Identification and distinction of non-small-cell lung cancer cells by intracellular SERS nanoprobes†
Yuqing Zhanga,
Xiaojia Ye*b,
Gengxin Xub,
Xiulong Jina,
Mengmeng Luanb,
Jiatao Louc,
Lin Wangc,
Chengjun Huangd and
Jian Ye*a
aSchool of Biomedical Engineering & Med-X Research Institute, Shanghai Jiao Tong University, 1954 Huashan Road, Shanghai 200030, China. E-mail: yejian78@sjtu.edu.cn; Tel: +86 021-62934760
bSchool of Mathematics and Information Science, Shanghai Lixin University of Commerce, 2800 Wenxiang Road, Shanghai 201620, China. E-mail: yxj@lixin.edu.cn; Tel: +86 021-67705383
cDepartment of Laboratory Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai 200030, China
dKey Laboratory of Microelectronics Devices and Integrated Technology, Institute of Microelectronics of Chinese Academy of Sciences, 3 Bei-Tu-Cheng West Road, Beijing 100029, China
First published on 7th January 2016
Abstract
Non-small-cell lung cancer (NSCLC) comprises ∼75% of all lung cancer and consists of several subtypes. Identification of lung cancer cell subtypes is important for choosing the appropriate therapy plan and reducing mortality. In this study, we have been able to identify and distinguish three subtypes of NSCLC cells (H1229, H460 and A549) and leukocytes on the single-cell level by combining surface-enhanced Raman scattering (SERS) spectroscopy and multivariate statistical methods. After the evaluation of three statistical methods, support vector machines (SVM) shows the best classification performance compared to hierarchical cluster analysis (HCA) and principal component analysis (PCA) methods based on a large amount of cell SERS spectra from Au nanoshells as intracellular nanoprobes. The SVM classification model provides a prediction accuracy of 88.75% for “unknown” independent cell types and an accuracy of ∼95% for the two subtypes mixed samples on a single-cell level. This method combining SERS and SVM could potentially be adapted to the distinction of other types of cancer cells and be applied for conducting non-invasive downstream cell identification after the capture of circulating tumor cells.
1. Introduction
Lung cancer is becoming a leading type of cancer case worldwide with two main categories: small cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC).1 The latter comprises ∼75% of all lung cancers and consists of adenocarcinoma, squamous cell carcinoma, large cell carcinoma, and rare types.2–4 As there is a significant difference in the diagnosis and treatment in each subtype,5 identification of lung cancer subtypes is important for choosing an appropriate treatment plan, reducing mortality and increasing the lifetime of patients. Currently, the evaluation of lung cancer subtypes via an invasive procedure that requires tissue specimens is not always feasible, because it is difficult to access small lung tumors and the sensitivity might be limited due to small tumor size.6At present, there are mainly two types of method to separate and identify cancer cells from blood: one based on physical properties such as size, density and deformability; the other based on biological properties such as protein expression.7,8 However, the methods in the former type including density gradient centrifugation and membrane filtration are difficulty to realize the identification and distinguishing of different types of cancer cells. The methods in the latter type, such as immunomagnetic separation, can only distinguish cancer cells with specific biomarkers and are limited by the spectral overlapping of fluorescent tags.9 Therefore, the development of new method for the non-invasive, highly sensitive and label-free identification and distinction of closely related cell phenotypes is in urgent need.10
Recently, surface-enhanced Raman scattering (SERS) spectroscopy, as a label-free and ultra-sensitive technique for chemical and biomedical analysis,11,12 is emerging as a new powerful tool for the analysis of individual cells, mainly due to its fingerprint spectral characteristic.10,13–15 As SERS can enhance the Raman signals of molecules close to metal surface by as much as 6 to 14 orders of magnitude,16 we can use near-infrared (NIR) light as excitation laser with low laser power and reduced photo-damage to analyze living cells.17,18 In SERS, Au nanoparticles are usually used as optical enhancing materials because of their chemical stability, good biocompatibility and strong enhancement capabilities. Au nanoparticles have been employed as intracellular probes facilitating cancer detection in blood plasma,19,20 cells,21,22 and tissues.23,24
The SERS spectra of cells are derived from cells themselves and no external label is required.25 SERS spectra of cells can be very complicated, containing spectral information from numerous biological molecules.26 Various types of cells have different SERS spectra due to their different biomolecular composition and structure, thus it can be used as the basis for distinguishing at a single-cell level.27,28 Since the spectral differences are often minute and difficult to identify, multivariate statistical methods such as principal component analysis (PCA) and hierarchical cluster analysis (HCA), have been applied to extract characteristic biochemical information presented in the spectra of different types of cells.10,15,28–31 However, few studies were reported about the application of Au nanoparticles as intracellular SERS probes with multivariate statistical methods for the identification and distinction of NSCLC cells with different subtypes.
In this study, the superparamagnetic Au nanoshells with strong NIR SERS effect were used as intracellular SERS nanoprobes, so that we can get strong signals from cells with lower laser power and less photo-damage. We combine SERS spectroscopy and multivariate statistical methods including HCA, PCA and support vector machines (SVM) to identify and distinguish three closely related NSCLC cell types (A549, H1299 and H460) and leukocytes. We have shown successful segregation of different types of cells using SVM analysis at a single-cell level, while HCA and PCA approach are more difficult to realize. A SVM classification model was built and tested for four independent cell types and mixture samples (two subtypes of NSCLC). The high prediction accuracies indicate that SERS spectra in combination with the SVM method can be a highly sensitive method for the distinction of NSCLC cells with different subtypes. Furthermore, if this method is combined with bio-chips, it will have potential to capture, detect and classify circulating tumor cells.
2. Materials and methods
2.1 Cell culture and sample preparation
The human NSCLC cell lines H1229, H460 and A549 were obtained from American Type Culture Collection (ATCC), and all reagents for the cell culture were purchased from Gibco. The cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. For Raman measurement purposes, the cells were grown on quartz coverslips to 80% confluence, and then incubated with Au nanoshells (Fe3O4@SiO2@Au) with a final concentration of 0.02 nM for six hours. The quartz coverslips were pretreated with 75% ethanol and then washed with sterile water, and Au nanoshells were just washed with sterile water before the incubation with cells. Fe3O4@SiO2@Au nanoparticles were synthesized with an average diameter of 180 nm and the detailed procedure was described in our previously published work.32 We separated leukocytes from peripheral blood of healthy donors following erythrocytes lysis as published before.33 Since the isolated leukocytes can't be cultured or adhere to the coverslips, the leukocytes suspensions were incubated with Au nanoshell particles with a final concentration of 0.02 nM for six hours.32Following the incubation, the cultured cells and leukocytes were washed extensively with phosphate buffered saline (PBS) and fixed with 4% paraformaldehyde for 10 min at room temperature. Then the excess paraformaldehyde was removed by deionized water. The leukocytes were transferred to quartz coverslips and air-dried together with other cultured cells for SERS measurements. For the mixed cell experiments, A549 were pre-incubated with Au nanoshells, then they were trypsinized and separated by an external magnetic field (Magical Trapper, Toyobo, nearly 200 mT) for 2 min after excess Au nanoshells were removed. For Raman experiments, the Au nanoshells labeled A549 were mixed with blank H1299 at two different ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9; then they were cultured on the coverslips and fixed with 4% paraformaldehyde.
9; then they were cultured on the coverslips and fixed with 4% paraformaldehyde.
2.2 SERS measurement
Raman spectrometer (SR-500, Andor, Northern Ireland) with 785 nm single mode diode laser (Innovative Photonic Solutions, America) was used to collect SERS spectra from the fixed cells on quartz coverslips. The laser power was 4.5 mW on the sample with a 50×/NA 0.5 objective (Leica, Germany). The scattered Raman signal was detected on a peltier-cooled, back-illuminated, deep-depletion CCD detector (Andor, Northern Ireland). Raman spectra of all individual cells were recorded over a spectra range of 550–1800 cm−1. For each Raman spectrum the collection time was 3 s. For all cells, three Raman spectra were obtained from three different parts of each cell and then averaged.2.3 Data processing and analysis
Spectral data from our experiments were pre-processed and analyzed using MATLAB R2011b, statistical analysis system (SAS), and R-Programme. For all analytical methods, the averaged Raman spectra from each individual cell were used. Statistical analysis system was used for HCA. HCA is a kind of cluster analysis method which aims to build a hierarchy of clusters in data mining and statistics.34 In our experiments, the hierarchy is built from the individual cell by progressively merging clusters. We analyzed the spectral data in the region 550–1800 cm−1 and the data were vector normalized. Then we used Euclidean distance method and Ward's algorithm to calculate the spectral differences and form clusters.26For PCA the same dataset with same pre-process was used. PCA was used to highlight the major variability existing in the spectral dataset. In this study, the PCA model was also calculated within the spectral region 550–1800 cm−1. Then PCA was used to reduce the dimension of the dataset and the first 7 principal components (PCs) described 95.01% of the variance of the dataset.
SVM with a linear kernel was used to build a differentiation model for the four different cell types. SVMs are a set of related methods for supervised learning, applicable to both classification and regression problems.34 Here, all individual spectra from 200 cells (50 leukocytes, 50 H1299, 50 H460, and 50 A549) were background corrected using the Daubechies wavelet transform (10 Daubechies, 7 transform levels, 10 iterations) and vector normalized. SVM algorithm was trained and tested by using leave-one-out cross validation. This procedure was repeated with each omitted spectrum, discriminating each spectrum in turn. Finally, a probability of prediction was calculated and expressed as a sensitivity and specificity for each group. All 200 spectra could be classified correctly, giving a prediction accuracy of 100%.
3. Results and discussion
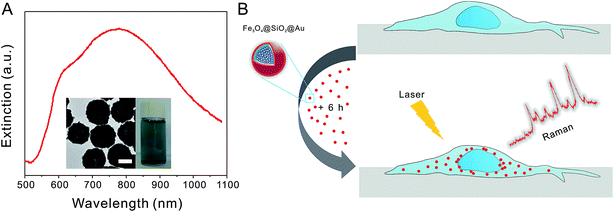
Fig. 1A shows the extinction spectrum of Au nanoshells in aqueous solution with a strong and broad peak (dipolar resonance mode) centered at ∼780 nm and a shoulder (quadrupolar resonance mode) at ∼610 nm. The TEM image (inset in Fig. 1A) shows that the average diameter of Au nanoshells is ∼180 nm. The Au nanoshells have shown the strong NIR SERS effect due to the large electric field enhancement at the dipolar plasmon resonance of the Au shells.32 In our study, the cells were grown on quartz coverslips due to the low background interference. After the cells were grown to 80% confluence, they were incubated with Au nanoshells with a final concentration of 0.02 nM for six hours, and then the Raman measurements were taken (see Fig. 1B). After the Au nanoshells are internalized by cells, they can greatly enhance the Raman signals of cell components near the shell surface. As various types of cells have different molecular compositions and structures, the SERS spectra can be used for distinguishing different cell types.27,28,35Fig. 2A shows the bright field images of representative examples of four types of cells (from top to bottom: H460, A549, H1299, and leukocytes) with SERS nanoprobes inside. In the bright field images, we could easily identify the nanoprobes with black colors in the cells. SERS nanoprobes were mainly located in the cytoplasm, and these results are consistent with previous reports about the cellular location of Au nanoparticles.26,36 Three closely related subtypes of NSCLC cells (H460, A549, and H1299) were adherent on the quartz coverslips, so they were slightly stretched. As leukocytes can't adhere to the coverslips, they were directly dropped onto the coverslip and appeared more or less round. On average the leukocytes are slightly smaller in size than the other cells. However, cancer cells have large heterogeneity including both size and morphology, and they are not universally larger than all leukocytes.37,38 Besides that, cells in blood are all suspended and their size difference will become smaller, therefore the size is not a reliable criterion used for classification.38
Averaged SERS spectra of four types of cells are depicted in Fig. 2B. The shaded areas represent the standard deviations of the means. The SERS spectra were obtained with lower laser power (4.5 mW) and shorter integration time (3 s) compared with normal Raman measurements.30 This can greatly reduce the photo-damage of cells, so that the constituents and viabilities of native cells themselves can remain almost unchanged.17 Therefore SERS has great potential to be used in live cell analysis. In this study, SERS spectra derived from specific cells were used to characterize the cellular biochemical composition of each individual type. 200 individual cells of four different types (50 H460, 50 A549, 50 H1299, 50 leukocytes) were tested and all SERS spectra were used for further statistical analysis. For the sake of convenient comparison, the intensity of the averaged SERS spectra for various types of cell was normalized to obtain the relative intensity from 0 to 1.10 The Raman spectra of the cells can act as molecular fingerprints containing information from various cellular components such as DNA, protein, lipids and carbohydrates.39 Molecular components of cells are very complex,21 and typical molecules of interest are often associated with RNA, DNA, carbohydrates, proteins, or lipids. With the help of SERS probes, these biochemical molecules may typically be assessed based on their individual Raman band assignments. By reference to the SERS band assignments in previous reports,10,26,40 we assigned the observed SERS bands in Table 1. The bands at around 747 cm−1 (T), 833 cm−1 (O–P–O backbone stretching), 1342 cm−1 (A, G), 1582 cm−1 (G, A) are assigned to nucleic acids. The bands at around 645 cm−1 (C–C twist in tyrosine), 833 cm−1 (tyrosine), 1002 cm−1 (phenylalanine), 1158 cm−1 (C–C and C–N stretch), 1252 cm−1 (amide III), 1307 cm−1 (C–N stretch), 1342 cm−1 (C–H deformation), 1414 cm−1 (aspartate and glutamate), 1543 cm−1 (tryptophan) are assigned to proteins and amino acids. And the band at around 1252 cm−1 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH deformation) is assigned to lipids. Although the SERS spectra of different types of cells look very alike at sometimes, they still have differences on some peak positions and peak intensities. In order to better compare and distinguish them, multivariate statistical methods including HCA, PCA and SVM were carried out.
CH deformation) is assigned to lipids. Although the SERS spectra of different types of cells look very alike at sometimes, they still have differences on some peak positions and peak intensities. In order to better compare and distinguish them, multivariate statistical methods including HCA, PCA and SVM were carried out.
| Raman shift (cm−1) | Nucleic acid | Protein | Lipids |
|---|---|---|---|
| a str, stretching; def, deformation; bk, backbone. | |||
| 645 | Tyrosine (C–C twist) | ||
| 747 | T | ||
| 833 | O–P–O str DNA bka | Tyrosine | |
| 1002 | Phenylalanine | ||
| 1158 | C–C str (and C–N str)a | ||
| 1252 | Amide III | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH defa CH defa |
|
| 1307 | C–N stra | ||
| 1342 | A, G | C–H defa | |
| 1414 | Aspartate, glutamate | ||
| 1543 | Tryptophan | ||
| 1582 | G, A | ||
HCA is a kind of cluster analysis method which aims to build a hierarchy of clusters in data mining and statistics,34 and this method has been successfully used in identification and differentiation of breast cancer cells, leukaemia cells and leukocytes with normal Raman measurements.26 In our study, HCA method was also used to form clusters according to the cell type. Euclidean distance method and Ward's algorithm were used to perform HCA and the spectra in the region 550–1800 cm−1 were vector normalized and averaged.26 Fig. 3 shows the dendrogram for all 200 cells. And we can see two well-separated major clusters: one for leukocytes and one for NSCLC cells. Within the NSCLC cell cluster, each cell type forms its own sub-cluster with some misclassifications. We can conclude from the dendogram that although vast majority of leukocytes were well separated, there were some misclassifications among the NSCLC cells. So the clustering result from the HCA can only separate leukocytes from lung cancer cells but cannot distinguish different lung cancer cells well because they are too closely related. The clustering result is similar to the previous study about lung cancer diagnosing.41 In that study, two large clusters were clearly visible in the dendrogram, but no sub-clustering was evident for different subtypes of lung cancer cells and even no cluster patterns emerged. The possible reasons are small number of samples and great similarity between the samples.41 Thus we speculate that HCA can achieve good results when distinguishing samples without high similarity, but several misclassifications may occur when distinguishing closely similar samples.
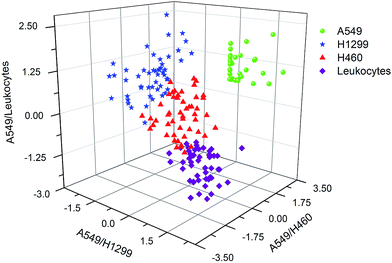
Next, we employed the PCA method to evaluate the possibility of distinguishing different types of NSCLC cells and leukocytes. PCA can select less number of significant components through linear transformation of multiple variables. As a classic analysis method, PCA has been widely used in multivariate statistical analysis for spectra.42,43 Herein, the spectral differences in the data sets comprising 200 spectra of four types of cells within the same spectral region from 550 to 1800 cm−1 (consisting of 1910 data points) were analyzed by PCA. First, we can roughly distinguish three types of cells (H1299, H460, leukocytes) using the PCA method with some overlaps between the H460 and leukocytes (see Fig. S1†). Fig. S1† shows the three-dimensional plot using the first, second and third principal components (PC1, PC2, and PC3). Different cell types are approximately separated as indicated by spectra assemblies with different colors and shapes. However, when it turned to four types of cells (A549, H1299, H460, leukocytes) that the distinguish effect became much worse (Fig. 4). In Fig. 4, using principal component scores PC1 (83.88% variation), PC2 (5.95% variation) and PC3 (1.83% variation), the scatter plots of SERS spectra for each cell line were projected into the two-dimensional and three-dimensional images. We can see that the SERS spectra are mainly divided into two groups, one is leukocytes and the other is NSCLC cells, which is caused by the intrinsic difference in biomolecular composition and concentration between normal and cancer cells. This result is consistent with the clustering result from the HCA as shown in Fig. 3. However, the plots groups of NSCLC cells are adjacent to each other and some even were overlapped with each other, which means they cannot be separated well. This result is similar to the previous studies.26,31 In those studies, PCA only separated normal cells from tumor cells but did not distinguish different tumor cells well. That means on high dimensional data of complex samples, especially when more samples are involved and the data have ambiguous distribution of noise features, PCA often cannot achieve good separation results.43
After the failure to discriminate subtypes of NSCLC cells using HCA and PCA method, SVM with a linear kernel were fed with this SERS dataset to create a classification model in order to find a more suitable classification method. Fig. 5 shows the analysis result using three out of the six decision values. It can be seen that four cell types are successfully separated as indicated by spectra assemblies with different colors and shapes. 200 spectra were used to train the SVM model with leave-one-out cross validation and all spectra could be classified correctly, giving an overall accuracy of 100% on a single-cell level. The details of the prediction accuracy of the classification model are presented in Table S1.†
 | ||
| Fig. 5 Graphical representation of the support vector machines with linear kernel for classification of four cell types. Three out of six decision values are plotted. | ||
Although the SVM model with 100% accuracy was built, these cells were all involved in building this model. It will be more practical if this model can predict some “unknown” cells which were not involved in building this model but within these four cell types. To further demonstrate the accuracy and practicability of this SVM model, this trained and validated SVM model was used to predict and classify a test set (including additional 80 cells: 20 A549, 20 H1299, 20 H460, 20 leukocytes). It should be mentioned that none of the cells was included in building the SVM model. The detailed predicted results for the individual cell types are shown in Table 2. A predicted accuracy of 88.75% can be achieved on a single-cell level. If we want a better distinguish effect, more samples are needed both for training and testing the model.44 For clinical purpose, the most relevant question is to identify the cancer cells from body's own healthy leukocytes.45 For this purpose the SVM model trained with spectra showed very good characteristics: all NSCLC cells were correctly identified as cancer cells with 100% accuracy.
| Predicted labels | |||||
|---|---|---|---|---|---|
| Leukocytes | A549 | H1299 | H460 | ||
| True labels | Leukocytes | 18 | 0 | 0 | 2 |
| A549 | 0 | 18 | 0 | 2 | |
| H1299 | 0 | 2 | 18 | 0 | |
| H460 | 0 | 2 | 1 | 17 | |
We further evaluated the performance of this SVM model when there were more than 1 subtype of NSCLC cells in the sample. The mixtures of two subtypes of NSCLC cell (A549 and H1299) with different ratios were used as the test sets. It should be emphasized again that none of these cells was included in building the SVM model. Although fluorescent biomarkers can help to distinguish cells in the mixture, they may also produce strong Raman background in the SERS measurements. In order to verify the accuracy of the SVM model in mixed samples, we mixed SERS probes labeled A549 and blank H1299 at different ratios, thus we can recognize the cell type directly from the bright-field images.
Fig. 6 shows the bright-field images from the mixtures of two subtypes of cell, where A549 can be recognized by SERS probes in the cells. Fig. 6A represents the group in which A549 and H1299 were mixed at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (A549 indicated by red arrows and H1299 by white arrows). Since the normal Raman signals of H1299 cells (no SERS probes inside) were very weak, we only detect the signal of A549 cells in mixed samples. Table 3A showed the predicted results based on the SVM model and an accuracy of 95% can be achieved on a single-cell level. Fig. 6B represents the group in which A549 and H1299 were mixed at a ratio of 1
1 (A549 indicated by red arrows and H1299 by white arrows). Since the normal Raman signals of H1299 cells (no SERS probes inside) were very weak, we only detect the signal of A549 cells in mixed samples. Table 3A showed the predicted results based on the SVM model and an accuracy of 95% can be achieved on a single-cell level. Fig. 6B represents the group in which A549 and H1299 were mixed at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, and a prediction accuracy of 100% can be achieved for A549 on a single-cell level (Table 3B). These results indicate the SVM model significantly improves the accuracy of prediction and classification of subtype of NSCLC cells in contrast to the HCA and PCA analysis methods.
9, and a prediction accuracy of 100% can be achieved for A549 on a single-cell level (Table 3B). These results indicate the SVM model significantly improves the accuracy of prediction and classification of subtype of NSCLC cells in contrast to the HCA and PCA analysis methods.
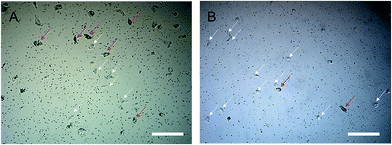 | ||
Fig. 6 Bright-field images of the mixed two subtypes of NSCLC cells cultured on quartz coverslips. The ratio of A549![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H1299 was (A) 1 H1299 was (A) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and (B) 1 1 and (B) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9. All scale bars are 200 μm. 9. All scale bars are 200 μm. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H1299 at the ratio of (A) 1
H1299 at the ratio of (A) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and (B) 1
1 and (B) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 using the SVM model
9 using the SVM model
| Predicted labels | |||
|---|---|---|---|
| A549 | H1299 | ||
| A | |||
| True labels | A549 | 19 | 1 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| B | |||
| True labels | A549 | 5 | 0 |
4. Conclusions
In this study, we have used SERS spectroscopy and multivariate statistical methods including HCA, PCA and SVM to identify and distinguish three subtypes of NSCLC cells and leukocytes on the single-cell level. With the help of statistical methods, a reproducible SVM model has been developed showing a much better classification performance compared to HCA and PCA method. The SVM classification model provides a predication accuracy of 88.75% for “unknown” independent cell types and an accuracy of ∼95% for the mixed samples on a single-cell level. This predication accuracy can be further improved by using larger amount of samples to train the model. The fast identification of subtype of NSCLC cells by SERS spectra with multivariate analysis can assist doctors to choose appropriate treatment plans for lung cancer patients as soon as possible. This method could also be adapted to the detection and distinction of other types of cancer cell. Moreover, the SERS nanoprobes used in this work are composed of superparamagnetic cores, which potentially further allows the isolation and enrichment of cancer cells in peripheral blood. We believe that future promising work may involve SERS and magnetic separation method combined with multivariate analysis as a rapid, simple, non-destructive and accurate diagnostic tool in early stage cancer detection and cancer-therapy monitoring.Acknowledgements
We gratefully acknowledge the National Natural Science Foundation of China (No. 81571763 and 21375087), the Ministry of Science and Technology of China (No. 2014DFA33010), the Science and Technology Commission of Shanghai Municipality (No. 13ZR1422100) and Shanghai Jiao Tong University (No. YG2014MS53 and SJTU-KUL Bilateral Program) for their financial support. C. H. thanks the financial support of the Presidential Foundation of IMECAS.References
- International Agency for Research on Cancer, Cancer Research UK (2012), World cancer factsheet, Cancer Research UK, London, 2012 Search PubMed.
- L. G. Collins, C. Haines, R. Perkel and R. E. Enck, Am. Fam. Physician, 2007, 75, 56–63 Search PubMed.
- I. Petersen and S. Petersen, Anal. Cell. Pathol., 2001, 22, 111–121 CrossRef CAS PubMed.
- L. Wang, T. Guo, Q. Lu, X. Yan, D. Zhong, Z. Zhang, Y. Ni, Y. Han, D. Cui, X. Li and L. Huang, ACS Appl. Mater. Interfaces, 2015, 7, 359–369 Search PubMed.
- T. Hensing, A. Chawla, R. Batra and R. Salgia, in Systems Analysis of Human Multigene Disorders, ed. N. Maltsev, A. Rzhetsky and T. C. Gilliam, Springer, New York, 2014, ch. 5, pp. 85–117 Search PubMed.
- J.-E. C. Holty, W. G. Kuschner and M. K. Gould, Thorax, 2005, 60, 949–955 CrossRef PubMed.
- C. Alix-Panabieres and K. Pantel, Lab Chip, 2014, 14, 57–62 RSC.
- S. Bhana, Y. Wang and X. Huang, Nanomedicine, 2015, 10, 1973–1990 CrossRef CAS PubMed.
- Z. A. Nima, M. Mahmood, Y. Xu, T. Mustafa, F. Watanabe, D. A. Nedosekin, M. A. Juratli, T. Fahmi, E. I. Galanzha and J. P. Nolan, Sci. Rep., 2014, 4, 4752 Search PubMed.
- C. Shi, X. Cao, X. Chen, Z. Sun, Z. Xiang, H. Zhao, W. Qian and X. Han, Biomaterials, 2015, 58, 10–25 CrossRef PubMed.
- J. Ye, F. Wen, H. Sobhani, J. B. Lassiter, P. V. Dorpe, P. Nordlander and N. J. Halas, Nano Lett., 2012, 12, 1660–1667 CrossRef CAS PubMed.
- J. F. Li, Y. F. Huang, Y. Ding, Z. L. Yang, S. B. Li, X. S. Zhou, F. R. Fan, W. Zhang, Z. Y. Zhou, Y. WuDe, B. Ren, Z. L. Wang and Z. Q. Tian, Nature, 2010, 464, 392–395 CrossRef CAS PubMed.
- A. Wang, W. Ruan, W. Song, L. Chen, B. Zhao, Y. M. Jung and X. Wang, J. Raman Spectrosc., 2013, 44, 1649–1653 CrossRef CAS.
- Y. Zhang, S. Liu, L. Wang, X. Qin, J. Tian, W. Lu, G. Chang and X. Sun, RSC Adv., 2012, 2, 538–545 RSC.
- M. Wang, X. Cao, W. Lu, L. Tao, H. Zhao, Y. Wang, M. Guo, J. Dong and W. Qian, RSC Adv., 2014, 4, 64225–64234 RSC.
- S. Nie and S. R. Emory, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- K. Kneipp, A. S. Haka, H. Kneipp, K. Badizadegan, N. Yoshizawa, C. Boone, K. E. Shafer-Peltier, J. T. Motz, R. R. Dasari and M. S. Feld, Appl. Spectrosc., 2002, 56, 150–154 CrossRef CAS.
- J. L. Deng, Q. Wei, M. H. Zhang, Y. Z. Wang and Y. Q. Li, J. Raman Spectrosc., 2005, 36, 257–261 CrossRef CAS.
- S. Feng, D. Lin, J. Lin, B. Li, Z. Huang, G. Chen, W. Zhang, L. Wang, J. Pan and R. Chen, Analyst, 2013, 138, 3967–3974 RSC.
- S. Feng, R. Chen, J. Lin, J. Pan, Y. Wu, Y. Li, J. Chen and H. Zeng, Biosens. Bioelectron., 2011, 26, 3167–3174 CrossRef CAS PubMed.
- H. Huang, W. Chen, J. Pan, Q. Chen, S. Feng, Y. Yu, Y. Chen, Y. Su and R. Chen, J. Spectrosc., 2011, 26, 187–194 CrossRef CAS.
- X. Wu, L. Luo, S. Yang, X. Ma, Y. Li, C. Dong, Y. Tian, L. E. Zhang, Z. Shen and A. Wu, ACS Appl. Mater. Interfaces, 2015, 7, 9965–9971 CAS.
- S. Feng, J. Lin, Z. Huang, G. Chen, W. Chen, Y. Wang, R. Chen and H. Zeng, Appl. Phys. Lett., 2013, 102, 043702 CrossRef.
- M. J. Sailor and J. H. Park, Adv. Mater., 2012, 24, 3779–3802 CrossRef CAS PubMed.
- K. Galler, K. Bräutigam, C. Groβe, J. Popp and U. Neugebauer, Analyst, 2014, 139, 1237–1273 RSC.
- U. Neugebauer, J. H. Clement, T. Bocklitz, C. Krafft and J. Popp, J. Biophotonics, 2010, 3, 579–587 CrossRef CAS PubMed.
- I. Notingher, G. Jell, P. L. Notingher, I. Bisson, O. Tsigkou, J. M. Polak, M. M. Stevens and L. L. Hench, J. Mol. Struct., 2005, 744–747, 179–185 CrossRef CAS.
- T. Bocklitz, M. Putsche, C. Stüber, J. Käs, A. Niendorf, P. Rösch and J. Popp, J. Raman Spectrosc., 2009, 40, 1759–1765 CrossRef CAS.
- E. Ly, O. Piot, A. Durlach, P. Bernard and M. Manfait, Analyst, 2009, 134, 1208–1214 RSC.
- A. Huefner, W.-L. Kuan, R. A. Barker and S. Mahajan, Nano Lett., 2013, 13, 2463–2470 CrossRef CAS PubMed.
- S. Mert and M. Çulha, Appl. Spectrosc., 2014, 68, 617–624 CrossRef CAS PubMed.
- X. Jin, H. Li, S. Wang, N. Kong, H. Xu, Q. Fu, H. Gu and J. Ye, Nanoscale, 2014, 6, 14360–14370 RSC.
- J. H. Clement, M. Schwalbe, N. Buske, K. Wagner, M. Schnabelrauch, P. Görnert, K. O. Kliche, K. Pachmann, W. Weitschies and K. Höffken, J. Cancer Res. Clin. Oncol., 2006, 132, 287–292 CrossRef CAS PubMed.
- O. Maimon and L. Rokach, Data mining and knowledge discovery handbook, Springer, 2005 Search PubMed.
- P. Rösch, M. Harz, M. Schmitt and J. Popp, J. Raman Spectrosc., 2005, 36, 377–379 CrossRef.
- A. Shamsaie, M. Jonczyk, J. Sturgis, J. Paul Robinson and J. Irudayaraj, J. Biomed. Opt., 2007, 12, 020502 CrossRef PubMed.
- S. Riethdorf, H. Fritsche, V. Müller, T. Rau, C. Schindlbeck, B. Rack, W. Janni, C. Coith, K. Beck, F. Jänicke, S. Jackson, T. Gornet, M. Cristofanilli and K. Pantel, Clin. Cancer Res., 2007, 13, 920–928 CrossRef CAS PubMed.
- W. Chen, S. Weng, F. Zhang, S. Allen, X. Li, L. Bao, R. H. W. Lam, J. A. Macoska, S. D. Merajver and J. Fu, ACS Nano, 2013, 7, 566–575 CrossRef CAS PubMed.
- H. Yamakoshi, K. Dodo, M. Okada, J. Ando, A. Palonpon, K. Fujita, S. Kawata and M. Sodeoka, J. Am. Chem. Soc., 2011, 133, 6102–6105 CrossRef CAS PubMed.
- P. Lu, J. Wang, J. Lin, J. Lin, N. Liu, Z. Huang, B. Li, H. Zeng and R. Chen, J. Biomed. Opt., 2014, 20, 051005 CrossRef PubMed.
- P. Lewis, K. Lewis, R. Ghosal, S. Bayliss, A. Lloyd, J. Wills, R. Godfrey, P. Kloer and L. Mur, BMC Cancer, 2010, 10, 640 CrossRef CAS PubMed.
- U. Thissen, M. Pepers, B. Üstün, W. J. Melssen and L. M. C. Buydens, Chemom. Intell. Lab. Syst., 2004, 73, 169–179 CrossRef CAS.
- C. Hu, J. Wang, C. Zheng, S. Xu, H. Zhang, Y. Liang, L. Bi, Z. Fan, B. Han and W. Xu, Med. Phys., 2013, 40, 063501 CrossRef PubMed.
- C. Beleites, U. Neugebauer, T. Bocklitz, C. Krafft and J. Popp, Anal. Chim. Acta, 2013, 760, 25–33 CrossRef CAS PubMed.
- U. Neugebauer, C. Kurz, T. Bocklitz, T. Berger, T. Velten, J. Clement, C. Krafft and J. Popp, Micromachines, 2014, 5, 204–215 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra21758j |
| This journal is © The Royal Society of Chemistry 2016 |