Biocompatible graphene nanosheets grafted with poly(2-hydroxyethyl methacrylate) brushes via surface-initiated ARGET ATRP†
Jin Shaab,
Yuan Gaoa,
Tong Wuab,
Xin Chenab,
Travis Cordieb,
Haili Zhaoa,
Linsheng Xiea,
Yulu Ma*a and
Lih-sheng Turng*b
aEngineering Center of Efficient Green Process Equipment and Energy Conservation, Ministry of Education, East China University of Science and Technology, Shanghai, 200237, China. E-mail: myl@ecust.edu.cn
bWisconsin Institute for Discovery, University of Wisconsin–Madison, Madison, Wisconsin 53715, USA. E-mail: turng@engr.wisc.edu
First published on 30th March 2016
Abstract
Using robust chemistry to graft polymer brushes on graphene nanosheets would promote the development of graphene nanomaterials as a versatile platform for biomedical applications. Based on surface-initiated activators regenerated by the electron transfer atom transfer radical polymerization (ARGET ATRP) technique, the study developed a protocol to prepare well-defined poly(2-hydroxyethyl methacrylate) (HEMA) brushes on chemically reduced graphene oxide surfaces. ATR-FTIR, XPS and TEM characterizations demonstrate tin(II) 2-ethylhexanoate to be an efficient reducing agent that provides controlled polymerization with a significant decreased Cu catalyst usage (down to about 20 ppm), and prevents trace amounts of elemental Cu residue on the graphene surface. Fetal bovine serum protein absorption assay reveals the effect of brush backbone structure change to tune the interfacial interaction between graphene nanosheets and proteins. Further, NIH-3T3 fibroblast cell and human umbilical vein endothelial cell viability assays indicate that the obtained graphene nanosheets meet the biocompatibility requirements to support fibroblast cells, even human cells, attach and proliferate. The approach and the graphene–polymer brush hybrid developed in this work should open new opportunities for a broader range of biomedical applications of carbon nanomaterials.
Introduction
Graphene nanosheets and related derivatives have shown outstanding potential in biomedical applications as a novel biocompatible and versatile platform for biosensing, bioimaging, drug delivery, and cell culture scaffolds.1–5 With the rapid development of synthesis and functionalization approaches, stably functionalized graphene derivatives, e.g. graphene oxide (GO),6 chemically reduced graphene oxide (rGO),7 and fluorinated graphene,8 have been exploited as graphene-based scaffolds and demonstrated to be biocompatible and promising for cellular regulation.9–11 As cell behaviors, including adhesion, proliferation, differentiation and signalling, are inherently sensitive to graphene surface's chemistry and topography features,12–14 understanding and controlling the graphene interfaces by robust surface chemistry would benefit the future biomedical and clinical applications of graphene-based nanomaterials.Polymer brushes are widely used to tailor the surface properties of materials.15,16 Via surface-initiated atom transfer radical polymerization (SI-ATRP) technique, functional polymer brushes can be reached on scaffold substrate surfaces to engineer the interaction between the substrate and cells.17–21 Though numerous studies have reported constructing polymer brushes on graphene nanosheet surface via SI-ATRP method,22–27 polymer brushes grafted graphene nanosheets haven't been exploited as scaffold material. One possible issue obstructs the exploitation can be attributed to the relative high transition metal catalyst usage in ATRP (on the order of 0.1 to 1 mol% relative to the monomer). Because of the ionization binding between graphene surface functional groups and divalent metal ions,24,28 the metal catalyst residual on graphene surface would be hazard for the further biomedical application of the obtained graphene–polymer brushes hybrid.29
Here we report the effort to graft poly(2-hydroxyethyl methacrylate) (poly(HEMA)) brushes onto chemically reduced graphene oxide (rGO) by surface-initiated ATRP reaction, based on activators regenerated by the electron transfer (ARGET) technique. Poly(HEMA) brushes are well-known as the ubiquitous platform for nanomedicine,19,30,31 that can be grafted onto carbon nanomaterial to achieve scaffold substrates for cell proliferation and bone formation.32 In present of appropriate reducing agents, small amount Cu(II) complex catalyst can initiate and maintain a controllable polymerization process, resulting in neglectable Cu element residual on the graphene surface. Furthermore, fetal bovine serum (FBS) protein absorption assay and cell viability experiments were performed to testify the biocompatibility and cell behavior regulation capacity of the obtained hybrid nanomaterials.
Experimental
Materials and detailed experimental procedures see ESI.† G-HEMAT and G-HEMAA were applied to denote the samples prepared with Sn(EH)2 and ascorbic acid as reducing agents. Random copolymer poly(2-hydroxyethyl methacrylate)-(2-carboxyethyl acrylate) (poly(HEMA-CA)) brushes grafted graphene nanosheets (G-HECA) was achieved by using 2-hydroxyethyl methacrylate (HEMA) and 2-carboxyethyl acrylate (CA) as comonomers. Different comomomer mol ratios result in different carboxyl group contents in the copolymer brushes, as shown in Scheme 1. When the mole ratio of HEMA and CA starts at 4 (m/n = 4), the achieved sample is denoted as G-HECA1; when the mole ratio of HEMA and CA starts at 2 (m/n = 2), the achieved sample is denoted as G-HECA2. Attenuated total reflection infrared (ATR-FTIR) spectra were measured using a Bruker Tensor 27 FT-IR spectrometer. All spectra were obtained with a gallium ATR crystal in the range of 1000 to 4000 cm−1. Thirty-two scans were taken on each sample to achieve the spectrum with a resolution of 4 cm−1. The raw data was processed using OPUS 6.5 software with an ATR correction and an atmosphere correction. Raman spectra were measured using a Raman microspectrometer (Thermo Scientific) with a 10 mW 631 nm wavelength incident laser light, a CCD detector, and a confocal depth resolution of 2 μm. The laser beam was focused on the sample using an optical microscope with a ×50 objective lens. OMNIC software was used to process the raw data and apply quantitative analysis. The surface morphology of spray coated graphene film were observed by scanning electron microscopy (SEM) (JEOL 6500, Nikon) with an accelerating voltage of 10 kV. Before SEM imaging, the samples were flow dried with a N2 stream and sputtered with gold for 40 s. The chemical composition information was obtained via X-ray photoelectron spectroscopy (XPS). The measurements were performed using a Thermo K-alpha XPS spectrometer with a micro-focused monochromated Al Kα X-ray source. For the XPS broad survey scan of the specimen, two scans were performed with a constant analyzer energy of 150.0 eV and a pass energy of 1.00 eV. For high-resolution region spectra, five scans were performed with a constant analyzer energy of 50.0 eV and a pass energy of 0.10 eV. The baselines were modeled with Shirley functions and the region scans were deconvoluted in freeware CasaXPS software with Monte Carlo error analysis. The binding energies were referenced to the C 1s line at 285.0 eV from adventitious carbon. Thermal stability was determined using thermogravimetric analysis (TGA Q50, AT Instruments) over a temperature range of 30 °C to 750 °C with a heating rate at 10 °C min−1 under a N2 atmosphere (the flow rate of the protection gas was 5 ml min−1). X-ray diffraction (XRD) curves were recorded on a D8 Discover diffractometer (Bruker) with a Cu target (λ = 0.1540 nm) at room temperature. The diffraction pattern was taken from 10° to 75° in the step scan mode, and it was recorded using a scintillation counter detector with a voltage of 40 kV and a current of 40 mA. Ultraviolet-visible (UV-visible) spectra were recorded on a NanoDrop 2000 Spectrophotometer. The saturated supernatants were achieved from ultrasonic dispersion of extra samples in water, ethanol, and DMF followed by centrifugation at 1500 rpm for 30 min. Optical images of cells were taken using a Nikon microscope (TS100 with ECLIPSE ME600L camera) with a ×10 objective lens, and fluorescent images were taken using a Nikon A1R confocal microscope. A Philips CM-100 transmission electron microscope (TEM) (Philips/FEI Corporation, Holland) at an accelerating voltage of 100 kV was used to study the microstructures of graphene nanosheets. The samples were prepared by depositing GO, G-HEMAT and G-HEMAA dilute DMF suspensions on Formvar-carbon coated grids.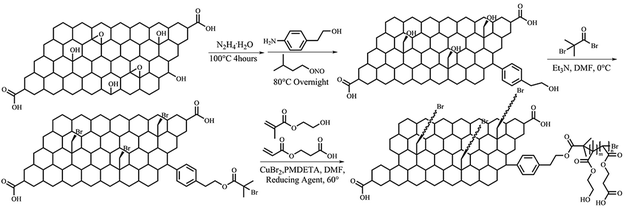 | ||
| Scheme 1 Schematic route to prepare the graphene nanosheets grafted with poly(HEMA) brushes via surface-initiated ARGET ATRP. | ||
Results and discussion
Preparation and characterization
The graphene nanosheets grafted with poly(HEMA) brushes were prepared by surface-initiated ARGET ATRP reaction using G-initiator as the initiator and CuBr2/PMDETA as the catalyst (Scheme 1). The synthetic route include three steps: (1) introducing the hydroxyl group onto the graphene surface, (2) grafting of the ATRP initiator to the reduced graphene surface, and (3) in situ polymerization in a heterogeneous ARGET ATRP condition. The ATRP system component calculation was conducted based on molar ratio [G-initiator]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [Cu(II)]
[Cu(II)]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [PMDETA]
[PMDETA]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [monomer]
[monomer]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [reducing agent] = 50
[reducing agent] = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000
5000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. Relative to the initial amount of Cu(II), the extra two-fold PMDETA addition ensures the coordination of the PMDETA with Cu(II) in the presence of large excess of monomer. It also avoided the loss of the Cu/PMDETA complex caused by competitive complexation of PMDETA with the stronger Lewis acid formed in the reactivation process.33–35 Compared to normal ATRP, which starts in the presence of a >1000 ppm (>0.1 mol% vs. monomer) catalyst, ARGET ATRP technique provides continuous controlled polymerization with a significant decrease in the Cu/PMDETA complex usage (down to about 20 ppm). The reducing agent brings a constant regeneration of the Cu(I) activator species, which compensates for any loss of Cu(I) by unavoidable radical termination reactions. The reducing agent can also influence the polymerization process due to the different concentrations and related reduction rates. Here, the amount of Cu(II) added to the reaction is much smaller than the amount of reducing agent needed to balance the termination process. The extra reducing agent allows a tolerance of air or other radical traps in the system. Various reducing agents have been reported for successful ARGET ATRP,33–36 while the most widely used and environmentally acceptable reducing agents are Sn(EH)2 and ascorbic acid, which have been approved by the Food and Drug Administration (FDA). A comparative experiment between two different reducing agents was performed and the products were characterized by ATR-FTIR, XPS, TEM and TGA.
100. Relative to the initial amount of Cu(II), the extra two-fold PMDETA addition ensures the coordination of the PMDETA with Cu(II) in the presence of large excess of monomer. It also avoided the loss of the Cu/PMDETA complex caused by competitive complexation of PMDETA with the stronger Lewis acid formed in the reactivation process.33–35 Compared to normal ATRP, which starts in the presence of a >1000 ppm (>0.1 mol% vs. monomer) catalyst, ARGET ATRP technique provides continuous controlled polymerization with a significant decrease in the Cu/PMDETA complex usage (down to about 20 ppm). The reducing agent brings a constant regeneration of the Cu(I) activator species, which compensates for any loss of Cu(I) by unavoidable radical termination reactions. The reducing agent can also influence the polymerization process due to the different concentrations and related reduction rates. Here, the amount of Cu(II) added to the reaction is much smaller than the amount of reducing agent needed to balance the termination process. The extra reducing agent allows a tolerance of air or other radical traps in the system. Various reducing agents have been reported for successful ARGET ATRP,33–36 while the most widely used and environmentally acceptable reducing agents are Sn(EH)2 and ascorbic acid, which have been approved by the Food and Drug Administration (FDA). A comparative experiment between two different reducing agents was performed and the products were characterized by ATR-FTIR, XPS, TEM and TGA.
ATR-FTIR spectra of GO, G-OH, G-initiator, G-HEMAA and G-HEMAT are shown in Fig. 1(a). The characteristic peaks of GO include: –OH stretching vibration at 3454 cm−1, C–H stretching vibration at 2930 cm−1 and 2850 cm−1, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration at 1736 cm−1, aromatic C
O stretching vibration at 1736 cm−1, aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibration at 1641 cm−1, C–O–H bending vibration at 1412 cm−1, and O–C stretching vibration at 1062 cm−1. After hydrazine reduction and hydroxyl functionalization, most characteristic peaks corresponding to –COOH, –C–OH disappear. Only the weak –OH stretching vibration from 3454 cm−1 to 3024 cm−1, stretching vibration of aromatic C
C stretching vibration at 1641 cm−1, C–O–H bending vibration at 1412 cm−1, and O–C stretching vibration at 1062 cm−1. After hydrazine reduction and hydroxyl functionalization, most characteristic peaks corresponding to –COOH, –C–OH disappear. Only the weak –OH stretching vibration from 3454 cm−1 to 3024 cm−1, stretching vibration of aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C at 1654 cm−1 and 1560 cm−1, and aromatic in-plane C–H vibration at 1037 cm−1 and 1008 cm−1 are observed. The spectrum from the ATRP initiator-grafted graphene nanosheets exhibits two sharp peaks at 1600 cm−1 (C
C at 1654 cm−1 and 1560 cm−1, and aromatic in-plane C–H vibration at 1037 cm−1 and 1008 cm−1 are observed. The spectrum from the ATRP initiator-grafted graphene nanosheets exhibits two sharp peaks at 1600 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration) and 1508 cm−1 (–CH2 or –CH3 deformation bending vibration), and one broad peak at 1228 cm−1 (C–O stretching vibration), indicating the presence of the ester bond formed by reactions between hydroxyl groups and α-bromoisobutyryl bromide. Multiple characteristic peaks are observed in the spectrum of G-HEMAA and G-HEMAT (note that the samples were exhaustively washed and dialyzed to remove free monomer residents prior to FTIR testing). The strong peaks at 1732 cm−1 (C
O stretching vibration) and 1508 cm−1 (–CH2 or –CH3 deformation bending vibration), and one broad peak at 1228 cm−1 (C–O stretching vibration), indicating the presence of the ester bond formed by reactions between hydroxyl groups and α-bromoisobutyryl bromide. Multiple characteristic peaks are observed in the spectrum of G-HEMAA and G-HEMAT (note that the samples were exhaustively washed and dialyzed to remove free monomer residents prior to FTIR testing). The strong peaks at 1732 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration), 1454 cm−1 (C–O–H bending vibration), 1389 cm−1 (–O–C
O stretching vibration), 1454 cm−1 (C–O–H bending vibration), 1389 cm−1 (–O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O symmetric stretching vibration), and 1228 cm−1 (C–O stretching vibration) reveal the presence of O–C
O symmetric stretching vibration), and 1228 cm−1 (C–O stretching vibration) reveal the presence of O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups. The peak at 3360 cm−1, related to –OH stretching vibration, becomes broader and stronger. And peaks at 1658 cm−1 and 1585 cm−1 (aromatic C
O groups. The peak at 3360 cm−1, related to –OH stretching vibration, becomes broader and stronger. And peaks at 1658 cm−1 and 1585 cm−1 (aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibration), 1512 cm−1 (–CH2 or –CH3 deformation bending vibration), and medium peaks at 2930 cm−1 and 2850 cm−1 (C–H stretching vibration at) are also observed. Moreover, stronger intensity of the carbonyl peak at 1732 cm−1 and hydroxy peak at 3300 cm−1 are observed in comparison between the G-HEMAA and G-HEMAT curves. These characteristic peaks demonstrate the success attachment of poly(HEMA) chain on the graphene surface.
C stretching vibration), 1512 cm−1 (–CH2 or –CH3 deformation bending vibration), and medium peaks at 2930 cm−1 and 2850 cm−1 (C–H stretching vibration at) are also observed. Moreover, stronger intensity of the carbonyl peak at 1732 cm−1 and hydroxy peak at 3300 cm−1 are observed in comparison between the G-HEMAA and G-HEMAT curves. These characteristic peaks demonstrate the success attachment of poly(HEMA) chain on the graphene surface.
 | ||
| Fig. 1 (a) ATR-FTIR spectra of GO, G-OH, G-initiator, G-HEMAA and G-HEMAT; (b) XPS survey scans of GO, G-OH, G-initiator, G-HEMAA and G-HEMAT; (c–e) TEM images of GO, G-HEMAT and G-HEMAA. | ||
As shown in Fig. 1(b), XPS survey scan was applied to characterize the bulk surface chemical composition of GO, G-OH, G-initiator, G-HEMAA and G-HEMAT (Table 1). GO is mainly composed of C and O indicating rich oxygen-containing groups on the graphene surface (estimated to be 35.42%). After hydrazine reduction and hydroxyl functionalization, the oxygen content decreases to 17.02%. The increase in nitrogen content (8.31%) can be attributed to the formation of hydrazones during the reduction process.37 A small signal around 70 eV, ascribed to the Br 3d excitation, is observed in G-initiator, suggesting the attachment of the ATRP initiator onto the graphene surface. Moreover, G-HEMAA and G-HEMAT sample show lower C/O atomic ratio (3.74 and 3.23) than that of G-initiator (4.75), suggesting the attachment of higher oxygen-containing monomer on the graphene surface. The XPS survey scans present no significant Cu signal, indicating a lower than the limit-of-detection (LOD) Cu element residual on the graphene surface. However, about 0.1 mol% Sn element residual is observed on the G-HEMAT sample. It is because basal carboxyl groups on graphene surfaces tend to ionize and bind metal ions.24,28
| Samples | C (%) | O (%) | N (%) | Br (%) | Sn (%) |
|---|---|---|---|---|---|
| GO | 64.58 | 35.42 | |||
| G-OH | 74.67 | 17.02 | 8.31 | ||
| G-initiator | 76.84 | 16.17 | 6.27 | 0.73 | |
| G-HEMAA | 76.37 | 20.40 | 3.21 | 0.02 | |
| G-HECAT | 73.67 | 22.81 | 3.40 | 0.02 | 0.10 |
The dry state morphologies of GO, G-HEMAA and G-HEMAT nanosheets were studied by TEM. Fig. 1(c) presents individual and layered GO nanosheets with smooth surface and some folding edge. Fig. 1(d) and (e) present the morphology of G-HEMAT and G-HEMAA sample. Because of low surface grafting density, constructed brushes form non-uniformly distributed agglomerate structure (as shown in Fig. 1(e)). Whereas for G-HEMAT sample, contiguous aggregates uniformly distribute on graphene surface and create a thin polymeric interface layer (Fig. 1(d)), indicating higher poly(HEMA) brushes grafting density. The agglomerate structure morphologies are similar to the observation on polystyrene brushes grafted graphene.27,38 Same conclusion can also be conducted based on the results of TGA analysis (see ESI Fig. S1†). By using Sn(EH)2 as reducing agent, higher poly(HEMA) brush grafting density on the graphene surface can be reached. A possible explanation is that the higher reduction ability of ascorbic acid not only helps to rapidly convert Cu(II) to Cu(I) and diminish Cu(II) complex concentration in the system, but also increase the free radical concentration, which decrease the control on polymerization process.39 Besides, Sn(EH)2 present better solubility and compatibility in the heterogeneous condition. Thus, we use Sn(EH)2 as reducing agent and denote the obtained sample as G-HEMA in the sections below.
Copolymer poly(HEMA-CA) brushes with different monomer compositions were obtained on the graphene nanosheets. Fig. 2(a) shows the ATR-FTIR spectra of G-HEMA, G-HECA1 and G-HECA2 samples. With the CA monomer content increase, the intensity of characteristic peaks at 1732 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration) increases, while the intensity of peaks at 1512 cm−1 (–CH2 or –CH3 deformation bending vibration) decreases, indicating more terminal carboxyl group presence in the copolymer brush structure. XPS C 1s high-resolution region scan was performed for a comparative analysis of G-HEMA, G-HECA1 and G-HECA2. The spectra were deconvoluted in the freeware CasaXPS software with Monte Carlo simulation to evaluate the uncertainties of peak parameter determination, which were expressed in the form of standard error (StDev). The spectra are curve-fitted into four peak components with binding energies at about 284.9, 285.4, 286.4 and 288.9 eV, attributable to the C–C/C–H, C–N, C–O/C–Br, and O–C
O stretching vibration) increases, while the intensity of peaks at 1512 cm−1 (–CH2 or –CH3 deformation bending vibration) decreases, indicating more terminal carboxyl group presence in the copolymer brush structure. XPS C 1s high-resolution region scan was performed for a comparative analysis of G-HEMA, G-HECA1 and G-HECA2. The spectra were deconvoluted in the freeware CasaXPS software with Monte Carlo simulation to evaluate the uncertainties of peak parameter determination, which were expressed in the form of standard error (StDev). The spectra are curve-fitted into four peak components with binding energies at about 284.9, 285.4, 286.4 and 288.9 eV, attributable to the C–C/C–H, C–N, C–O/C–Br, and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O species, respectively. The relative intensity of the O–C
O species, respectively. The relative intensity of the O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak component in G-HEMA is 8.94 ± 1.15%, and the peak intensity increases to 12.35 ± 0.14% and 16.07 ± 0.12% in G-HECA1 and G-HECA2 due to CA monomer proportion increase. The ATR-FTIR spectra and XPS deconvolution analysis prove the feasibility of the protocol to tune the structure and monomer content of brush backbone, which implies the versatility of the approach in grafting multifunctional polymer brushes on the graphene surface.
O peak component in G-HEMA is 8.94 ± 1.15%, and the peak intensity increases to 12.35 ± 0.14% and 16.07 ± 0.12% in G-HECA1 and G-HECA2 due to CA monomer proportion increase. The ATR-FTIR spectra and XPS deconvolution analysis prove the feasibility of the protocol to tune the structure and monomer content of brush backbone, which implies the versatility of the approach in grafting multifunctional polymer brushes on the graphene surface.
Raman spectra measurements were conducted to characterize the structural information of different graphene specimens, as shown in Fig. 3. In Raman spectrum of GO, the prominent peaks at 1352 cm−1 and 1600 cm−1 correspond to the D and G bands. The G band arises primarily form the presence of an sp2 carbon network, while the D band originates from defects inherent in the graphite and the edge effect of graphite crystallites. After hydrazine reduction, the vibration frequency of the G band shifts back to the value of the pristine graphene G band (about 1590 cm−1), which indicates a restoration of electric conjugation within the graphitic network. The D band and G band position are almost same in G-HEMA, G-HECA1 and G-HECA2 spectra, except for a slight variance in the ratio of the D to G band intensity (ID/IG). For graphene nanosheets, the value of ID/IG increases along with the chemical treatments, reflecting the disorder of the graphene plane through the formation of covalent bonds. A simple method has been proposed to calculate in-plane crystallite size La:40,41 La (nm) = (2.4 × 10−10) × λ4(ID/IG)−1 (λ is the laser excitation wavelength, and ID and IG are the peak intensities of the D and G bands). It allows a indirect assessment of the graphene defects density in terms of crystallite size. Based on the data from Fig. 3, crystal sizes of GO, G-OH, G-initiator, G-HEMA, and G-HECA derivatives were calculated as 26.6, 29.5, 26.9, 24.4, 24.4, and 24.2 nm, respectively. The hydrazine reduction process partially restores the aromaticity of graphene nanosheets (La increases from 26.6 to 29.5 nm). The crystallite size reduction resulting from the polymer brushes covalent grafting appears to be limited (from 26.9 to 24.2 nm), in agreement with what the reference does.38 Moreover, the close La value of the G-HEMA, G-HECA1 and G-HECA2 implies some extent preservation of the graphitic nanocrystallite size even through different polymerization condition. The Raman and XRD (see ESI Fig. S2†) analysis indicate that the ARGET ATRP process induces no significant reduction in graphene nanosheets quality.
Protein absorption and cell attachment
Smooth and uniform thin film was achieved by spray coating graphene nanosheets/DMF solutions onto preheated coverslips, as shown in Fig. 4. The SEM images present the surface morphology of the obtained film: the GO film surface is smooth and few micron-scale aggregations are observed. The obtained G-HEMA, G-HECA1 and G-HECA2 film surfaces present rough topography, and much more small aggregations are observed should cause consequential influences on cell spatial distribution and differentiation.32 The aggregation is expected to arise from strong van der Waals interactions between the chemically reduced graphene nanosheets (see ESI, Fig. S2†). In the absence of external disturbances (e.g., from sonication) and sufficient electrostatic repulsion, graphene aggregation may have been retained during polymerization, yielding single- or multi-layer graphene nanosheets.42 However, the grafted polymer brushes have significant influence on the dispersibility of the hybrid materials in solvents. G-HEMA, G-HECA1 and G-HECA2 present limited dispersibility in water and ethanol, but form stable yellow solution in DMF due to the hydrophilic properties of HEMA brush backbone structure (see ESI, Fig. S3†). | ||
| Fig. 4 (a) Schematic representation of the spray coating; (b) SEM images of spray coated surfaces constituted by (i) GO, (ii) G-HEMA, (iii) G-HECA1, and (iv) G-HECA2. | ||
Interfacial protein absorption is critical for regulating cellular adhesion and proliferation behaviors on the graphene substrate surface. Thus FBS protein absorption experiment was performed as shown in Fig. 5. After 12 hour incubation, G-HECA1 and G-HECA2 sample present higher FBS absorption. Specifically, the amount of absorbed FBS protein on G-HECA2 is about three fold higher than that on G-HEMA, and about 50% higher than that on GO and glass, whereas G-HEMA presents the lowest protein absorption. The protein absorption on the material surface highly depends on the surface physicochemical characteristics and non-covalent interactions (electrostatic forces, hydrogen bonding, hydrophobic interactions, etc.) induced by the material's molecular structure.43 The carboxyl groups functionalization on the graphene surface introduces charged and electronegative regions and enabled the formation of hydrogen bonds with proteins. And there also have possible hydrophobic interactions between proteins and aromatic graphene nanocrystallites.44 The poly(HEMA) brushes backbone has extensive hydrogen bonding to water, and results in a partially structured hydration layer above the graphene surface. Absorbing a protein molecule requires the disruption of this structured water layer and therefore leads to an entropically unfavorable compression of the brushes layer toward the graphene surface. This hydration effect inhibits the FBS protein attachment on the graphene surface, and explains why G-HEMA sample present the lowest FBS protein absorption. In the GO, G-HECA1 and G-HECA2 comparison, all four kinds of non-covalent interactions, e.g. hydration effect, electrostatic forces, hydrogen bonding, and hydrophobic interaction, are taken into consideration. A possible reason for the highest FBS absorption on the highest-carboxyl-groups-content G-HECA2 may be due to the synergetic effect of electrostatic forces, hydrogen bonding, and hydrophobic interactions. And the FBS absorption difference between G-HECA1 and G-HECA2 should be induced by the competition of electrostatic repulsion and hydrogen bonding.7 The FBS protein absorption experiment proves the feasibility of using the obtained G-HEMA, G-HECA1 and G-HECA2 samples as scaffold substrates.
NIH-3T3 cells were seeded on the spray coating surfaces at density 2 × 104 cells per cm2, and cultured in high-glucose 3T3 medium for 3 days. An advantage of using spray coating method to prepare the scaffold substrate is the resultant transparency allows for direct microscopic observation. Bright field images of NIH-3T3 cells on different substrates are shown in Fig. 6. The observation indicates that NIH-3T3 cells adhere and spread well on the different substrates' surface. A high fraction of NIH-3T3 cells present adhering and spreading morphologies, whereas the rest maintain their circular phenotype.
 | ||
| Fig. 6 Optical bright field view of (a) dry coverslip coated with G-HEMA, NIH-3T3 cells growing on coverslip coated by (b) G-HEMA, (c) G-HECA1 and (d) G-HECA2 at day 3 of incubation. | ||
Cell viability and proliferation
After incubation on each substrate for 24 hours and 72 hours, the viability of NIH-3T3 cells was evaluated using Live/Dead staining with calcic-AM (to stain live cells with green color) and ethidium homodimer (to stain dead cells with red color). In Fig. 7(a), the confocal microscopy images reveal that most of the cells are alive, well adhere, and spread out. The proliferation of the adherent NIH-3T3 cells was further investigated by using a MTS cell proliferation assay kit. As shown in Fig. 7(b), the number of adherent NIH-3T3 cells on each substrate keeps increasing with the incubation time extending. At day 1 of incubation, the glass and G-HECA2 show higher cell attachment, while G-HEMA presents the lowest adherent cells number. At day 3 of incubation, the number of adherent cells on GO, G-HEMA and G-HECA1 substrates is lower than the others, especially for the G-HEMA substrate which still presents the lowest adherent cell number. At day 5 of incubation, NIH-3T3 cells on all graphene substrates have greatly proliferate, and the adherent cells number on GO, G-HEMA, and G-HECA1 are close. The cell viability experiment indicates that the resident 0.1 mol% Sn element on G-HEMA, G-HECA1 and G-HECA2 has no significant deleterious influence of on the NIH-3T3 cells behavior. In contrast, the number of adherent cells on different substrates present clear dependence relationship to the substrates' FBS proteins absorption capacity. Because of the strong hydration effect of grafted poly(HEMA) brushes, G-HEMA present a short-term non-fouling property that inhibits NIH-3T3 cells adhesion and proliferation at the first 3 day incubation. For G-HECA1 and G-HECA2, higher-carboxyl-group-content brush structure enhances NIH-3T3 cell adhesion and proliferation. Especially, G-HECA2 present the highest adherent cells number at day 5 of incubation. In addition, the cell shape indexes (CSI) were calculated to quantitate the cell morphology. The CSI is the ratio between the cell length and width, reflecting the circularity of a cell with a value between 0, which indicates a linear shape, and 1, which denotes a circle. In Fig. 7(c), the CSI value of NIH-3T3 cells on different substrates is close, in a range of 0.4 to 0.6, indicating elongated morphology characteristic of the cells. Similar cell attachment and proliferation results are also observed in human umbilical vein endothelial cells (HUVECs) culture experiment (see ESI, Fig. S4†). The cell viability experiments demonstrate that the graphene nanosheets grafted with poly(HEMA) brushes meet the minimum biocompatible requirements to support fibroblast cells, even human cells, attachment and proliferation. In summary, the ARGET ATRP approach provides a versatile way to obtain biocompatible and structure controllable graphene–polymer brushes hybrid.Conclusions
Based on surface-initiated activators regenerated by the electron transfer atom transfer radical polymerization (ARGET ATRP) technique, an approach has been developed to graft poly(HEMA) brushes onto graphene nanosheets by using tin(II) 2-ethylhexanoate as reducing agent. Uniformly distributed and structure controllable poly(HEMA) brushes are grafted without trace amounts of Cu(II) complex residual on graphene substrates. The brush backbone structure change presents great potential to tune the interfacial interaction between graphene nanosheets and proteins. And further cells viability assays indicate that the obtained graphene nanosheets meet the biocompatible requirements to support fibroblast cells, even human cells, attach and proliferate. The approach and the graphene–polymer brushes hybrid developed in this work should open new opportunities for broader biomedical applications of carbon nanomaterials.Acknowledgements
The authors sincerely acknowledge the support of Fundamental Research Funds for the Central Universities (22A201514030), China Postdoctoral Science Foundation (2015M571504), National Natural Science Foundation of China (5150306, 551273065) and the Wisconsin Institute for Discovery in University of Wisconsin–Madison.Notes and references
- C. Chung, Y. K. Kim, D. Shin, S. R. Ryoo, B. H. Hong and D. H. Min, Acc. Chem. Res., 2013, 46, 2211–2224 CrossRef CAS PubMed
.
- V. C. Sanchez, A. Jachak, R. H. Hurt and A. B. Kane, Chem. Res. Toxicol., 2012, 25, 15–34 CrossRef CAS PubMed
.
- Y. Zhang, T. R. Nayak, H. Hong and W. Cai, Nanoscale, 2012, 4, 3833–3842 RSC
.
- H. Shen, L. M. Zhang, M. Liu and Z. J. Zhang, Theranostics, 2012, 2, 283–294 CrossRef CAS PubMed
.
- M. S. Catherine, M. Joey, A. Farid, L. Alex, C. A. Rigoberto and F. R. Debora, Nanotechnology, 2012, 23, 395101 CrossRef PubMed
.
- S. Agarwal, X. Zhou, F. Ye, Q. He, G. C. Chen, J. Soo, F. Boey, H. Zhang and P. Chen, Langmuir, 2010, 26, 2244–2247 CrossRef CAS PubMed
.
- X. T. Shi, H. X. Chang, S. Chen, C. Lai, A. Khademhosseini and H. K. Wu, Adv. Funct. Mater., 2012, 22, 751–759 CrossRef CAS
.
- Y. Wang, W. C. Lee, K. K. Manga, P. K. Ang, J. Lu, Y. P. Liu, C. T. Lim and K. P. Loh, Adv. Mater., 2012, 24, 4285–4290 CrossRef CAS PubMed
.
- T. R. Nayak, H. Andersen, V. S. Makam, C. Khaw, S. Bae, X. Xu, P. L. Ee, J. H. Ahn, B. H. Hong, G. Pastorin and B. Ozyilmaz, ACS Nano, 2011, 5, 4670–4678 CrossRef CAS PubMed
.
- S. R. Ryoo, Y. K. Kim, M. H. Kim and D. H. Min, ACS Nano, 2010, 4, 6587–6598 CrossRef CAS PubMed
.
- S. Y. Park, J. Park, S. H. Sim, M. G. Sung, K. S. Kim, B. H. Hong and S. Hong, Adv. Mater., 2011, 23, 263–267 CrossRef PubMed
.
- G. Y. Chen, D. W. Pang, S. M. Hwang, H. Y. Tuan and Y. C. Hu, Biomaterials, 2012, 33, 418–427 CrossRef CAS PubMed
.
- W. C. Lee, C. H. Lim, H. Shi, L. A. Tang, Y. Wang, C. T. Lim and K. P. Loh, ACS Nano, 2011, 5, 7334–7341 CrossRef CAS PubMed
.
- M. M. Stevens and J. H. George, Science, 2005, 310, 1135–1138 CrossRef CAS PubMed
.
- R. Barbey, L. Lavanant, D. Paripovic, N. Schüwer, C. Sugnaux, S. Tugulu and H.-A. Klok, Chem. Rev., 2009, 109, 5437–5527 CrossRef CAS PubMed
.
- M. Krishnamoorthy, S. Hakobyan, M. Ramstedt and J. E. Gautrot, Chem. Rev., 2014, 114, 10976–11026 CrossRef CAS PubMed
.
- S. Tugulu, P. Silacci, N. Stergiopulos and H. A. Klok, Biomaterials, 2007, 28, 2536–2546 CrossRef CAS PubMed
.
- B. P. Harris, J. K. Kutty, E. W. Fritz, C. K. Webb, K. J. Burg and A. T. Metters, Langmuir, 2006, 22, 4467–4471 CrossRef CAS PubMed
.
- Y. Mei, T. Wu, C. Xu, K. Langenbach, J. Elliott, B. Vogt, K. Beers, E. Amis and N. Washburn, Langmuir, 2005, 21, 12309–12314 CrossRef CAS PubMed
.
- J. Sha, E. S. Lippmann, J. McNulty, Y. Ma and R. S. Ashton, Biomacromolecules, 2013, 14, 3294–3303 CrossRef CAS PubMed
.
- A. Hucknall, S. Rangarajan and A. Chilkoti, Adv. Mater., 2009, 21, 2441–2446 CrossRef CAS
.
- S. H. Lee, D. R. Dreyer, J. An, A. Velamakanni, R. D. Piner, S. Park, Y. Zhu, S. O. Kim, C. W. Bielawski and R. S. Ruoff, Macromol. Rapid Commun., 2010, 31, 281–288 CrossRef CAS PubMed
.
- G. Goncalves, P. A. A. P. Marques, A. Barros-Timmons, I. Bdkin, M. K. Singh, N. Emami and J. Gracio, J. Mater. Chem., 2010, 20, 9927–9934 RSC
.
- M. Mathesh, J. Q. Liu, N. D. Nam, S. K. H. Lam, R. K. Zheng, C. J. Barrow and W. R. Yang, J. Mater. Chem. C, 2013, 1, 3084–3090 RSC
.
- L. Ren, S. Huang, C. Zhang, R. Wang, W. W. Tjiu and T. Liu, J. Nanopart. Res., 2012, 14, 940–949 CrossRef
.
- J. M. Bak and H.-i. Lee, Polymer, 2012, 53, 4955–4960 CrossRef CAS
.
- J. M. Bak, T. Lee, E. Seo, Y. Lee, H. M. Jeong, B. S. Kim and H. I. Lee, Polymer, 2012, 53, 316–323 CrossRef CAS
.
- M. Fang, Z. Chen, S. Wang and H. Lu, Nanotechnology, 2012, 23, 085704 CrossRef PubMed
.
- S. Stohs, Free Radical Biol. Med., 1995, 18, 321–336 CrossRef CAS PubMed
.
- S. J. Jhaveri, M. R. Hynd, N. Dowell-Mesfin, J. N. Turner, W. Shain and C. K. Ober, Biomacromolecules, 2008, 10, 174–183 CrossRef PubMed
.
- K. Riehemann, S. W. Schneider, T. A. Luger, B. Godin, M. Ferrari and H. Fuchs, Angew. Chem., Int. Ed., 2009, 48, 872–897 CrossRef CAS PubMed
.
- T. R. Nayak, L. Jian, L. C. Phua, H. K. Ho, Y. Ren and G. Pastorin, ACS Nano, 2010, 4, 7717–7725 CrossRef CAS PubMed
.
- W. Jakubowski, K. Min and K. Matyjaszewski, Macromolecules, 2006, 39, 39–45 CrossRef CAS
.
- W. Jakubowski and K. Matyjaszewski, Angew. Chem., Int. Ed. Engl., 2006, 45, 4482–4486 CrossRef CAS PubMed
.
- K. Matyjaszewski, W. Jakubowski, K. Min, W. Tang, J. Huang, W. A. Braunecker and N. V. Tsarevsky, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15309–15314 CrossRef CAS PubMed
.
- K. Matyjaszewski, H. Dong, W. Jakubowski, J. Pietrasik and A. Kusumo, Langmuir, 2007, 23, 4528–4531 CrossRef CAS PubMed
.
- J. R. Lomeda, C. D. Doyle, D. V. Kosynkin, W. F. Hwang and J. M. Tour, J. Am. Chem. Soc., 2008, 130, 16201–16206 CrossRef CAS PubMed
.
- M. Fang, K. G. Wang, H. B. Lu, Y. L. Yang and S. Nutt, J. Mater. Chem., 2010, 20, 1982–1992 RSC
.
- K. Min, H. F. Gao and K. Matyjaszewski, Macromolecules, 2007, 40, 1789–1791 CrossRef CAS
.
- L. G. Cançado, K. Takai, T. Enoki, M. Endo, Y. A. Kim, H. Mizusaki, A. Jorio, L. N. Coelho, R. Magalhães-Paniago and M. A. Pimenta, Appl. Phys. Lett., 2006, 88, 163106 CrossRef
.
- M. A. Pimenta, G. Dresselhaus, M. S. Dresselhaus, L. G. Cancado, A. Jorio and R. Saito, Phys. Chem. Chem. Phys., 2007, 9, 1276–1291 RSC
.
- M. Fang, K. G. Wang, H. B. Lu, Y. L. Yang and S. Nutt, J. Mater. Chem., 2009, 19, 7098–7105 RSC
.
- T. Y. Chang, V. G. Yadav, S. De Leo, A. Mohedas, B. Rajalingam, C. L. Chen, S. Selvarasah, M. R. Dokmeci and A. Khademhosseini, Langmuir, 2007, 23, 11718–117125 CrossRef CAS PubMed
.
- J. Liu, S. Fu, B. Yuan, Y. Li and Z. Deng, J. Am. Chem. Soc., 2010, 132, 7279–7281 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: TGA thermograms and XRD of the different materials, UV-vis spectra of the different suspensions, and confocal microscopic observation of Live/Dead stained HUVECs growing on different substrates. See DOI: 10.1039/c6ra04223f |
| This journal is © The Royal Society of Chemistry 2016 |




