Combined experimental/theoretical study of d-glucosamine promoted Ullmann-type C–N coupling catalyzed by copper(i): does amino really count?†
Abstract
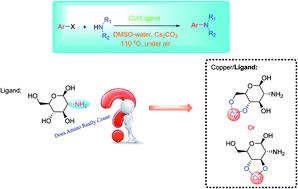
Ullmann type C–N coupling reaction catalyzed by copper(I) with D-glucosamine derivatives as promoters was studied by means of combined experimental/theoretical investigation. The catalytic role of D-glucosamine was addressed. In contrast with previous speculations, the amino group may not count in the catalytic cycle in which the oxidative addition/reductive elimination mechanism works. Experimental results are in good agreement with theoretical findings. Extensive work indicates the wide applicability of the C–N coupling strategy exploited in this work.

 Please wait while we load your content...
Please wait while we load your content...