Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A rapid and sensitive profiling of free fatty acids using liquid chromatography electrospray ionization tandem mass spectrometry (LC/ESI-MS/MS) after chemical derivatization†
Hyuck Jun
Mok‡
a,
Jae Won
Lee‡
a,
Raju
Bandu
a,
Hong Seok
Kang
b,
Kyun-Hwan
Kim
b and
Kwang Pyo
Kim
*a
aDepartment of Applied Chemistry, The Institute of Natural Science, College of Applied Science, Kyung Hee University, Yongin, 446-701, Republic of Korea. E-mail: kimkp@khu.ac.kr
bDepartment of Pharmacology, School of Medicine, Konkuk University, Seoul 143-701, Republic of Korea
First published on 22nd March 2016
Abstract
Free fatty acids (FFAs) have diverse roles in cellular energy and signaling and they are critical molecules in various biological states. Due to the poor ionization efficiency of FFAs under electrospray ionization mass spectrometry (ESI-MS) conditions, it is a challenging aspect to construct a robust platform for profiling of various FFAs in biological samples using liquid chromatography ESI-MS. In the present study, we applied trimethylsilyldiazomethane (TMSD) derivatization to improve ionization efficiencies in the profiling of FFAs. Multiple reaction monitoring (MRM) was used for the selective quantification of methylated FFAs. The optimal TMSD methylation was validated for a reliable FFA profiling. Furthermore, the high-throughput analysis of FFAs was successfully performed in short analysis and derivatization times. To verify the utility and effectiveness of the developed method, we compared both methylation and nonmethylation (intact FFA) data in the profiling of FFAs in mice liver and plasma. It is noteworthy that the methylation derivatization provided better results in FFA profiling. Further, we performed statistical data analysis where HBV and mock mice tissues were discriminated when the methylated FFAs data were used. In the lipidomics field, the present method can also be applied for the profiling of FFAs in biological samples for biomarker discovery. The present validated LC/ESI-MS/MS assay method may also be used for FFA profiling modeling studies in other biomedical samples.
Introduction
Free fatty acids (FFAs), categorized as a class of lipids,1 provide an important energy source as nutrients,2 and they also function as signalling molecules in various cellular processes including insulin secretion.3 FFAs are biomedical indicators of the abnormal lipid metabolism in various metabolic diseases including diabetes mellitus.4 Several researchers have applied FFA profiling to find biomarkers for the diagnosis and characterization of various metabolic diseases.5–7 In some studies, FFAs were also profiled to find the potential biomarkers of Alzheimer's disease,8 coronary heart disease9 and other diseases.Mass spectrometry (MS) has been widely used for the profiling of FFAs in biological samples.10,11 Gas chromatography (GC) coupled to electron ionization (EI) MS was usually applied to analyse fatty acid methyl ester (FAME) which is one of the most common fatty acid derivatives.12–14 By using this technique, the separation of positional and geometrical (cis/trans) isomers was also achieved in approximately 20 min.12 In addition to GC-MS, liquid chromatography (LC)-MS has also been established as an effective analytical technique in the profiling of FFAs with short run times.15 The use of electrospray ionization (ESI) MS alone, which is a soft ionization technique, only provides the information of molecular ions. Thus, tandem MS (MS/MS) is generally applied for the sensitive and selective analysis of FFAs.16,17 FFAs have been analysed in the negative ion mode and there is a limitation to the ionization efficiency. In the previous study, barium acetate was used as the cationization agent for the sensitive profiling of FFAs in the positive ion multiple reaction monitoring (MRM) mode using triple quadrupole mass spectrometer.18 Some researchers, Lee et al., and Pettinella et al., have demonstrated that trimethylaminoethyl ester iodide derivatization provided good sensitivity in the analysis of FFAs.19,20 Zhou et al. has also demonstrated that isotope-labelling derivatization by using 2,4-dimethoxy-6-piperazin-1-yl pyrimidine (DMPP) could improve ionization efficiency for analysing of FFAs.21 Yang et al. derivatized FFAs with 2-bromo-1-methylpyridinium iodide and 3-carbinol-1-methylpyridinium iodide, forming 3-acyloxymethyl-1-methylpyridinium iodide (AMMP). They observed that the detection sensitivity was approximately 2500-folds higher in positive-ion mode ionization than negative-ion mode.22 Similarly, Bollinger et al. have reported a method on conversion of FFAs carboxylic acid group in to an amide bearing permanent positive charge, N-(4-aminomethylphenyl)pyridinium (AMPP) and that method showed approximately 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-folds increased detection sensitivity in positive ion mode ionization when compared to negative ion mode.23 However, those methods are complex and time-consuming processes to derivatize FFAs.
000-folds increased detection sensitivity in positive ion mode ionization when compared to negative ion mode.23 However, those methods are complex and time-consuming processes to derivatize FFAs.
In particular, a rapid and sensitive profiling method is required for the analysis of FFAs in clinical samples. The chemical derivatization can significantly increase the sensitivity and specificity of GC-MS and LC/ESI-MS method to analyse highly acidic compounds such as phosphopeptides, phospholipids and FFAs.24–29 Particularly, trimethylsilyldiazomethane (TMSD) derivatization method is extremely simple and it was applied for analysis of phosphopeptides26 and phospholipids29 to improve detection sensitivity. Recently, Lee et al. also demonstrated TMSD derivatization method that could improve peak shape and detection limits of many lipid classes.29 In the present study, LC/ESI-MS/MS method with TMSD methylation was used for the profiling of FFAs. The utility and effectiveness of the developed method was verified by the profiling of FFAs in plasma and liver samples.
Liver is a core organ which plays a key role in lipid metabolism and has various enzymes related to lipid synthesis such as fatty acid synthase (FAS), a key enzyme for FFA.30 Hepatitis B virus can cause various liver diseases such as cirrhosis and hepatocellular carcinoma.31 It is known that HBV virus disrupts various lipid synthesis-related enzymes in liver.32–34 Therefore, we have applied our developed method to HBV and mock mice tissues to identify FFA changes.
Experimental
Materials
HPLC-grade acetonitrile, methanol, water and isopropanol were purchased from J.T. Baker (Avantor Performance Material, Inc, PA, USA). Trimethylsilyldiazomethane (TMSD) reagent was purchased from Sigma-Aldrich, St. Louis, MO, USA. The FFAs, palmitic acid (16:0), oleic acid (18:1) and arachidonic acid (20:4) were purchased from Sigma-Aldrich St. Louis, MO, USA. The FFA standard, arachidonic acid-d8 (20:4-d8) was purchased from Cayman chemical, Ann Arbor, MI. The 26 G needle with 3 mL syringes were purchased from KOVAX syringe, Korea vaccine CO, Seoul, Korea. Zoletil and xylazine were purchased from Virbac S.A, France and Sigma-Aldrich Co. LLC., USA, respectively. Dulbecco's phosphate-buffered saline (DPBS) was purchased from Thermo Fisher Scientific Inc., USA.Animal samples
All mice used for our study are maintained in Konkuk University of Seoul, Republic of Korea according to standard animal care protocols and fed normal laboratory chow and tap RO water. All mice related experiments were carried out in accordance with Konkuk University IACUC (Institutional Animal Care and Use Committees). The four week aged female mice (C57BL/6) were purchased from Nara biotech co., ltd (Seoul, Korea). Animals were housed at temperature 23 ± 2 °C, with relative humidity of 50 ± 10% and light controlled environment. To obtain plasma samples, mice were anesthetized by intraperitoneal injection of 200 μL of zoletil–xylazine (40 mg mL−1)–DPBS solution (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90; v/v) and plasma was collected by cardiac puncture and stored at −80 °C. In addition, livers particularly right lateral lobe were also excised from the same set of mice and washed with PBS to remove the plasma and connective tissue. After that they blotted dry with filter paper wiper and then stored at −80 °C until analysis.
90; v/v) and plasma was collected by cardiac puncture and stored at −80 °C. In addition, livers particularly right lateral lobe were also excised from the same set of mice and washed with PBS to remove the plasma and connective tissue. After that they blotted dry with filter paper wiper and then stored at −80 °C until analysis.
For applying the developed FFAs profiling method to diseased animal models, we made acute hepatitis B infected mice using male BALB/c mice (six weeks old, 18–20 g). The hepatitis B virus infection model of mice and its mock were used. In this model, HBV (hepatitis B virus) plasmid DNA (pHBV 1.2) and control vector (pGEM-4z) were used. A total of 25 μg of plasmid DNA diluted in PBS which is equivalent to 10% of mice body were hydrodynamically injected into mice tails. To make the injection easier, the mice were placed in cylinder-shaped restraining device to prevent their moving and vasodilate veins of their tails by using its light-bulb device. The injection was performed with high pressure within 5 seconds by using a 26 G needle with 3 mL syringe as described by Park E. S. et al.35 The mice were sacrificed 3 days after hydrodynamic injection and liver were extracted as described above.
Sample preparation
Each lipid standard was dissolved in methanol and stored at −20 °C. We compared three common lipid extraction methods to select the better one for analysing FFAs.First, we applied Bligh and Dyer method which is the most common lipid extraction method.36 In this method, a 750 μL of chloroform/methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2; v/v) was added to 50 μL of plasma samples and 50 mg of liver tissues which were taken in different Eppendorf tubes (1.5 mL). Next, a 10 μL of arachidonic acid-d8 (internal standard (IS)) solution was added to above plasma and tissue samples at a concentration of 10 ng μL−1. In the case of liver tissues, an additional step of homogenization was performed after the addition of chloroform and methanol. After vortexing for 1 min, samples were incubated in ice for 10 min. After that a 250 μL of chloroform and 450 μL of water were added to both plasma and tissue sample tubes. After the centrifugation (13
2; v/v) was added to 50 μL of plasma samples and 50 mg of liver tissues which were taken in different Eppendorf tubes (1.5 mL). Next, a 10 μL of arachidonic acid-d8 (internal standard (IS)) solution was added to above plasma and tissue samples at a concentration of 10 ng μL−1. In the case of liver tissues, an additional step of homogenization was performed after the addition of chloroform and methanol. After vortexing for 1 min, samples were incubated in ice for 10 min. After that a 250 μL of chloroform and 450 μL of water were added to both plasma and tissue sample tubes. After the centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 × g, 2 min at 4 °C), the organic phases were collected into different Eppendorf tubes.
800 × g, 2 min at 4 °C), the organic phases were collected into different Eppendorf tubes.
Secondly, we performed extremely simple lipid extraction method where we used only methanol as extraction solvent. For about 50 μL of plasma sample, 10 μL of IS followed by 1 mL of ice cold methanol was added. After that, the sample mixture was vortexed for 30 s and incubated for 10 min in ice. After centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 5 min at 4 °C), about 1 mL of supernatant was collected into another Eppendorf tube. In the case of liver tissue, approximately 50 mg of tissue was homogenized in 1 mL of ice cold methanol and 10 μL of IS solution was added to it. Further, tissue samples were mixed for 30 s and incubated for 10 min in ice. The centrifugation was performed at 10
000 × g, 5 min at 4 °C), about 1 mL of supernatant was collected into another Eppendorf tube. In the case of liver tissue, approximately 50 mg of tissue was homogenized in 1 mL of ice cold methanol and 10 μL of IS solution was added to it. Further, tissue samples were mixed for 30 s and incubated for 10 min in ice. The centrifugation was performed at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 5 min at 4 °C and the supernatants were collected in to 1.5 mL collection tubes.37
000 × g for 5 min at 4 °C and the supernatants were collected in to 1.5 mL collection tubes.37
Lastly, Dole's extraction method was used for FFA analysis in plasma and liver tissues.38 Plasma and liver tissues were mixed separately with 500 μL of the Dole's mixture (methanol, n-hexane, phosphoric acid (2 mol L−1), 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v)) and IS solution. An additional step of homogenization was performed in case of liver tissues, as described above in the context of Bligh & Dyer and methanol extraction methods. After vortexing, samples were incubated for 10 min at room temperature. Then 300 μL of water and 200 μL of n-hexane were added and centrifuged for 5 min at 13
1 (v/v)) and IS solution. An additional step of homogenization was performed in case of liver tissues, as described above in the context of Bligh & Dyer and methanol extraction methods. After vortexing, samples were incubated for 10 min at room temperature. Then 300 μL of water and 200 μL of n-hexane were added and centrifuged for 5 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. The organic supernatants were collected into 1.5 mL tubes. All extracted FFA samples were divided into two; one of them was used for negative ion mode MS analysis and another one was used for TMSD methylation. All samples were stored at −80 °C before analysis. For TMSD methylation, the samples were dried in SpeedVac concentrator and reconstituted with 100 μL methanol.
000 rpm. The organic supernatants were collected into 1.5 mL tubes. All extracted FFA samples were divided into two; one of them was used for negative ion mode MS analysis and another one was used for TMSD methylation. All samples were stored at −80 °C before analysis. For TMSD methylation, the samples were dried in SpeedVac concentrator and reconstituted with 100 μL methanol.
TMSD methylation
A simple and rapid TMSD methylation derivatization method was used for the analysis of FFAs in the plasma and liver samples. A solution of TMSD (2 mol L−1) in hexane (50 μL) was mixed in equal proportions with sample and standard (v/v). After vortexing for 30 s, mixture was incubated at 30 °C for 10 min under optimized conditions which has been described in the succeeding text. For quenching the derivatization reaction, glacial acetic acid (6 μL) was added as described previously.29LC-ESI/MS/MS equipment and conditions
The HPLC analysis was performed on an Agilent 1290 infinity series HPLC instrument (Agilent Technologies, USA) equipped with binary pump (G4220A, USA), an autosampler (G4226A, USA), a column compartment (G1316C, USA) and a thermostat (G1330B, USA). The temperature of column oven and autosampler was set at 40 °C and 4 °C, respectively. For the separation of FFAs, Hypersil GOLD column (2.1 × 100 mm ID; 1.9 μm Thermo scientific) was used. The mobile phase solvent A consisted of a acetonitrile/methanol/water mixture (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) with 0.1% (v/v) formic acid and 20 mmol L−1 ammonium formate; and the mobile phase solvent B consisted of isopropanol with 0.1% (v/v) formic acid and 20 mmol L−1 ammonium formate. The flow rate of the mobile phase was 0.25 mL min−1 and the injection volume was 3 μL. A 20 min lipid elution gradient was performed as follows: first 10 min, solvent composition was set at 95% A and 5% B to elute FFAs; followed by a linear gradient to solvent 90% A and 10% B for 2 min and kept for 3 min for elution of other lipids. Finally, the column was equilibrated at 5% solvent B for 5 min before reuse.
2) with 0.1% (v/v) formic acid and 20 mmol L−1 ammonium formate; and the mobile phase solvent B consisted of isopropanol with 0.1% (v/v) formic acid and 20 mmol L−1 ammonium formate. The flow rate of the mobile phase was 0.25 mL min−1 and the injection volume was 3 μL. A 20 min lipid elution gradient was performed as follows: first 10 min, solvent composition was set at 95% A and 5% B to elute FFAs; followed by a linear gradient to solvent 90% A and 10% B for 2 min and kept for 3 min for elution of other lipids. Finally, the column was equilibrated at 5% solvent B for 5 min before reuse.
LC-MS analysis was performed on a triple quadrupole mass spectrometer (QQQ LC-MS 6490 series, Agilent Technologies, USA) equipped with an ESI source which provides high sensitivity by iFunnel technology that consists of three components: a hexabore capillary, Agilent Jet Stream technology, and a dual ion funnel. The typical operating source conditions for MS scan in the positive and negative ion ESI mode were optimized as follows: capillary voltage 4000 V, nozzle voltage 500 V. The nebulizer was set at 40 psig and the nitrogen drying gas was set at a flow rate of 13 L min−1 and the temperature was maintained at 250 °C. For collision-induced dissociation (CID) experiments, the precursor ion of each FFA species was selected using the quadrupole analyser and the product ions were analysed using another quadrupole analyser. Ultra-pure nitrogen was used as collision gas. The collision energies for methylated and nonmethylated FFAs (intact FFA) were also optimized by using FFA standards (8 eV for intact FFAs and 10 eV for methylated FFAs). All the spectra of FFAs were recorded under optimized experimental conditions and the quantitative analysis was performed in multiple reaction monitoring (MRM) mode using computed transitions for methylated and intact FFAs (Table 1).
| Species | Ion mode | MRM transitions | CE (eV) | |
|---|---|---|---|---|
| Q1 m/z | Q3 m/z | |||
| a Internal standard (IS). | ||||
| Intact FFAs | ||||
| C8:0 | Negative | 143 | 99 | −8 |
| C10:0 | Negative | 171 | 127 | −8 |
| C12:0 | Negative | 199 | 155 | −8 |
| C14:0 | Negative | 227 | 183 | −8 |
| C14:1Δ9 | Negative | 225 | 181 | −8 |
| C16:0 | Negative | 255 | 211 | −8 |
| C16:1Δ9 | Negative | 253 | 209 | −8 |
| C18:0 | Negative | 283 | 239 | −8 |
| C18:1n9 | Negative | 281 | 237 | −8 |
| C18:2n6 | Negative | 279 | 235 | −8 |
| C18:3n3 | Negative | 277 | 233 | −8 |
| C18:3n6 | Negative | 277 | 233 | −8 |
| C18:4 | Negative | 275 | 231 | −8 |
| C20:0 | Negative | 311 | 267 | −8 |
| C20:1Δ11 | Negative | 309 | 265 | −8 |
| C20:2Δ11,14 | Negative | 307 | 263 | −8 |
| C20:3n3 or n6 | Negative | 305 | 261 | −8 |
| C20:4n6 | Negative | 303 | 259 | −8 |
| (d8) C20:4n6 Isa | Negative | 311 | 267 | −8 |
| C20:5n3 | Negative | 301 | 257 | −8 |
| C22:0 | Negative | 339 | 295 | −8 |
| C22:1n9 | Negative | 337 | 293 | −8 |
| C22:2Δ13,16 or n6 | Negative | 335 | 291 | −8 |
| C22:3 | Negative | 333 | 289 | −8 |
| C22:4n6 | Negative | 331 | 287 | −8 |
| C22:5n3 | Negative | 329 | 285 | −8 |
| C22:6n3 | Negative | 327 | 283 | −8 |
| C24:0 | Negative | 367 | 323 | −8 |
| C24:1Δ15 or n9 | Negative | 365 | 321 | −8 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Methylated FFAs | ||||
| Methylated-C8:0 | Positive | 159 | 127 | 10 |
| Methylated-C10:0 | Positive | 187 | 155 | 10 |
| Methylated-C12:0 | Positive | 215 | 183 | 10 |
| Methylated-C14:0 | Positive | 243 | 211 | 10 |
| Methylated-C14:1Δ9 | Positive | 241 | 209 | 10 |
| Methylated-C16:0 | Positive | 271 | 239 | 10 |
| Methylated-C16:1Δ9 | Positive | 269 | 237 | 10 |
| Methylated-C18:0 | Positive | 299 | 267 | 10 |
| Methylated-C18:1n9 | Positive | 297 | 265 | 10 |
| Methylated-C18:2n6 | Positive | 295 | 263 | 10 |
| Methylated-C18:3n3 or n6 | Positive | 293 | 261 | 10 |
| Methylated-C18:4 | Positive | 291 | 259 | 10 |
| Methylated-C20:0 | Positive | 327 | 295 | 10 |
| Methylated-C20:1Δ11 | Positive | 325 | 293 | 10 |
| Methylated-C20:2Δ11,14 | Positive | 323 | 291 | 10 |
| Methylated-C20:3n3 | Positive | 321 | 289 | 10 |
| Methylated-C20:3n6 | Positive | 321 | 289 | 10 |
| Methylated-C20:4n6 | Positive | 319 | 287 | 10 |
| Methylated-(d8) C20:4n6 Isa | Positive | 327 | 295 | 10 |
| Methylated-C20:5n3 | Positive | 317 | 285 | 10 |
| Methylated-C22:0 | Positive | 355 | 323 | 10 |
| Methylated-C22:1n9 | Positive | 353 | 321 | 10 |
| Methylated-C22:2Δ13,16 or n6 | Positive | 351 | 319 | 10 |
| Methylated-C22:3 | Positive | 349 | 317 | 10 |
| Methylated-C22:4n6 | Positive | 347 | 315 | 10 |
| Methylated-C22:5n3 | Positive | 345 | 313 | 10 |
| Methylated-C22:6n3 | Positive | 343 | 311 | 10 |
| Methylated-C24:0 | Positive | 383 | 351 | 10 |
| Methylated-C24:1Δ15 or n9 | Positive | 381 | 349 | 10 |
Result and discussion
FFA profiling by LC/MS/MS with TMSD methylation
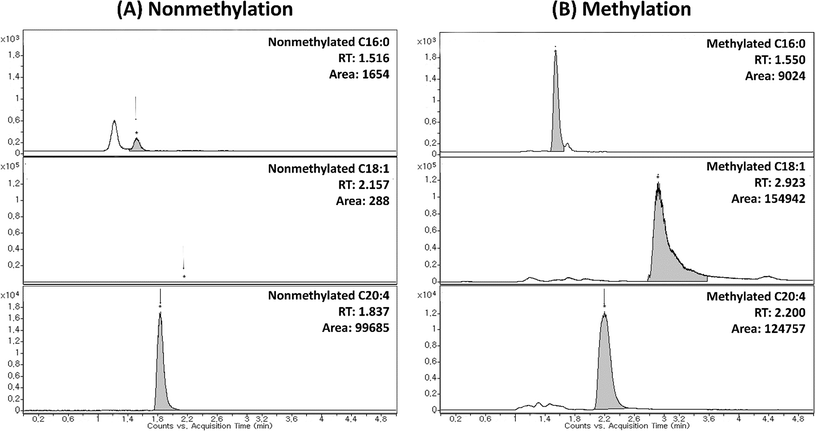
In this study, TMSD methylation was applied to increase the sensitivity of FFA analysis. To the best of our knowledge, this is the first attempt to analyse FFAs in plasma and liver samples with TMSD methylation by LC/MS/MS. Thus, we established the MRM transition (m/z value of precursor ion (Q1) > m/z value of product ion (Q3)) for the profiling of methylated FFAs. The ESI-MS conditions were also optimized in the analysis of methylated FFA standards (palmitic acid-saturated FFA, oleic acid-monounsaturated FFA and arachidonic acid-polyunsaturated FFA). The methylated FFAs were analysed as [M + H]+ ions in positive ion mode whereas intact FFAs were predominately analysed as [M − H]− ions in negative ion mode. The m/z value of the molecular ions of methylated FFAs showed the mass shift of 15 Da to their intact molecular weights which corresponds to the addition of methyl group (Fig. 1). The fragmentation pattern of methylated and intact FFAs were confirmed by using tandem MS analysis. In the negative ion mode, intact FFAs showed similar fragmentation behaviour that was reported earlier by Nagy et al., and other researchers.15,39–42 The diagnostic product ion observed in the negative ion mode MS spectra of FFAs is [M − H-44]− which corresponds to the neutral loss of CO2. On the other hand, the positive ion mode analysis of methylated FFAs showed [M + H-32]+ ion as the base peak which corresponds to the loss of CH3OH. The peak corresponds to the loss CH3OH was selected as Q3 transition in the MRM analysis of all FFAs (Fig. 1). For the profiling of various FFAs, we fixed the MRM transitions of each species based on their abundant characteristic product ions of [M − H − CO2]− and [M + H − CH3OH]+ for intact and methylated FFAs, respectively.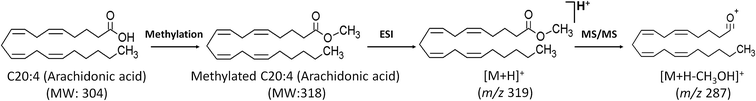 | ||
| Fig. 1 Formation of TMSD derivative of arachidonic acid (C20:4) and its proposed fragmentation pattern. | ||
Further, we also optimized the tested time (10, 15, 20, 25 and 30 min) and temperatures (10, 20, 30, 40, and 50 °C) for TMSD methylation of FFAs. In these experiments, we analysed FFA standards, C16:0 (palmitic acid-saturated FFA), C18:1 (oleic acid-monounsaturated FFA) and C20:4 (arachidonic acid-polyunsaturated FFA) with different derivatization time and temperatures. The arachidonic acid-d8 standard was used as an IS for this analysis. According to different derivatization time and temperature, the efficiency of methylation was estimated by the peak area of FFA standards normalized by IS. 10 min of derivatization time was enough for methylation of FFAs and 30 °C of reaction temperature showed the best efficiency for the derivatization (Table 2). Thus, 10 min and 30 °C were selected as the optimized conditions for methylation of FFAs.
| Species | RTa (min) | Temperatureb (°C) | Reaction time (min) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | 30 | 40 | 50 | 60 | 10 | 15 | 20 | 25 | 30 | ||
| a Retention time. b Average of relative peak area (compound/internal standard) (n = 3). c The values of percentages are in mean ± SD (n = 3). | |||||||||||
| Methylated palmitic acid | 1.6 | 1.61 ± 0.02 c | 1.75 ± 0.02 | 1.83 ± 0.25 | 1.64 ± 0.1 | 1.66 ± 0.3 | 0.2 ± 0.02 | 0.28 ± 0.04 | 0.26 ± 0.03 | 0.52 ± 0.28 | 0.29 ± 0.02 |
| Methylated oleic acid | 2.8 | 7.2 ± 0.53 | 7.4 ± 0.13 | 7.14 ± 0.33 | 7.8 ± 0.19 | 6.96 ± 0.05 | 5.53 ± 0.30 | 5.27 ± 0.19 | 5.21 ± 0.02 | 4.89 ± 0.13 | 4.81 ± 0.12 |
| Methylated arachidonic acid | 2.15 | 0.47 ± 0.02 | 0.46 ± 0.01 | 0.47 ± 0.02 | 0.44 ± 0.02 | 0.41 ± 0.01 | 0.48 ± 0.01 | 0.45 ± 0.02 | 0.42 ± 0.00 | 0.39 ± 0.00 | 0.37 ± 0.01 |
The derivatization efficiency of TMSD methylation for FFA profiling is given in Table 3. The methylated and intact FFAs showed difference in ionization efficiency which may presumably be due to their structural differences. To evaluate the efficiency of TMSD methylation, we compared the peak areas of intact FFAs in both methylated and nonmethylated mixtures. The TMSD methylation efficiency of each FFA was described as follows: the percentage of methylated FFAs ((peak area of compound in nonmethylated FFAs − peak area of nonmethylated compound in methylated FFAs)/peak area of a compound in a nonmethylated FFAs × 100 (%)) (Table 3). All FFA standards showed high TMSD derivatization efficiency as shown in Table 3 (the lowest efficiency was 82.9%).
| Species | The percentage of methylated compoundsa (%) |
|---|---|
| a ((Peak area of compound in nonmethylated FFAs − peak area of nonmethylated compound in methylated FFAs)/peak area of a compound in a nonmethylated FFAs) × 100 (%). b The values of percentages are in mean ± SD (n = 6). | |
| Palmitic acid | 96.15 ± 5.93b |
| Oleic acid | 82.90 ± 5.69 |
| Arachidonic acid | 99.95 ± 0.01 |
Validation study
The performance of TMSD methylation was validated to confirm whether the methylation is applicable to FFA profiling in biological samples (Table 4). First, the reproducibility of methylation was estimated by calculating the relative standard deviation (% RSD) of 1 ng of FFA standards in intra- and inter-day experiments. In the analysis of four FFA standards, the % RSDs of the retention time and peak areas showed less than 15.1% and 1.6% RSD in both intra- and inter-day studies, respectively (Table 4). Second, the sensitivity of the FFA profiling with and without methylation were also compared. The limitation of detection (LOD) and coefficient (R2) were calculated in terms of methylated and nonmethylated conditions. As a result, both methylated and intact FFA standards showed good coefficient (higher than 0.975). However, in the case of LOD, methylated standards showed better sensitivity than nonmethylated standards. It can be that noted that the intact oleic acid showed 25 fold higher LOD than the corresponding methylated standard (Table 4). Furthermore, sensitivity was significantly increased in case of palmitic acid and oleic acid standards (Fig. 2). Even though the intact arachidonic acid showed same LOD and sensitivity as similar to its methylated form, its average peak area was smaller than TMSD (Fig. 2). The results of lower LOD, good coefficient and low % RSD of inter and intraday variation supported the reproducibility and reliability of our developed method. The matrix effect has been evaluated, where the matrix factor was ranged from 0.86–1.05. These data indicate that there was no significant endogenous interference. The stabilities of samples were within the limits and the mean percentage changes were less than ±10% from their nominal concentrations. Our results indicate that the samples were stable at 4 °C for 24 h in autosampler at −80 °C for 30 days. The samples were stable at room temperature for 6 h.| Species | RTa (min) | Intraday variationb | Interday variationc | Coefficient (R2) | LOD (ng) | Linearity range (ng) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| RT | Peak area | RT | Peak area | Intact | Methylated | Intact | Methylated | |||
| a Average of retention time (n = 9). b % RSD (n = 9) of intraday variation. c % RSD (n = 27) of interday variation (3 days). | ||||||||||
| Palmitic acid | 1.59 | 1.6 | 5.6 | 1.1 | 15.1 | 0.9931 | 0.9869 | 1600 | 800 | 800–25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Oleic acid | 2.8 | 0.2 | 5 | 0.4 | 7.1 | 0.9819 | 0.9927 | 1000 | 40 | 40–25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Arachidonic acid | 2.15 | 0.4 | 4.6 | 0.4 | 4.8 | 0.9909 | 0.9943 | 100 | 100 | 100–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| (d8) arachidonic acid-IS | 2.09 | 0.9 | 1.7 | 0.6 | 15 | 0.991 | 0.975 | 100 | 100 | 100–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Comparison of extraction methods of FFA
For the optimization of extraction method, we compared three common lipid extraction methods (Method 1: Bligh and dyer; Method 2: extraction with methanol; Method 3: Dole's procedure) with liver sample from C57BL/6 mice. A 50 mg each of liver tissue was used for each experiment to compare the extraction efficiency. As shown in Fig. 3, the overall area of FFAs extracted from methods 1 and 2 showed higher value than method 3. When we compared the extraction methods 1 and 2, they showed similar results in terms of the number of identified FFAs (14 FFAs in both methods) and relative area (area of FFA/area of IS) of FFAs. As these extraction methods are very complex and tedious time consuming processes which may cause the experimental handling error, we moved to the simple extraction method which involves methanol as extraction solvent. Finally, methanol extraction method was selected due to its simplicity and good performance (Fig. 3).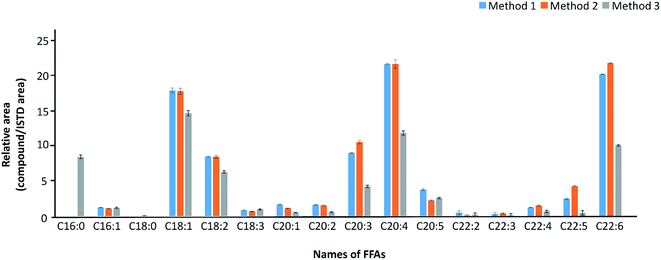 | ||
| Fig. 3 Comparison among three common FFA extraction methods (Method 1: Bligh & dyer; Method 2: extraction using methanol; Method 3: Dole's mixture). | ||
Application of TMSD methylation for profiling of FFAs in mice liver and plasma
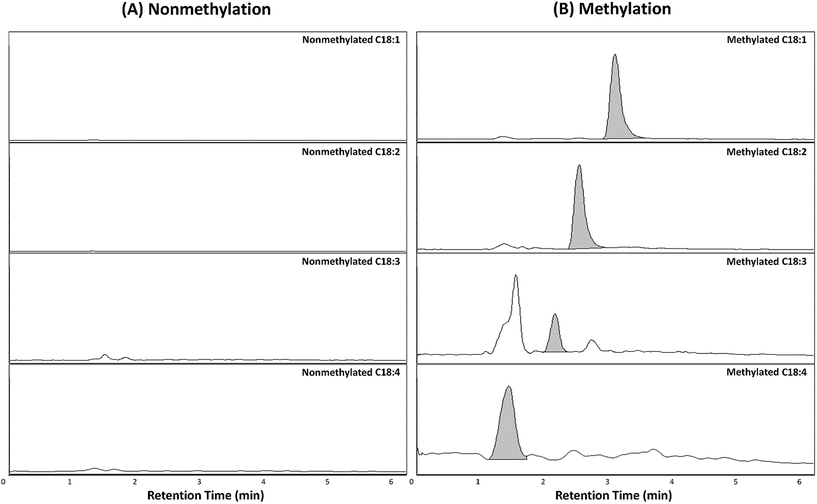
In order to validate the utility of TMSD methylation for the profiling of FFAs in biological samples, we applied our validated method for analysing mice liver and plasma (C57BL/6 mice). The arachidonic acid-d8 (20:4) was used as the IS for the normalization of each FFA species. The relative area (area of FFA/area of IS) of FFA species was characterized for the detailed phenotype of mice samples. Each sample (plasma and tissues) was analysed four times (n = 4). Furthermore, the results of FFA profiling with and without methylation were compared. Compared to intact FFAs, the RT of methylated FFAs was slightly delayed due to the formation of methyl ester structure (Fig. 2 and 4). It can be noted that the methylated FFAs were eluted within 5 min even though their RTs were delayed. Thus, it is appropriate to perform the high-throughput profiling of FFAs. Upon methylation, 11 FFAs in mice plasma and 16 FFAs in mice liver were successfully analysed. On the other hand, in the case of intact FFAs, six FFAs in mice liver and plasma were detected (Table 5). It is noteworthy that five methylated FFAs (C14:1, 18:1, 18:2, 18:3, and 18:4) were additionally analysed in mice plasma when compared to intact FFAs. Furthermore, ten FFAs (C16:1, 18:0, 18:1, 18:2, 18:3, 18:4, 20:1, 20:2, 22:2, and 22:3) were only detected by TMSD methylation in mice liver. The average relative areas of methylated FFAs are much higher than the intact FFAs (Fig. 4 and Table 5). All the detected FFAs were also validated by tandem mass spectrometric experiments (Fig. 5 and S1†).| Species | RTa (min) | Liverb | Plasmab | |||
|---|---|---|---|---|---|---|
| Intact | Methylated | Intact | Methylated | Intact | Methylated | |
| a Average of retention time. b Average of relative peak area (compound/internal standard) ± S.D (n = 4). c The values of percentages are in mean ± SD (n = 4). | ||||||
| C8:0 | — | — | — | — | — | — |
| C10:0 | — | — | — | — | — | — |
| C12:0 | — | — | — | — | — | — |
| C14:0 | — | — | — | — | — | — |
| C14:1Δ9 | — | 1.6 | — | — | — | 0.48 ± 0.05c |
| C16:0 | — | 3 | — | — | — | — |
| C16:1Δ9 | — | 2.3 | — | 1.09 ± 0.12 | — | — |
| C18:0 | — | 3.7 | — | 0.09 ± 0.10 | — | — |
| C18:1n9 | — | 3 | — | 17.7 ± 1.65 | — | 1.47 ± 0.22 |
| C18:2n6 | — | 2.5 | — | 8.44 ± 0.82 | — | 0.72 ± 0.13 |
| C18:3n3 or n6 | — | 2.1 | — | 0.68 ± 0.10 | — | 0.23 ± 0.04 |
| C18:4 | — | 1.4 | — | 0.91 ± 0.11 | — | 1.10 ± 0.15 |
| C20:0 | — | — | — | — | — | — |
| C20:1Δ11 | — | 3.9 | — | 1.13 ± 0.09 | — | — |
| C20:2Δ11,14 | — | 3.1 | — | 1.54 ± 0.21 | — | — |
| C20:3n3 or n6 | 2.2 | 2.6 | 0.26 ± 0.01 | 10.5 ± 0.94 | 0.01 ± 0.00 | 0.19 ± 0.04 |
| C20:4n6 | 1.9 | 2.3 | 22.2 ± 1.42 | 21.5 ± 2.39 | 1.03 ± 0.01 | 0.15 ± 0.04 |
| C20:5n3 | 1.7 | 1.8 | 5.43 ± 0.25 | 2.26 ± 0.18 | 0.16 ± 0.01 | 23.0 ± 3.07 |
| C22:0 | — | — | — | — | — | — |
| C22:1n9 | — | — | — | — | — | — |
| C22:2Δ13,16 or n6 | — | 4 | — | 0.14 ± 0.03 | — | — |
| C22:3 | — | 3.3 | — | 0.41 ± 0.05 | — | — |
| C22:4n6 | 2.2 | 2.7 | 0.19 ± 0.02 | 1.54 ± 0.20 | 0.02 ± 0.00 | 0.20 ± 0.00 |
| C22:5n3 | 1.9 | 2.4 | 3.40 ± 0.22 | 4.28 ± 0.44 | 0.30 ± 0.01 | 6.26 ± 1.10 |
| C22:6n3 | 1.7 | 2 | 52.7 ± 1.98 | 21.7 ± 2.32 | 2.59 ± 0.11 | 1.55 ± 0.24 |
| C24:0 | — | — | — | — | — | — |
| C24:1Δ15 or n9 | — | — | — | — | — | — |
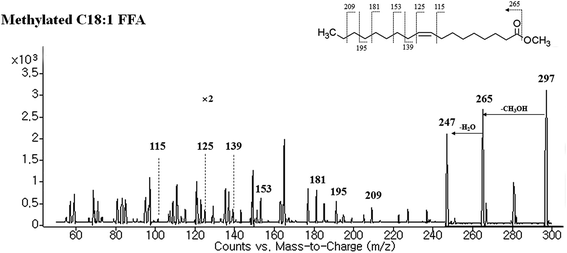 | ||
| Fig. 5 Tandem mass spectra of representative methylated C18:1 FFA (the m/z 297 ([M + H]+) ion corresponds to methylated C18:1 FFA (oleic acid) in positive ion mode). | ||
Application of TMSD methylation in the FFA profiling in HBV infected mice
We also applied the developed method for profiling of FFAs in three samples (HBV infected samples and its mock mice liver tissue). In these experiments, each tissue was analysed four times (n = 4). The peak area of individual FFA species was divided by internal standard for normalization and relative quantification.29,43 The normalized FFA data of mice samples were subjected to principal component analysis (PCA) and hierarchical clustering analysis (HCA) (heat map, dendro gram) which were performed through the web-based data analysis software, MetaboAnalyst 3.0. In addition, the uses of FFA profiling with and without methylation were also compared in this study. When the data used that was obtained by nonmethylation, HBV and mock mice tissues were not well separated in the PCA score plot and HCA (Fig. 6A). On the other hand, HBV and mock mice tissues were clearly distinguished when the data of methylated FFAs was used in the statistical analysis (Fig. 6B). As it can be seen from Fig. 6B, many FFAs were decreased in HBV induced mice and finally we found some significantly changed FFAs (C16, C16:1, C20:1, C20:2, C20:3, C20:4, C20:5, C22:1, C22:3, C22:4 and C22:5; P-value < 0.05) in methylated conditions. These results indicated that the use of methylation can provide the comprehensive information of FFA profiles with higher intensity and more number of identifications than the nonmethylation.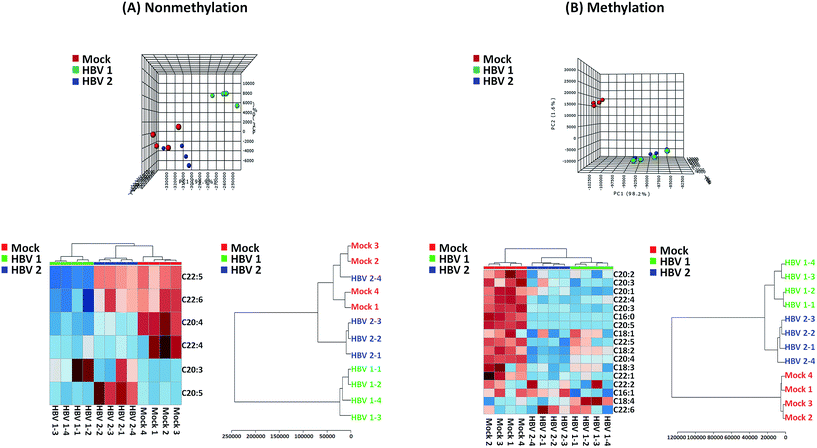 | ||
| Fig. 6 Principal component analysis (PCA) and hierarchical clustering analysis (heat map, dendro gram) of (A) intact and (B) methylated FFAs data in mock and HBV mice (triplicate MS run). | ||
Conclusions
In this study, the LC/MS/MS with simple and rapid methylation method was used for the profiling of FFAs. The proposed method is simple for profiling of methylated and various other FFA species within a short analysis time (5 min) and derivatization time (10 min). We observed that our method has high derivatization efficiency after the optimization of TMSD methylation by using FFA standards (palmitic acid-saturated FFA, oleic acid-monounsaturated FFA, arachidonic acid-polyunsaturated FFA). The results also revealed that methylated FFAs showed higher detection sensitivity, reproducibility and reliability in the positive ion mode than intact FFAs in the negative ion mode. The data showed that our methylation method provided in detail information of FFAs with higher intensity and more number of identifications than the nonmethylation. Besides, we also applied the developed method for profiling of FFAs in animal models. The utility of FFA profiling with methylation was also proved by PCA and HCA of HCV infected and control liver tissues. In the lipidomics, this method can be used for analysing even low abundant FFA species qualitatively and quantitatively. This method can also be applied to many biological and clinical samples to discover FFA biomarker. Furthermore, this study also explained the utility and effectiveness of methylation derivatization in profiling of FFAs in biological samples (mice liver and plasma).Acknowledgements
This work was supported by the Bio and Medical Technology Development Program (project no. 2012M3A9B6055305) and supported by Proteogenomic Research Program through the National Research Foundation of Korea funded by the Korean Ministry of Education, Science and Technology (to K. P. K.), Korea and supported by Proteogenomic Research Program.Notes and references
- E. Fahy, S. Subramaniam, H. A. Brown, C. K. Glass, A. H. Merrill Jr, R. C. Murphy, C. R. Raetz, D. W. Russell, Y. Seyama, W. Shaw, T. Shimizu, F. Spener, G. van Meer, M. S. VanNieuwenhze, S. H. White, J. L. Witztum and E. A. Dennis, J. Lipid Res., 2005, 46, 839–861 CrossRef CAS PubMed
.
- S. K. Gebauer, T. L. Psota, W. S. Harris and P. M. Kris-Etherton, Am. J. Clin. Nutr., 2006, 83, 1526S–1535S CAS
.
- Y. Itoh, Y. Kawamata, M. Harada, M. Kobayashi, R. Fujii, S. Fukusumi, K. Ogi, M. Hosoya, Y. Tanaka, H. Uejima, H. Tanaka, M. Maruyama, R. Satoh, S. Okubo, H. Kizawa, H. Komatsu, F. Matsumura, Y. Noguchi, T. Shinohara, S. Hinuma, Y. Fujisawa and M. Fujino, Nature, 2003, 422, 173–176 CrossRef CAS PubMed
.
- P. Bohov, V. Balaz, E. Sebokova and I. Klimes, Ann. N. Y. Acad. Sci., 1997, 827, 561–567 CrossRef CAS PubMed
.
- L. Liu, Y. Li, C. Guan, K. Li, C. Wang, R. Feng and C. Sun, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 2817–2825 CrossRef CAS PubMed
.
- B. B. Tan, Y. Z. Liang, L. Z. Yi, H. D. Li, Z. G. Zhou, X. Y. Ji and J. H. Deng, Metabolomics, 2010, 6, 219–228 CrossRef CAS
.
- L. Z. Yi, J. He, Y. Z. Liang, D. L. Yuan and F. T. Chau, FEBS Lett., 2006, 580, 6837–6845 CrossRef CAS PubMed
.
- D. C. Wang, C. H. Sun, L. Y. Liu, X. H. Sun, X. W. Jin, W. L. Song, X. Q. Liu and X. L. Wan, Neurobiol. Aging, 2012, 33, 1057–1066 CrossRef CAS PubMed
.
- X. T. Zheng, J. Shen, Q. Liu, S. F. Wang, Y. Y. Cheng and H. B. Qu, J. Pharm. Biomed. Anal., 2009, 49, 481–486 CrossRef CAS PubMed
.
- W. J. Griffiths, Mass Spectrom. Rev., 2003, 22, 81–152 CrossRef CAS PubMed
.
- C. Afonso, A. Riu, Y. Xu, F. Fournier and J. C. Tabet, J. Mass Spectrom., 2005, 40, 342–349 CrossRef CAS PubMed
.
- J. Ecker, M. Scherer, G. Schmitz and G. Liebisch, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2012, 897, 98–104 CrossRef CAS PubMed
.
- L. Akoto, R. J. Vreuls, H. Irth, R. Pel and F. Stellaard, J. Chromatogr. A, 2008, 1186, 365–371 CrossRef CAS PubMed
.
- D. Fabbri, V. Baravelli, G. Chiavari and S. Prati, J. Chromatogr. A, 2005, 1100, 218–222 CrossRef CAS PubMed
.
- K. Nagy, A. Jakab, J. Fekete and K. Vekey, Anal. Chem., 2004, 76, 1935–1941 CrossRef CAS PubMed
.
- R. C. Murphy, J. Fiedler and J. Hevko, Chem. Rev., 2001, 101, 479–526 CrossRef CAS PubMed
.
- J. L. Kerwin, A. M. Wiens and L. H. Ericsson, J. Mass Spectrom., 1996, 31, 184–192 CrossRef CAS
.
- N. Zehethofer, D. M. Pinto and D. A. Volmer, Rapid Commun. Mass Spectrom., 2008, 22, 2125–2133 CrossRef CAS PubMed
.
- S. H. Lee, C. Pettinella and I. A. Blair, Curr. Drug Metab., 2006, 7, 929–937 CrossRef CAS PubMed
.
- C. Pettinella, S. H. Lee, F. Cipollone and I. A. Blair, J. Chromatogr. B, 2007, 850, 168–176 CrossRef CAS PubMed
.
- T. Zhou, J. Leng, Y. Peng, L. Zhang and Y. Guo, J. Sep. Sci., 2016, 39, 873–879 CrossRef CAS PubMed
.
- W.-C. Yang, J. Adamec and F. E. Regnier, Anal. Chem., 2007, 79, 5150–5157 CrossRef CAS PubMed
.
- J. G. Bollinger, G. S. Naika, M. Sadilek and M. H. Gelb, J. Lipid Res., 2013, 54, 3523–3530 CrossRef CAS PubMed
.
- J. M. Halket, D. Waterman, A. M. Przyborowska, R. K. P. Patel, P. D. Fraser and P. M. Bramley, J. Exp. Bot., 2005, 56, 219–243 CrossRef CAS PubMed
.
- N. Aldai, B. E. Murray, A. I. Najera, D. J. Troy and K. Osoro, J. Sci. Food Agric., 2005, 85, 1073–1083 CrossRef CAS
.
- J. Clark, K. E. Anderson, V. Juvin, T. S. Smith, F. Karpe, M. J. Wakelam, L. R. Stephens and P. T. Hawkins, Nat. Methods, 2011, 8, 267–272 CrossRef CAS PubMed
.
-
W. W. Christie, Gas Chromatography and Lipids, 1989, pp. 161–184 Search PubMed
.
- W. A. Devane, L. Hanus, A. Breuer, R. G. Pertwee, L. A. Stevenson, G. Griffin, D. Gibson, A. Mandelbaum, A. Etinger and R. Mechoulam, Science, 1992, 258, 1946–1949 CAS
.
- J. W. Lee, S. Nishiumi, M. Yoshida, E. Fukusaki and T. Bamba, J. Chromatogr. A, 2013, 1279, 98–107 CrossRef CAS PubMed
.
- P. Nguyen, V. Leray, M. Diez, S. Serisier, J. L. Bloc'h, B. Siliart and H. Dumon, J. Anim. Physiol. Anim. Nutr., 2008, 92, 272–283 CrossRef CAS PubMed
.
- M. Schwalbe, O. Ohlenschlager, A. Marchanka, R. Ramachandran, S. Hafner, T. Heise and M. Gorlach, Nucleic Acids Res., 2008, 36, 1681–1689 CrossRef CAS PubMed
.
- H. Zhang, H. Li, Y. Yang, S. Li, H. Ren, D. Zhang and H. Hu, J. Proteome Res., 2013, 12, 2967–2979 CrossRef CAS PubMed
.
- M. Hajjou, R. Norel, R. Carver, P. Marion, J. Cullen, L. E. Rogler and C. E. Rogler, J. Med. Virol., 2005, 77, 57–65 CrossRef CAS PubMed
.
- K. Kim, K. H. Kim, H. H. Kim and J. Cheong, Biochem. J., 2008, 416, 219–230 CrossRef CAS PubMed
.
- E. S. Park, Y. K. Park, C. Y. Shin, S. H. Park, S. H. Ahn, D. H. Kim, K. H. Lim, S. Y. Kwon, K. P. Kim, S. I. Yang, B. L. Seong and K. H. Kim, Hepatology, 2013, 58, 762–776 CrossRef CAS PubMed
.
- E. G. Bligh and W. J. Dyer, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef CAS PubMed
.
- Z. Zhao and Y. Xu, J. Lipid Res., 2010, 51, 652–659 CrossRef CAS PubMed
.
- M. Puttmann, H. Krug, E. Vonochsenstein and R. Kattermann, Clin. Chem., 1993, 39, 825–832 CAS
.
- J. Sajiki and J. Yonekubo, Chemosphere, 2002, 46, 345–354 CrossRef CAS PubMed
.
- A. Carrier and J. Parent, Rapid Commun. Mass Spectrom., 2001, 15, 1681–1684 CrossRef CAS PubMed
.
- F. Valianpour, J. J. Selhorst, L. E. van Lint, A. H. van Gennip, R. J. Wanders and S. Kemp, Mol. Genet. Metab., 2003, 79, 189–196 CrossRef CAS PubMed
.
- D. Perret, A. Gentili, S. Marchese, M. Sergi and L. Caporossi, Rapid Commun. Mass Spectrom., 2004, 18, 1989–1994 CrossRef CAS PubMed
.
- X. Han, K. Yang and R. W. Gross, Mass Spectrom. Rev., 2012, 31, 134–178 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra01344a |
| ‡ Author contribution: Hyuck Jun Mok and Jae Won Lee contributed equally. |
| This journal is © The Royal Society of Chemistry 2016 |