), 294 (3.40) nm; IR (neat) νmax 3406, 1742, 1725, 1710 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS m/z: [M + Na]+ calcd for C26H30O10Na, 525.1737; found, 525.1736.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 207 (3.91), 224 (3.71), 280 (3.31) nm; IR (neat) νmax 3377, 1739, 1722, 1711 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS m/z: [M + Na]+ calcd for C26H30O10Na, 525.1737; found, 525.1738.
ε) 207 (3.91), 224 (3.71), 280 (3.31) nm; IR (neat) νmax 3377, 1739, 1722, 1711 cm−1; 1H and 13C NMR data, see Table 2; HRESIMS m/z: [M + Na]+ calcd for C26H30O10Na, 525.1737; found, 525.1738.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 217 (3.80), 269 (3.42), 329 (2.76) nm; IR (neat) νmax 3368, 1734, 1700 cm−1; 1H and 13C NMR data, see Tables S3 and S4;† HRESIMS m/z: [M + Na]+ calcd for C23H28O7Na, 439.1733; found, 439.1733.
ε) 217 (3.80), 269 (3.42), 329 (2.76) nm; IR (neat) νmax 3368, 1734, 1700 cm−1; 1H and 13C NMR data, see Tables S3 and S4;† HRESIMS m/z: [M + Na]+ calcd for C23H28O7Na, 439.1733; found, 439.1733.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 222 (3.71), 282 (3.01) nm; IR (neat) νmax 1736 cm−1; 1H and 13C NMR data, see Tables S3 and S4;† HRESIMS m/z: [M + Na]+ calcd for C25H34O7Na, 469.2202; found, 469.2202.
ε) 222 (3.71), 282 (3.01) nm; IR (neat) νmax 1736 cm−1; 1H and 13C NMR data, see Tables S3 and S4;† HRESIMS m/z: [M + Na]+ calcd for C25H34O7Na, 469.2202; found, 469.2202.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 222 (4.29), 282 (3.96) nm; IR (neat) νmax 1733 cm−1; 1H and 13C NMR data, see Tables S3 and S4;† HRESIMS m/z: [M + Na]+ calcd for C24H32O7Na, 455.2046; found, 455.2046.
ε) 222 (4.29), 282 (3.96) nm; IR (neat) νmax 1733 cm−1; 1H and 13C NMR data, see Tables S3 and S4;† HRESIMS m/z: [M + Na]+ calcd for C24H32O7Na, 455.2046; found, 455.2046.Determination of the absolute configuration of C-14 in 1
According to the published procedure,29 a mixture of compound 1 (0.5 mg) and Mo2(OAc)4 (0.7 mg) in AR grade DMSO was subjected to CD measurement. The mixture was kept for 30 min to form a stable chiral metal complex, after which the CD spectrum was recorded. The observed sign of the diagnostic band at around 303 nm in the induced CD spectrum was correlated to the absolute configuration of C-14 in 1. Compound 1: CD (c 1.70 × 10−3 mol L−1, DMSO) Δε −3.8 (303 nm).17Preparation of the (R)- and (S)-MTPA ester derivative of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 30
30
Pyridine (100 μL) and (R)-(−)-MTPACl (30 μL) were added to a CH2Cl2 solution (100 μL) of 6 (2.3 mg). The reaction mixture was stirred at room temperature for 20 h. After removal of the solvent, the mixture was purified by PTLC using ethyl acetate/petroleum ether (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) (2 runs) to afford the (S)-MTPA ester (3.3 mg). Compound 6 (2.3 mg) was treated in a similar way with (S)-(+)-MTPACl and, after purification by PTLC, (R)-MTPA ester (3.2 mg) was obtained.
5) (2 runs) to afford the (S)-MTPA ester (3.3 mg). Compound 6 (2.3 mg) was treated in a similar way with (S)-(+)-MTPACl and, after purification by PTLC, (R)-MTPA ester (3.2 mg) was obtained.
Methylation of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 31
31
Sodium hydride (10.0 mg, 2 eq.) was slowly added to a solution of 6 (33.4 mg) and methyl iodide (60 μL, 10 eq.) in THF (1 mL) at 0 °C. The reaction mixture was allowed to warm up to room temperature. After 24 h, the reaction mixture was diluted with ethyl acetate (10 mL) and then quenched with ice water (10 mL). The organic layer was washed with saturated aqueous Na2S2O3 (3 × 5 mL) and saturated aqueous NaHCO3 (5 mL), respectively, and dried over anhydrous Na2SO4. After filtration and subsequent removal of the solvent, the reaction mixture was purified by CC over silica gel using ethyl acetate/hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) to provide 6a (10.7 mg) as a major product. Compound 6a (9.5 mg) was treated in a similar way with sodium hydride (in oil, 8.5 mg) and methyl iodide (13 μL), after work-up and purification by PTLC using acetone/hexane (1
4) to provide 6a (10.7 mg) as a major product. Compound 6a (9.5 mg) was treated in a similar way with sodium hydride (in oil, 8.5 mg) and methyl iodide (13 μL), after work-up and purification by PTLC using acetone/hexane (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) (3 runs), 6b (1.3 mg) and 9a (0.8 mg) were obtained.
4) (3 runs), 6b (1.3 mg) and 9a (0.8 mg) were obtained.
Methylation of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 32
32
Methyl iodide (10 μL) was added to a mixture of 9 (3.0 mg) and K3CO3 (4.8 mg) in acetone (1 mL) and the mixture was stirred at room temperature for 2 days. Water (5 mL) was added and the mixture was extracted with ethyl acetate. After removal of the solvent, the residue was purified by PTLC using ethyl acetate/CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19) to yield 9a (1.6 mg).
19) to yield 9a (1.6 mg).
Antimicrobial assays
The antimicrobial activity was determined as described by the Clinical and Laboratory Standards Institute.33 For antibacterial assay against S. aureus and methicillin-resistant S. aureus, pure compounds were dissolved in 10% DMSO and diluted two-fold to give final concentrations ranged from 128–0.25 μg mL−1. The bacterial suspensions of the mid exponential growth phase were adjusted to the standard of McFarland number 0.5 with 0.85% NaCl solution or normal saline solution (NSS) to achieve a concentration of approximately 1.5 × 108 colony forming unit (CFU) mL−1 and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 with MHB. One hundred microliters of the culture, containing approximately 106 CUF mL−1 of the microorganism, was inoculated in 50 μL of MHB supplemented with 50 μL of the compounds. The microtiter plates were incubated at 35 °C for 15 h and then 30 μL of 0.09% resazurin was added into each well. Plates were further incubated for 3 h. Vancomycin and 1% DMSO were used as positive and negative controls, respectively. The experiment was carried out in triplicate. After incubation, the lowest concentration of the bioactive compounds that inhibited bacterial growth (blue or purple colour) was recorded as the MIC. The antifungal activities of pure compounds against C. neoformans were performed in a similar way to bacteria but Sabouraud Dextrose Broth (SDB) was used as culture medium. The tested plates were incubated at 25 °C for 48 h, and then 10 μL of 0.18% resazurin was added. Plates were further incubated for 24 h. Amphotericin B was used as a positive control.
200 with MHB. One hundred microliters of the culture, containing approximately 106 CUF mL−1 of the microorganism, was inoculated in 50 μL of MHB supplemented with 50 μL of the compounds. The microtiter plates were incubated at 35 °C for 15 h and then 30 μL of 0.09% resazurin was added into each well. Plates were further incubated for 3 h. Vancomycin and 1% DMSO were used as positive and negative controls, respectively. The experiment was carried out in triplicate. After incubation, the lowest concentration of the bioactive compounds that inhibited bacterial growth (blue or purple colour) was recorded as the MIC. The antifungal activities of pure compounds against C. neoformans were performed in a similar way to bacteria but Sabouraud Dextrose Broth (SDB) was used as culture medium. The tested plates were incubated at 25 °C for 48 h, and then 10 μL of 0.18% resazurin was added. Plates were further incubated for 24 h. Amphotericin B was used as a positive control.
Antimycobacterial assay
Antimycobacterial activity was determined against gfp recombinant Mycobacterium tuberculosis H37Ra (H37Ra gfp) using a green fluorescent protein (GFP)-based fluorescent detection method.34 Briefly, each 5 μL of test compounds serially diluted in 5% DMSO was mixed with 45 μL of H37Ra gfp suspended in Middlebrook 7H9 medium (approximately 2 × 104 CFU per well). The assay was performed in duplicate in 384-well plate format. Plates were incubated at 37 °C for 7 days and the fluorescence signals were measured using SpectraMax M5 microplate reader (Molecular devices, USA) in the bottom-reading mode at the excitation and emission wavelengths of 485 and 535 nm. Fluorescence signals at day zero were used as background. The percentage of growth inhibition was calculated from the mean of fluorescence unit of the treated (FUT) and untreated cells (FUC) as the following equation: % inhibition = [1 − (FUT/FUC)] × 100. The lowest concentration that inhibited cell growth by 90% was recorded as the minimum inhibitory concentration (MIC). Standard drugs rifampin, streptomycin, isoniazid, ofloxacin, and ethambutol were used as positive controls and DMSO (0.5%) was used as a negative control.Antimalarial assay
The activity was evaluated against the parasite Plasmodium falciparum K1 (multidrug resistant stain) using the microculture radioisotope technique based on the method described by Desjardins et al.35 Briefly, 25 μL of test solution in RPMI-1640 was mixed in microplate wells with 200 μL of a 1.5% erythrocyte suspension with 1% parasitaemia in the early ring stage and incubated in a CO2 incubator (5% O2, 5% CO2 and 90% N2) at 37 °C for 24 h. Subsequently, 25 μL of 3H-hypoxanthine in culture medium (0.5 μCi) was added into each well. The plates were further incubated for an additional 24 h. Levels of labeled hypoxanthine incorporation indicating parasite growth were determined by the TopCount microplate scintillation counter (Packard, USA). The percentage of parasite growth was calculated from the signal count per minute of treated (CPMT) and untreated conditions (CPMU) by the following formula: % parasite growth = CPMT/CPMU × 100. Inhibition concentration recorded as IC50 value was the concentration which indicates 50% reduction in parasite growth. Dihydroartemisinin and mefloquine were used as the standard antimalarial drugs.Cytotoxicity assays
The activity assay against African green monkey kidney cell lines (Vero, ATCC CCL-81) was performed in triplicate employing the method described by Hunt and co-workers.36 The GFP-expressing Vero cell line was generated in-house by stably transfecting the Vero cells with pEGFP-N1 plasmid (Clontech) and maintained in minimal essential medium supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 1 mM sodium pyruvate, 1.5 g L−1 sodium bicarbonate and 0.8 mg mL−1 geneticin at 37 °C in an incubator with 5% CO2. Vero cell suspension (45 μL, 3.3 × 104 cells per mL) was added into each well of 384-well plates containing 5 μL of test compounds. Plates were incubated in a CO2 incubator at 37 °C for 4 days. Fluorescence signals were measured using SpectraMaxM5 microplate reader (Molecular Devices, USA) and the percentages of cytotoxicity were calculated in the same manner as for the anti-mycobacterial assay. IC50 values were derived from dose–response curves using 6 concentrations of 2-fold serially diluted test compounds by the SOFTMax Pro software. Ellipticine was used as a positive control.The anti-proliferative activities against human oral epidermoid carcinoma (KB) cell lines (ATCC CCL-17) and human breast adenocarcinoma (MCF-7) cell lines (ATCC HTB-22) were evaluated using the resazurin microplate assay.37 Briefly, 5 μL of test compound diluted in 5% DMSO was mixed with 45 μL of KB suspension (2.2 × 104 cells per mL) and MCF-7 (3.3 × 104 cells per mL) in a 384-well plate and incubated at 37 °C in 5% CO2 incubator 3 days. After incubation, cell growth was determined by adding 12.5 μL of 62.5 μg mL−1 resazurin solution and incubated further for 4 h. Fluorescence signals were measured using SpectraMax M5 microplate reader (Molecular devices, USA) in the bottom-reading mode at the excitation and emission wavelengths of 530 and 590 nm. The percentage of growth inhibition was calculated from the mean of fluorescence unit of the treated (FUT) and untreated cells (FUC) as the following equation: % inhibition = [1 − (FUT/FUC)] × 100. The IC50 value was obtained from the dose response curve using the SOFTMax Pro software. Ellipticine and doxorubicin were used as positive controls for KB cells whereas tamoxifen and doxorubicin were positive controls for MCF-7 cells. DMSO (0.5%) was used as a negative control.
Acknowledgements
C. D. is grateful to the Office of the Higher Education Commission, Thailand, under the Strategic Scholarships Fellowships Frontier Research Networks (specific for the Southern region) for the Joint PhD Program Thai Doctoral degree program (CHE-SSR-Ph.D.-THA) and Princess of Naradhiwas University for a scholarship. V. R. thanks the Thailand Research Fund (TRF) for the research grant (Grant No. DBG5680001). The Center of Excellence for Innovation in Chemistry (PERCH-CIC), and Prince of Songkla University are acknowledged for partial support. We thank Department of Chemistry, Faculty of Science Mahidol University for the optical rotation measurement of some compounds. Finally, the National Center for Genetic Engineering and Biotechnology (BIOTEC) is acknowledged for antimycobacterial, antimalarial, anticancer and cytotoxic assays.Notes and references
- J. Peng, X. Zhang, L. Du, W. Wang, T. Zhu, Q. Gu and D. Li, J. Nat. Prod., 2014, 77, 424 CrossRef CAS PubMed.
- T. H. Quang, N. T. T. Ngan, W. Ko, D.-C. Kim, C.-S. Yoon, J. H. Sohn, J. H. Yim, Y.-C. Kim and H. Oh, Bioorg. Med. Chem. Lett., 2014, 24, 5787 CrossRef CAS PubMed.
- C. Tansakul, V. Rukachaisirikul, A. Maha, T. Kongprapan, S. Phongpaichit, N. Hutadilok-Towatana, K. Borwornwiriyapan and J. Sakayaroj, Nat. Prod. Res., 2014, 28, 1718 CrossRef CAS PubMed.
- K. Trisuwan, V. Rukachaisirikul, K. Borwornwiriyapan, S. Phongpaichit and J. Sakayaroj, Tetrahedron Lett., 2014, 55, 1336 CrossRef CAS.
- V. Rukachaisirikul, S. Satpradit, S. Klaiklay, S. Phongpaichit, K. Borwornwiriyapan and J. Sakayaroj, Tetrahedron, 2014, 70, 5148 CrossRef CAS.
- J. Xue, Y. Fu, P. Wu, L. Xu, R. Huang, X. Wei and H. Li, J. Antibiot., 2015, 68, 598 CrossRef CAS PubMed.
- S. Komai, T. Hosoe, T. Itabashi, K. Nozawa, T. Yaguchi, K. Fukushima and K. Kawai, J. Nat. Med., 2006, 60, 185 CrossRef CAS.
- K. Suzuki, K. Nozawa, S. Udakawa, S. Nakajima and K. Kawai, Phytochemistry, 1991, 30, 2096 CrossRef CAS.
- H. Oh, T. O. Kwon, J. B. Gloer, L. Marvanová and C. A. Shearer, J. Nat. Prod., 1999, 62, 580 CrossRef CAS PubMed.
- Y. Zhang, X.-M. Li, Z. Shang, C.-S. Li, N.-Y. Ji and B.-G. Wang, J. Nat. Prod., 2012, 75, 1888 CrossRef CAS PubMed.
- H. Luo, X.-M. Li, C.-S. Li and B.-G. Wang, Phytochem. Lett., 2014, 9, 22 CrossRef CAS.
- H. Hayashi, Y. Oka, K. Kai and K. Akiyama, Biosci., Biotechnol., Biochem., 2012, 76, 745 CrossRef CAS PubMed.
- H. Hayashi, Y. Oka, K. Kai and K. Akiyama, Biosci., Biotechnol., Biochem., 2012, 76, 1765 CrossRef CAS PubMed.
- S. Nakanishi, S. Toki, Y. Saitoh, E. Tsukuda, K. Kawahara, K. Ando and Y. Matsuda, Biosci., Biotechnol., Biochem., 1995, 59, 1333 CrossRef CAS PubMed.
- H. Nishida, H. Tomoda, J. Cao, S. Okuda and S. Ōmura, J. Antibiot., 1991, 44, 144 CrossRef CAS PubMed.
- S. Komai, T. Hosoe, T. Itabashi, K. Nozawa, K. Okada, G. M. de C. Takaki, T. Yaguchi, K. Takizawa, K. Fukushima and K. Kawai, Chem. Pharm. Bull., 2005, 53, 1114 CrossRef CAS PubMed.
- L. D. Bari, G. Pescitelli, C. Pratelli, D. Pini and P. Salvadori, J. Org. Chem., 2001, 66, 4819 CrossRef PubMed.
- I. Ohtani, T. Kusumi, Y. Kashman and H. Kakisawa, J. Am. Chem. Soc., 1991, 113, 4092 CrossRef CAS.
- D.-L. Zhao, C.-L. Shao, Q. Zhang, K.-L. Wang, F.-F. Guan, T. Shi and C.-Y. Wang, J. Nat. Prod., 2015, 78, 2310 CrossRef CAS PubMed.
- L. Wen, Y.-C. Lin, Z.-G. She, D.-S. Du, W.-L. Chan and Z.-H. Zheng, J. Asian Nat. Prod. Res., 2008, 10, 133 CrossRef CAS PubMed.
- H. Nishida, H. Tomoda, S. Okuda and S. Ōmura, J. Org. Chem., 1992, 57, 1271 CrossRef CAS.
- S. Klaiklay, V. Rukachaisirikul, K. Tadpetch, Y. Sukpondma, S. Phongpaichit, J. Buatong and J. Sakayaroj, Tetrahedron, 2012, 68, 2299 CrossRef CAS.
- C. Wu, Y. Zhao, R. Chen, D. Liu, M. Liu, P. Proksch, P. Guo and W. Lin, RSC Adv., 2016, 6, 21969 RSC.
- S. Sureram, C. Kesornpun, C. Mahidol, S. Ruchirawat and P. Kittakoop, RSC Adv., 2013, 3, 1781 RSC.
- D.-L. Zhao, C.-L. Shao, Q. Zhang, K.-L. Wang, F.-F. Guan, T. Shi and C.-Y. Wang, J. Nat. Prod., 2015, 78, 2310 CrossRef CAS PubMed.
- A. A. Sy-Cordero, M. Figueroa, H. A. Raja, M. E. M. Aviňa, M. P. Croatt, A. F. Adcock, D. J. Kroll, M. C. Wani, C. J. Pearce and N. H. Oberlies, Tetrahedron, 2015, 71, 8899 CrossRef CAS PubMed.
- M. Chen, L. Han, C.-L. Shao, Z.-G. She and C.-Y. Wang, Chem. Biodiversity, 2015, 12, 443 CAS.
- J. Arunpanichlert, V. Rukachaisirikul, Y. Sukpondma, S. Phongpaichit, S. Tewtrakul, N. Rungjindamai and J. Sakayaroj, Chem. Pharm. Bull., 2010, 58, 1033 CrossRef CAS PubMed.
- V. Rukachaisirikul, N. Rungsaiwattana, S. Klaiklay, S. Phongpaichit, K. Borwornwiriyapan and J. Sakayaroj, J. Nat. Prod., 2014, 77, 2375 CrossRef CAS PubMed.
- C. Daengrot, V. Rukachaisirikul, C. Tansakul, T. Thongpanchang, S. Phongpaichit, K. Bowornwiriyapan and J. Sakayaroj, J. Nat. Prod., 2015, 78, 615 CrossRef CAS PubMed.
- Y. Onishi, Y. Nishimoto, M. Yasuda and A. Baba, Chem. Lett., 2011, 40, 1223 CrossRef CAS.
- H. Miyatake-Ondozabal and A. G. M. Barrett, Org. Lett., 2010, 12, 5573 CrossRef CAS PubMed.
- S. Phongpaichit, N. Rungjindamai, V. Rukachaisirikul and J. Sakayaroj, FEMS Immunol. Med. Microbiol., 2006, 48, 367 CrossRef CAS PubMed.
- C. Changsen, S. G. Franzblau and P. Palittapongarnpim, Antimicrob. Agents Chemother., 2003, 47, 3682 CrossRef CAS PubMed.
- R. E. Desjardins, C. J. Canfield, J. D. Haynes and J. D. Chulay, Antimicrob. Agents Chemother., 1979, 16, 710 CrossRef CAS PubMed.
- L. Hunt, M. Jordan, M. De Jesus and F. M. Wurm, Biotechnol. Bioeng., 1999, 65, 201 CrossRef CAS PubMed.
- J. O'Brien, I. Wilson, T. Orton and F. Pognan, Eur. J. Biochem., 2000, 267, 5421 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: 1H and 13C NMR spectra for penicillanthone (1), penicillidic acids A–C (2–4), tenellic acid A methyl ester (14), penicillide methyl ester A (6b) and penicillide methyl ester B (9a), optical rotations of known compounds, CD data of 7, 15–17, and 1H and 13C NMR data of 14, 6b and 9a. See DOI: 10.1039/c6ra04401h |
| This journal is © The Royal Society of Chemistry 2016 |