Influence of selected inorganic counter-ions on the structure and antimicrobial properties of silver(I) complexes with imidazole-containing ligands†
Urszula
Kalinowska-Lis
*a,
Aleksandra
Felczak
b,
Lilianna
Chęcińska
c,
Magdalena
Małecka
c,
Katarzyna
Lisowska
b and
Justyn
Ochocki
a
aDepartment of Bioinorganic Chemistry, Medical University of Lodz, Muszyńskiego 1, 90-151 Łódź, Poland. E-mail: urszula.kalinowska-lis@umed.lodz.pl; Fax: +48 42 677 92 20; Tel: +48 42 677 92 20
bDepartment of Industrial Microbiology and Biotechnology, Faculty of Biology and Environmental Protection, University of Lodz, 12/16 Banacha Street, 90-237 Łódź, Poland
cDepartment of Theoretical and Structural Chemistry, Faculty of Chemistry, University of Lodz, Pomorska 163/165, 90-236 Łódź, Poland
First published on 16th November 2015
Abstract
Water-soluble silver(I) complexes containing the 4(5)-(hydroxymethyl)imidazole ligand with the formula [Ag(4-CH2OHimH)2]X (where X = NO3−, ClO4−, CF3COO−, BF4− and SO3CH3−) were synthesized. The complexes were characterized by NMR (1H and 13C) and IR spectroscopy, ESI-MS spectrometry and EA. The molecular structures of three complexes were confirmed by X-ray crystallography. The antimicrobial activity of the silver(I) complexes were evaluated against six strains of microorganisms: Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli, Pseudomonas aeruginosa, Proteus hauseri and Candida albicans. All tested silver complexes displayed excellent antibacterial properties against Gram-positive bacteria. Among them, the complex containing the trifluoroacetate counter-ion exhibited the highest antibacterial activity, with MIC values 2 to 3-fold lower than that required by silver sulfadiazine.
Introduction
In the face of increasing difficulties in the treatment of infections caused by resistant pathogenic microorganisms, new broad-spectrum antimicrobial agents are required. The medication of infectious diseases is increasingly more often associated with high levels of morbidity and fatality rates as well as increasing costs.1,2The study on novel antimicrobial agents amongst the silver complex group seems to be proved as silver(I) complexes are known to possess substantial antimicrobial activity3–5 and have been demonstrated to be less toxic to the human body than other metal complexes.6 Silver sulfadiazine (SSD or AgSD) is the most popular topical drug used in burn infections and other serious external infections.7,8 Unfortunately, the drug is not devoid of side effects, including toxicity to fibroblasts and keratinocytes that delays the wound healing process as well as associated gastrointestinal reactions and blood dyscrasias.9–12
In addition, the silver cation is an active ingredient of healthcare products such as wound dressings, urinary tract and venous catheters and mats.3,13–16
Imidazole-containing drugs (ketoconazole and clotrimazole) are commonly used to treat infections caused by fungi or yeasts. A slightly different group of imidazole-based medications is based on nitroimidazole with different side chains (metronidazole, tinidazole, ornidazole, and secnidazole), these being compounds with excellent activity against anaerobic micro-organisms and protozoa.17
Recently, a great deal of research interest has been directed toward the discovery of new antimicrobial molecules amongst imidazole-based derivatives.18,19
Numerous Ag(I) complexes containing imidazole derivatives have shown significant broad spectrum antimicrobial activity.20–27 Interestingly, although free imidazole ligands display no activity against pathogenic microorganisms, their activity increases when complexed with silver ions. The components of the silver(I) complexes probably act synergistically.
Lately, a silver(I) complexes containing N-heterocyclic carbenes (NHCs) have gained special attention as potential antimicrobial agents.28–33 The research by Wiley J. Youngs and coworkers had a high impact on the development of this group of compounds. They established syntheses of (benz)imidazole-derived NHC–silver(I) acetates, called SCC1 and SCC10, which were highly effective against pathogens recovered from the respiratory track of patients with cystic fibrosis.34–38 In the meantime, another researchers synthesized a various Ag–NCHs complexes by a modification of substituents on the imidazolium center to improve their antimicrobial efficacy.39–41
Our previous article reports a study of the antimicrobial activity of a series of silver(I) complexes of metronidazole and selected counter-ions. The complex containing a methanesulphonate counter-ion was found to be the most active against tested Gram-positive strains (S. epidermidis ATCC 12228 and S. aureus ATCC 6538) as well as yeast C. albicans ATCC 10231. However, it was not so active against tested Gram-negative strains. Nevertheless, the silver(I)–metronidazole complexes containing perchlorate and tetrafluoroborate counter-ions were found to be very effective against tested Gram-negative bacteria.42
The aim of the present study is to determine the antimicrobial activity of a second series of novel silver(I) complexes containing 4(5)-(hydroxymethyl)imidazole [4(5)-CH2OHimH] (Scheme 1) and selected counter-ions. It also analyses the impact of particular counter-ions (NO3−, ClO4−, CF3COO−, BF4− and CH3SO3−) on the biological properties of tested complexes. The study describes the synthesis of five novel silver(I) complexes of 4-CH2OHimH and selected counter-ions. The complexes are characterized by (1H and 13C) NMR and IR spectroscopy as well as ESI-MS spectrometry, three of which being described by X-ray measurements of single crystals.
Experimental
Reagents and physical measurements
4(5)-(Hydroxymethyl)imidazole [4(5)-CH2OHimH] and inorganic salts AgX (X = NO3−, ClO4−, CF3COO−, BF4− and SO3CH3−) were purchased from Sigma-Aldrich.Elemental analyses (C, H and N) were performed using a EuroVector 3018 analyzer. 1H and 13C NMR spectra were recorded on a Bruker Avance III 600 MHz spectrometer using D2O as a solvent. Infrared (IR) spectra were recorded in the region 4000–400 cm−1 on a Bruker IFS66 spectrophotometer using KBr pellets. Electrospray mass spectra (ESI-MS) were collected in positive ion mode on a Varian 500-MS LC ion trap using water as a solvent.
Preparation and characterization of [Ag(4-CH2OHimH)2]NO3 (1)
Silver nitrate (1 mmol, 0.170 g) was dissolved in ca. 10 ml of water and added to an aqueous solution (ca. 10 ml) of 4(5)-(hydroxymethyl)imidazole (2 mmol, 0.196 g). The reaction mixture was stirred for ca. 30 min at 60 °C. Then the mixture was evaporated to dryness under reduced pressure (water bath: 70 °C) giving a beige oil. The oil was dissolved in hot ethanol (60 °C). A small amount of beige impurities was filtered and removed. The clear ethanol solution was left in the refrigerator to slow evaporation. The white product which precipitated was washed with anhydrous ethyl ether and air-dried.Yield: 0.304 g (83%); MW = 366.12. Anal. calcd for C8H12N5O5Ag ([Ag(4-CH2OHimH)2]NO3): C, 26.24; H, 3.31; N, 19.13%. Found: C, 26.25; H, 3.32; N, 19.11%. ESI-MS (H2O) m/z (relative intensity): 303.1 (100) [Ag(4-CH2OHimH)2]+, 205.0 (49) [Ag(4-CH2OHimH)]+. 1H NMR (600 MHz, D2O): δ 4.64 (s, 4H, 2 × CH2–O), 7.25 (s, 2H, 2 × H(5)), 7.97 (s, 2H, 2 × H(2)). 13C NMR (151 MHz, D2O): δ 138.0 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N), 137.1 (C5) (weak int.), 117.4 (C4), 56.3 (CH2–O). IR (KBr) νmax (cm−1): (O–H, C–H) 3306(s, br), 2872(m); (C
C–N), 137.1 (C5) (weak int.), 117.4 (C4), 56.3 (CH2–O). IR (KBr) νmax (cm−1): (O–H, C–H) 3306(s, br), 2872(m); (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1766(w), 1654(w), 1591(m); (C–H) 1491(m); (NO3) 1383(vs); (C–H) 1280(m), 1243(w), 1207(w); (C–O) 1106(w), 1089(m), 1027(s), 983(s); (im ring) 833(s), 749(m), 659(m), 620(s) (vs: very strong; s: strong; m: medium; w: weak, br: broad).
C) 1766(w), 1654(w), 1591(m); (C–H) 1491(m); (NO3) 1383(vs); (C–H) 1280(m), 1243(w), 1207(w); (C–O) 1106(w), 1089(m), 1027(s), 983(s); (im ring) 833(s), 749(m), 659(m), 620(s) (vs: very strong; s: strong; m: medium; w: weak, br: broad).
Preparation and characterization of complexes 2–4
4(5)-(Hydroxymethyl)imidazole [4(5)-CH2OHimH] (2 mmol, 0.196 g) was dissolved in ca. 10 ml of ethanol at room temperature. An ethanol solution (ca. 10 ml) of AgX (1 mmol) (X = ClO4− (0.207 g), CF3COO− (0.221 g), BF4− (0.195 g)) was added to the solution of 4(5)-CH2OHimH and stirred for 10 min at 60 °C. The reaction mixture was evaporated under reduced pressure (water bath: 60 °C) to half the initial volume. A small amount of beige impurities was filtered and removed. The ethanol solution was left in the refrigerator to crystallize. After a few days, colourless crystals suitable for X-ray determination were collected and washed with anhydrous diethyl ether.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N), 137.1 (C5) (weak int.), 117.6 (C4), 56.4 (CH2–O). IR (KBr) νmax (cm−1): (O–H, C–H) 3270(s, br), 2876(m); (C
C–N), 137.1 (C5) (weak int.), 117.6 (C4), 56.4 (CH2–O). IR (KBr) νmax (cm−1): (O–H, C–H) 3270(s, br), 2876(m); (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1631(w), 1590(m); (C–H) 1492(m), 1448(m); (C–H) 1281(m), 1242(w); (ClO4, C–O) 1146(vs), 1111(vs), 1089(vs), 1022(m), 982(s); (im ring) 835(m), 749(m), 660(m), 636(vs) (vs: very strong; s: strong; m: medium; w: weak, br: broad).
C) 1631(w), 1590(m); (C–H) 1492(m), 1448(m); (C–H) 1281(m), 1242(w); (ClO4, C–O) 1146(vs), 1111(vs), 1089(vs), 1022(m), 982(s); (im ring) 835(m), 749(m), 660(m), 636(vs) (vs: very strong; s: strong; m: medium; w: weak, br: broad).
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) F3COO) (weak int.), 138.1 (N
F3COO) (weak int.), 138.1 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N), 137.1 (C5) (weak int.), 117.5 (C4), 115.5 (CF3
C–N), 137.1 (C5) (weak int.), 117.5 (C4), 115.5 (CF3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OO) (weak int.), 56.3 (CH2–O). IR (KBr) νmax (cm−1): (O–H, C–H) 3386(s, br), 2932(w), 2869(w); (C
OO) (weak int.), 56.3 (CH2–O). IR (KBr) νmax (cm−1): (O–H, C–H) 3386(s, br), 2932(w), 2869(w); (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) 1682 (vs) (C
O) 1682 (vs) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1632(w), 1595(w); (C–H) 1499(m), 1434(w), 1354(m), 1283(m); (CF3COO) 1207(vs), 1137(s); (C–O) 1088(m), 1021(s), 981(s); (im ring) 840(s), 803(s), 750(w), 724(s), 627(s) (vs: very strong; s: strong; m: medium; w: weak, br: broad).
C) 1632(w), 1595(w); (C–H) 1499(m), 1434(w), 1354(m), 1283(m); (CF3COO) 1207(vs), 1137(s); (C–O) 1088(m), 1021(s), 981(s); (im ring) 840(s), 803(s), 750(w), 724(s), 627(s) (vs: very strong; s: strong; m: medium; w: weak, br: broad).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N), 137.2 (C5) (weak int.), 117.7 (C4) (weak), 56.4 (CH2–O). IR (KBr) νmax (cm−1): (O–H, C–H) 3117(s), 2870(s), 2794(s), 2613(m); (C
C–N), 137.2 (C5) (weak int.), 117.7 (C4) (weak), 56.4 (CH2–O). IR (KBr) νmax (cm−1): (O–H, C–H) 3117(s), 2870(s), 2794(s), 2613(m); (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1656(w), 1631(m,) 1572(m); (C–H) 1499(s), 1462(s), 1450(m), 1358(s), 1284(s), 1238(w); (BF4, C–O) 1198(m), 1086(vs), 1021(s), 981(s), 953(s); (im ring) 848(s), 805(m), 751(m), 665(w), 627(s) (vs: very strong; s: strong; m: medium; w: weak).
C) 1656(w), 1631(m,) 1572(m); (C–H) 1499(s), 1462(s), 1450(m), 1358(s), 1284(s), 1238(w); (BF4, C–O) 1198(m), 1086(vs), 1021(s), 981(s), 953(s); (im ring) 848(s), 805(m), 751(m), 665(w), 627(s) (vs: very strong; s: strong; m: medium; w: weak).
Preparation and characterization of [Ag(4-CH2OHimH)2]SO3CH3 (5)
AgSO3CH3 (1 mmol, 0.203 g) was dissolved in ca. 20 ml of ethanol at 70 °C. A solution of 4(5)-CH2OHimH (2 mmol, 0.196 g) in hot ethanol (ca. 10 ml) was added to the previous solution. The resulting mixture was stirred for 15 min at 70 °C. A small amount of a beige solid was filtered off and the solution was evaporated to one-third of the initial volume and left to crystallize. The white solid which precipitated the following day was washed with anhydrous diethyl ether.Yield: 0.252 g. MW = 399.22. Anal. calcd for C9H15N4O5SAg ([Ag(4-CH2OHimH)2]SO3CH3): C, 27.08; H, 3.79; N, 14.04%. Found: C, 27.01; H, 3.80; N, 13.98%. ESI-MS (H2O) m/z (relative intensity): 303.1 (100) [Ag(4-CH2OHimH)2]+, 205.0 (19) [Ag(4-CH2OHimH)]+. 1H NMR (600 MHz, D2O): δ 2.80 (s, 3H, CH3), 4.63 (s, 4H, 2 × CH2–O), 7.24 (s, 2H, 2 × H(5)), 7.92 (s, 2H, 2 × H(2)). 13C NMR (151 MHz, D2O): δ 138.1 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N), 137.1 (weak int.) (C5), 117.4 (C4) (weak), 56.3 (CH2–O), 38.6 (CH3). IR (KBr) νmax (cm−1): (O–H, C–H) 3118(s), 2932(s), 2870(m), 2794(m); (C
C–N), 137.1 (weak int.) (C5), 117.4 (C4) (weak), 56.3 (CH2–O), 38.6 (CH3). IR (KBr) νmax (cm−1): (O–H, C–H) 3118(s), 2932(s), 2870(m), 2794(m); (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C
N, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1631(m), 1572(m); (C–H) 1499(m), 1450 (m), 1357(s), 1284(s), 1239(w); (SO3) 1195(vs) (C–O) 1088(m), 1058(vs), 1021(s), 981(s); (im ring) 847(m), 785(s), 665(w), 627(s) (vs: very strong; s: strong; m: medium; w: weak).
C) 1631(m), 1572(m); (C–H) 1499(m), 1450 (m), 1357(s), 1284(s), 1239(w); (SO3) 1195(vs) (C–O) 1088(m), 1058(vs), 1021(s), 981(s); (im ring) 847(m), 785(s), 665(w), 627(s) (vs: very strong; s: strong; m: medium; w: weak).
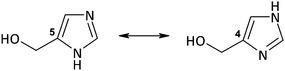
4(5)-CH2OHimH
(Given for comparative purposes). MW = 98.12. 1H NMR (600 MHz, D2O): δ 4.59 (s, 2H, CH2–O), 7.13 (s, 1H, H(4(5))), 7.73 (s, 1H, H(2)). 13C NMR (151 MHz, D2O): δ 136.9 (C4(5)) (weak int.), 136.3 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N), 117.3 (C5(4)), 56.1 (CH2–O).
C–N), 117.3 (C5(4)), 56.1 (CH2–O).
Light stability of the complexes 1–5 and salts S1–S5
Cotton pads were impregnated with 0.025 mol l−1 aqueous solutions of complexes 1–5 and their appropriate silver salts S1–S5 and then exposed to indirect light in air atmosphere at room temperature. The stability was monitored visually within 120 h (5 days). The photos of the samples are given in the ESI† (Fig. S1).X-ray crystal structure determination
The X-ray data for compounds 2–4 were collected from single crystals using an Agilent SuperNova diffractometer equipped with an Atlas detector at T = 100(2) K with monochromatic MoKα radiation (λ = 0.71073 Å). Multi-scan absorption correction was applied to all data.43 All structures were solved by direct methods using SHELX and further refined on F2 using SHELXL-2014/7.44 All non-hydrogen atoms were refined anisotropically.The fluorine atoms (F1, F2, and F3) in the CF3COO− anion in structure 3 were found to be disordered and as a consequence refined over two positions with a ratio of 0.56(2): 0.42(2). To the disordered components, appropriate restraints of geometry and displacement parameters using SADI and EADP commands were applied.
In structure 4, the hydroxymethyl group is disordered over two positions with a ratio of 0.731(4): 0.269(4).
In all structures, heteroaromatic NH and hydroxyl OH atoms were located on the Fourier difference map and refined with isotropic thermal parameters.
The position of remaining hydrogen atoms were calculated from the known geometry (C–H bond lengths at 0.95 and 0.99 Å for aromatic CH and methylene CH2 atoms, respectively) and treated as riding, where the isotropic thermal parameters of these hydrogen atoms were fixed as Uiso(H) = 1.2Ueq(C) for H atoms. However, atomic coordinates for H4A1, H4A2, H4B1, and H4B2 in the disordered hydroxymethyl group of structure 4 and hydroxyl H1A and H1B atoms were refined with C–H and O–H distances restrained to 0.99 and 0.84 Å, respectively. Basic experimental details and crystallographic data are presented in Table 1.
| 2 | 3 | 4 | |
|---|---|---|---|
| Empirical formula | C32H48N16O24Cl4Ag4 | C20H24N8O8F6Ag2 | C16H24N8O4B2F8Ag2 |
| Formula weight | 1614.14 | 834.21 | 781.79 |
| Crystal system | Triclinic | Monoclinic | Triclinic |
| Space group |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P21/c |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| a (Å) | 10.6600(2) | 7.7766(2) | 7.9688(3) |
| b (Å) | 10.7580(2) | 15.2479(3) | 9.2044(2) |
| c (Å) | 13.6855(3) | 11.6266(2) | 9.3306(3) |
| α (°) | 106.201(2) | 90.000 | 73.719(3) |
| β (°) | 95.799(2) | 97.599(2) | 76.471(3) |
| γ (°) | 119.067(2) | 90.000 | 74.445(2) |
| V (Å3) | 1263.90(6) | 1366.54(5) | 623.48(4) |
| Z | 1 | 2 | 1 |
| T (K) | 100(2) | 100(2) | 100(2) |
| F(000) | 800 | 824 | 384 |
| D x (g cm−3) | 2.121 | 2.027 | 2.082 |
| μ (mm−1) | 1.84 | 1.53 | 1.67 |
| Scan method | ω scan | ω scan | ω scan |
| θ range (°) | 2.2–30.0 | 2.2–27.0 | 2.3–30.0 |
| Measured reflections | 16910 | 15057 | 6974 |
| Unique reflections | 7321 | 2983 | 3626 |
| Observed reflections [I > 2σ(I)] | 7007 | 2950 | 3532 |
| Completeness to θmax (%) | 99.4 | 100 | 99.7 |
| No. of parameters/restraints | 394/0 | 219/16 | 211/18 |
| R [I > 2σ(I)] | 0.017 | 0.029 | 0.021 |
| wR (all data) | 0.046 | 0.067 | 0.050 |
| S | 1.10 | 1.14 | 1.05 |
| Largest diff. peak (e Å−3) | 0.61 | 1.97 | 0.69 |
| Largest diff. hole (e Å−3) | −0.51 | −0.36 | −0.94 |
Antimicrobial activity determination
The antibacterial and antifungal properties of the synthesized complexes, free ligand and silver sulfadiazine were evaluated against two Gram-positive strains: Staphylococcus aureus ATCC 6538 and Staphylococcus epidermidis ATCC 12228, three Gram-negative bacteria: Pseudomonas aeruginosa ATCC 15442, Escherichia coli ATCC 25922, and Proteus hauseri ATCC 13315 and yeast Candida albicans ATCC 10231. The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)/minimum fungicidal concentration (MFC) were determined by a modified broth microdilution method according to the recommendation of Clinical & Laboratory Standards Institute(M07-A8) for bacteria and Standards of European Committee on Antimicrobial Susceptibility Testing (EDef7.1.) for fermentative yeasts with modifications. The MIC and MBC values were expressed in μmol L−1.Results and discussion
Synthesis and structural characterization
Silver(I) complexes 1–5 containing the 4-(hydroxymethyl)-imidazole ligand were obtained by stirring and heating the ligand (2 moles) with the appropriate silver salts AgX (X = NO3−, ClO4−, CF3COO−, BF4− and SO3CH3−) (1 mol) in ethanol or water solution for a short time.Colourless products were isolated in ca. 70–80% yields. All products were soluble in water, methanol and hot ethanol. The novel complexes were characterized using spectroscopic methods including (1H and 13C) NMR and IR spectroscopy, ESI-MS spectrometry as well as X-ray diffraction analysis.
As shown in Scheme 1, the ligand 4(5)-CH2OHimH exists in two tautomeric forms. The form containing the hydroxymethyl group in position 4 is preferable in complexes 2–4, as unambiguously confirmed by X-ray determination. Although the ligand is able to form five-membered chelate rings with metal ions,45,46 it acts as a monodentate ligand coordinated through the nitrogen atom (N3) in complexes 2–4. The same way of coordination was observed in isostructural Cd(II)47 and Zn(II)48 complexes of 4-CH2OHimH and tri-tert-butoxysilanethiolate, in which the oxygen atom of the hydroxymethyl group also does not act as an electron donor and the hydroxyl group of 4(5)-CH2OHimH forms an intramolecular hydrogen bond with sulfur O–H⋯S. The coordination mode of a very similar ligand, 4(5)-CH2OH-5(4)-CH3imH, in the Co(II), Cu(II) and Ni(II) complexes, where the ligand presents a dualistic nature, appears to be somewhat different. Two of these ligands act as monodentate (N3) and two other as bidentate (O,N3).49,50
The 1H NMR spectra of complexes 1–5 and the free ligand are given in the ESI† as Fig. S2. The spectra are virtually identical as they differ from each other only by the counter-ions. In the spectrum of the free ligand 4(5)-CH2OHimH, three singlets at 7.73 ppm, 7.13 ppm and 4.59 ppm are observed and can be attributed to the respective proton signals H(2), H(4(5)) and CH2O. Although the ligand exists in tautomeric forms (Scheme 1), the presence of these two forms are not visible in the 1H NMR spectrum of the D2O solvent. The NH proton of the imidazole ring is not observed in the spectrum because it is exchanged for deuterium, the same as the hydroxyl proton. Accordingly, it is impossible to state which tautomeric form (4 or 5) exists in the complexes. Finally, the X-ray crystal structures of complexes 2–4 and a literature review47,48 indicate that tautomeric form 4 is also preferable in complexes 1 and 5. A comparison of the spectra of 1–5 with the spectrum of the free ligand revealed slight shifts of the signals to a higher field, ca. 0.24 ppm, 0.16 ppm and 0.08 ppm. The greatest shift is observed for the signal of H(2) proton located in the neighborhood of the coordination site (N3). Weak displacement of the signals of the Ag(I) complexes with respect to the free ligand seems to be specific to silver complexes in solution.51–53
The 13C NMR spectra of 1–5 and the free ligand are very similar. The signals of the carbons C(2) of 1–5 are shifted at ca. 1.7 ppm compared to the ligand. The signals of further carbon atoms are nearly unaffected, ca. 0.1–0.3 ppm. The spectrum of 5 contains an additional peak at 38.6 ppm attributed to the CH3SO3− group, and the spectrum of 3 contains two weak peaks at 162.9 ppm and 115.5 ppm assigned to the CF3COO− counter-ion.
Light stability study
As silver complexes 1–5 are potential antimicrobial agents for external application on infected skin in the form of creams, gels or impregnated wound dressings, the stability of complexes 1–5 when exposed to indirect light at room temperature was tested. Cotton pads were impregnated with 0.025 mol l−1 aqueous solutions of 1–5 and their appropriate silver salts S1–S5 for comparison purposes. The stability of the samples was monitored visually within 120 h (Fig. S1, ESI†).The treatment of infections with the silver nitrate salt has a negative side, namely its solutions make the skin and dressings dark. The light stability test on cotton pads will show whether solutions of 1–5 cause the cotton pads to blacken.
The results were generally positive, as complexes 1–5 had clearly better light stability than their appropriate silver salts S1–S5. Cotton pads treated with complexes 1–5 started to become gently beige only after 44 h. The reference silver salts S1–S5 caused the pads to become dark brown after that period. Salts S1–S5 induced greater darkening of the pads after 4 h of exposure than complexes 1–5 after 44 h. All the cotton pads impregnated by 1–5 appeared to be similar and were coloured grey-beige after 5 days of light exposure.
Crystal and molecular structures 2–4
| Bond lengths | |||||||
|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | |||||
| Symmetry codes: (2) (i) 1 − x, −y, 1 − z; (ii) 2 − x, 1 − y, −z; (viii) 2 − x, 2 − y, −z; (3) (i) 1 − x, 1 − y, 1 − z; (4) (i) 1 − x, 1 − y, 1 − z. | |||||||
| Ag1–N1 | 2.1024(11) | Ag2–N5 | 2.0959(10) | Ag1–N1 | 2.106(2) | Ag1–N1 | 2.0900(15) |
| Ag1–N3 | 2.0963(10) | Ag2–N7 | 2.0919(10) | Ag1–N3 | 2.108(2) | Ag1–N3 | 2.0938(16) |
| Ag1⋯O2i | 2.9091(11) | Ag2⋯O11ii | 3.0123(13) | Ag1⋯O2 | 3.089(2) | Ag1⋯O1Ai | 3.079(3) |
| Ag1–Ag1i | 3.2205(2) | Ag2⋯Ag2viii | 3.4282(3) | Ag1⋯O3i | 2.875(2) | Ag1⋯Ag1i | 3.2967(3) |
| Ag1–Ag1i | 3.2044(4) | ||||||
| Bond angles | |||||||
| N1–Ag1–N3 | 166.21(4) | N5–Ag2–N7 | 172.87(4) | N1–Ag1–N3 | 166.46(9) | N1–Ag1–N3 | 169.28(6) |
| Ag1i–Ag1–N1 | 101.29(3) | Ag1i–Ag1–N1 | 114.09(6) | Ag1i–Ag1–N1 | 80.69(4) | ||
| Ag1i–Ag1–N3 | 83.52(3) | Ag1i–Ag1–N3 | 73.21(6) | Ag1i–Ag1–N3 | 103.53(4) | ||
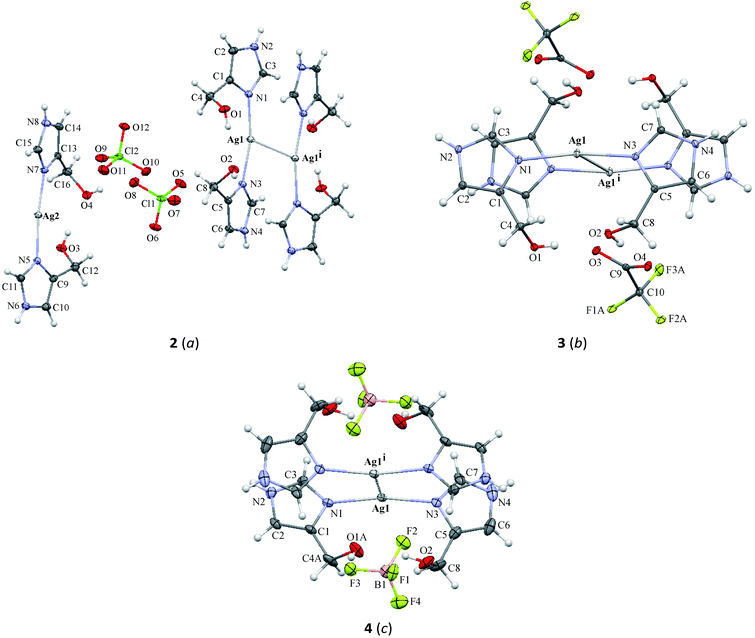
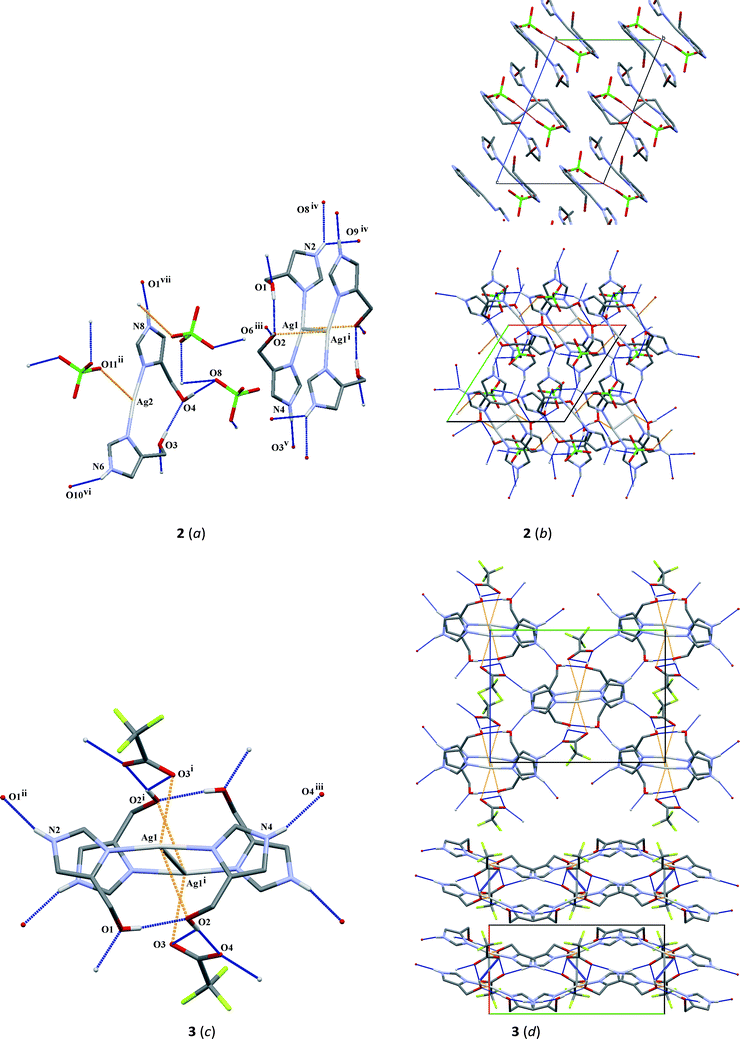
The crystal structure of complex 2 crystallizes in the P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group with a monomer, half of the centrosymmetric dimer and two ClO4− ions in the asymmetric unit. In the monomer, the Ag2 atom is coordinated approximately linearly by two nitrogen atoms (N5, N7) from imidazole rings with the N5–Ag2–N7 angle equal to 172.87(4)°. However, the coordination sphere is complemented by the short interaction between the Ag2 and O11ii atoms (symmetry code (ii) 2 − x, 1 − y, −z) (Fig. 2a and Table 2). In the crystal lattice, two Ag2 ions only weakly interact, the distance separating them being 3.4282(3) Å, which is on the border of the sum of the van der Waals radii for silver(I).
space group with a monomer, half of the centrosymmetric dimer and two ClO4− ions in the asymmetric unit. In the monomer, the Ag2 atom is coordinated approximately linearly by two nitrogen atoms (N5, N7) from imidazole rings with the N5–Ag2–N7 angle equal to 172.87(4)°. However, the coordination sphere is complemented by the short interaction between the Ag2 and O11ii atoms (symmetry code (ii) 2 − x, 1 − y, −z) (Fig. 2a and Table 2). In the crystal lattice, two Ag2 ions only weakly interact, the distance separating them being 3.4282(3) Å, which is on the border of the sum of the van der Waals radii for silver(I).
The centrosymmetric dimer includes two [Ag(L)2]+ (L = 4(5)-CH2OHimH) units with an Ag1–Ag1i short length of 3.2205(2) Å (symmetry code (i) 1 − x, −y, 1 − z). This is in agreement with other Ag–Ag lengths observed for silver complexes.24,42,54,55 Each Ag1 atom is coordinated by two imidazole nitrogens, N1 and N3, with the N1–Ag1–N3 angle of 166.21(3)°, which is smaller than in the monomer. The environment of the central ion is completed by a short Ag1⋯O2i (symmetry code (i) 1 − x, −y, 1 − z) (Fig. 2a and Table 2).
In the crystal packing of complex 2, oxygen atoms from hydroxymethyl groups act as donors in several O–H⋯O hydrogen bonds and together with N–H⋯O hydrogen bonds (Table 3) form a 3-D network. In addition, it is stabilized by π⋯π interactions between particular imidazole rings with the centroid separation distances ranging from 3.529(1) to 3.904(1) Å (Table 4). Interestingly, looking down the a-axis, complex 2 can be observed in a form of a layered structure in the crystal packing. Between layers created by π⋯π stacking interactions, ClO4− anions are located with short O6⋯O6 (2 − x, 2 − y, 1 − z) and O12⋯O12 (1 − x, −y, −z) interactions with corresponding distances: 2.841(2) and 2.897(2) Å, respectively (Fig. 2b).
| H-bond | D–H | H⋯A | D⋯A | ∠D–H⋯A |
|---|---|---|---|---|
| Symmetry codes: (2) (iii) x, y − 1, z; (iv) x − 1, y − 1, z; (v) 2 − x, 2 − y, 1 − z; (vi) 1 + x, 1 + y, z; (vii) 1 − x, −y, −z; (3) (ii) x, 3/2 − y, z − 1/2; (iii) x, 1/2 −y, z − 1/2; (4) (ii) x, y, 1 + z; (iii) x − 1, 1 + y, z; (iv) 1 − x, 1 − y, −z. | ||||
| 2 | ||||
| O1–H1A⋯O2 | 0.82(2) | 2.01(2) | 2.823(1) | 173(2) |
| O2–H2A⋯O6iii | 0.79(2) | 2.10(2) | 2.870(1) | 168(2) |
| N2–H20⋯O8iv | 0.87(2) | 2.47(2) | 2.179(2) | 140(2) |
| N2–H20⋯O9iv | 0.87(2) | 2.31(2) | 2.921(2) | 128(2) |
| N4–H40⋯O3v | 0.85(2) | 2.02(2) | 2.794(1) | 151(2) |
| O3–H3A⋯O4 | 0.76(3) | 2.00(3) | 2.755(1) | 169(3) |
| O4–H4A⋯O8 | 0.79(2) | 2.25(2) | 3.014(2) | 165(2) |
| N6–H60⋯O10vi | 0.83(2) | 2.15(2) | 2.935(1) | 157(2) |
| N8–H80⋯O1vii | 0.88(2) | 1.97(2) | 2.777(2) | 153(2) |
| 3 | ||||
| O1–H1A⋯O2 | 0.72(4) | 1.97(5) | 2.690(3) | 172(5) |
| O2–H2A⋯O3 | 0.81(4) | 2.67(4) | 3.222(3) | 127(4) |
| O2–H2A⋯O4 | 0.81(4) | 1.90(4) | 2.709(3) | 177(4) |
| N2–H20⋯O1ii | 0.84(2) | 1.91(3) | 2.733(3) | 166(4) |
| N4–H40⋯O4iii | 0.80(4) | 2.01(4) | 2.787(3) | 164(4) |
| 4 | ||||
| O2–H2A⋯O1A | 0.84 | 1.92 | 2.741(3) | 164 |
| O1A–H1A⋯F3 | 0.84 | 2.01 | 2.832(3) | 165 |
| N2–H20⋯O2ii | 0.80(3) | 2.05(3) | 2.776(3) | 152(3) |
| N4–H40⋯F4iii | 0.82(4) | 2.23(4) | 2.936(3) | 143(3) |
| N4–H40⋯F1iv | 0.82(4) | 2.45(4) | 2.994(2) | 124(3) |
| Cg(I)⋯Cg(J) | CgI⋯CgJ | CgI_(J) | CgJ_(I) | ∠α |
|---|---|---|---|---|
| Symmetry codes: (2) (i) 1 − x, −y, 1 − z; (iv) x − 1, y − 1, z; (viii) 2 − x, 2 − y, −z; (3) (i) 1 − x, 1 − y, 1 − z; (iv) 2 − x, 1 − y, 1 − z; (4) (i) 1 − x, 1 − y, 1 − z; (v) −x, 1 − y, 1 − z. | ||||
| 2 | ||||
| Cg1⋯Cg2i | 3.529(1) | −3.476(1) | −3.379(1) | 15.3(1) |
| Cg1⋯Cg3iv | 3.904(1) | −3.222(1) | 3.283(1) | 2.1(1) |
| Cg3⋯Cg4viii | 3.584(1) | 3.485(1) | 3.470(1) | 17.6(1) |
| 3 | ||||
| Cg1⋯Cg2i | 3.666(2) | −3.460(1) | −3.255(1) | 8.2(2) |
| Cg1⋯Cg2iv | 4.153(2) | 3.635(1) | 3.313(1) | 8.2(1) |
| 4 | ||||
| Cg1⋯Cg2i | 3.540(1) | 3.402(1) | 3.481(1) | 14.2(1) |
| Cg2⋯Cg2v | 4.234(1) | −3.283(1) | −3.283(1) | 0.0(1) |
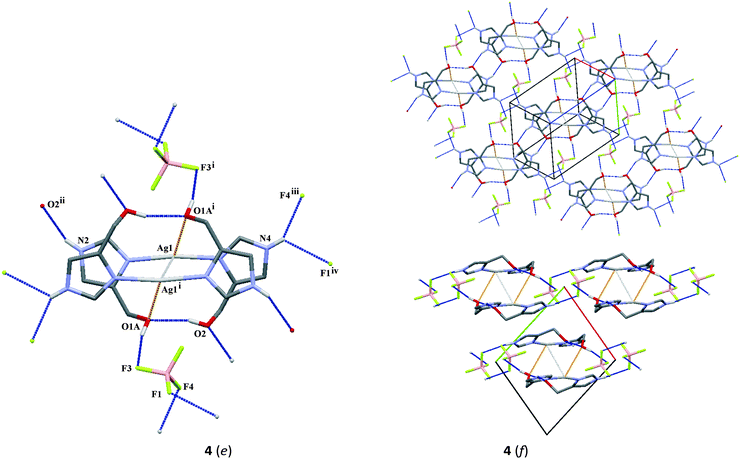
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) ) crystal structure of complex 4 consists of the discrete centrosymmetric dimeric cation [Ag2(L)4]2+ consisting of two[Ag(L)2]+ units, where Ag1⋯Ag1i (symmetry code (i) 1 − x, 1 − y, 1 − z) interaction is formed with a distance of 3.2967(3) Å and two BF4− anions. The environment around every silver atom is defined by two nitrogen atoms (N1 and N3) of imidazole ligands with a nearly linear coordination. The N1–Ag1–N3 angle is 169.28(6)° being between analogous angles in 2 and 3. The silver donor bond lengths are similar to those in 2 and slightly shorter than corresponding bonds in 3. The coordination sphere around the silver atom is completed by the oxygen atom O1Ai (symmetry code (i) 1 − x, 1 − y, 1 − z) from the hydroxymethyl group.
) crystal structure of complex 4 consists of the discrete centrosymmetric dimeric cation [Ag2(L)4]2+ consisting of two[Ag(L)2]+ units, where Ag1⋯Ag1i (symmetry code (i) 1 − x, 1 − y, 1 − z) interaction is formed with a distance of 3.2967(3) Å and two BF4− anions. The environment around every silver atom is defined by two nitrogen atoms (N1 and N3) of imidazole ligands with a nearly linear coordination. The N1–Ag1–N3 angle is 169.28(6)° being between analogous angles in 2 and 3. The silver donor bond lengths are similar to those in 2 and slightly shorter than corresponding bonds in 3. The coordination sphere around the silver atom is completed by the oxygen atom O1Ai (symmetry code (i) 1 − x, 1 − y, 1 − z) from the hydroxymethyl group.
In the case of the crystal packing of 4, the molecules are linked by a combination of N–H⋯O/F and Ag⋯O interactions (Fig. 2f and Table 3). The formation of the molecular framework can be described as 2-D sheets perpendicular to the [110] direction, consisting of double ribbons which are formed by N2–H20⋯O2ii (symmetry code (ii) x, y, 1 + z) hydrogen bonds and Ag1⋯O1Ai (symmetry code (i) 1 − x, 1 − y, 1 − z) interaction (lower Fig. 2f). Neighboring ribbons are linked by π⋯π interactions with separation distances between centroids of 3.540(1) and 4.234(1) Å, respectively, for Cg1⋯Cg2 (1 − x, 1 − y, 1 − z) and Cg2⋯Cg2 (−x, 1 − y, 1 − z;). In the crystal packing, further O2–H2A⋯O1A intramolecular interaction as well as other O1A–H1A⋯F3, N4–H40⋯F4iii (symmetry code (iii) x − 1, 1 + y, z) and N4–H40⋯F1iv (symmetry code (iv) 1 − x, 1 − y, −z) intermolecular interactions are observed.
Antimicrobial activity
Newly synthesized silver(I) complexes with 4-(hydroxymethyl)-imidazole 1–5 were assessed for antimicrobial activity against six strains of microorganisms: Staphylococcus aureus, Staphylococcus epidermidis, Escherichia coli, Pseudomonas aeruginosa, Proteus hauseri and Candida albicans. Their antimicrobial properties were compared with the biological activity of the free ligand 4(5)-(hydroxymethyl)imidazole and silver sulfadiazine (AgSD), the most widely used silver drug. The studies revealed that the tested imidazole ligand lack antibacterial and antifungal activities at concentrations up to 500 mg L−1. The obtained MIC (minimum inhibitory concentration) (Fig. 3) and MBC (minimum bactericidal concentration) (Table 5) values indicated that complexes 1–5 exhibit excellent activity against tested Gram-positive bacteria (S. aureus and S. epidermidis). All the complexes inhibited the growth of Gram-positive bacteria at concentrations much lower than those required by silver sulfadiazine (AgSD). Among them, the highest antibacterial properties were expressed by complex 3, containing trifluoroacetate as a counter-ion. The complex inhibited the growth of S. aureus and S. epidermidis at a concentration 2 to 3-fold lower than that established for AgSD. Moreover, all the tested silver compounds showed better bactericidal activity (Table 5) against Gram-positive strains than reference drug silver sulfadiazine. In the case of S. aureus, the MBC values of complex 3 and AgSD were 71 μM and 252 μM, respectively.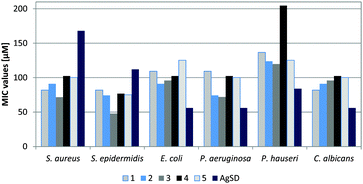 | ||
| Fig. 3 Antibacterial activity of silver(I) complexes containing 4(5)-(hydroxymethyl)imidazole, given as MIC (minimum inhibitory concentration). The MIC values of the free ligand are >500 mg L−1. | ||
| Tested compound | MBC [μM] | |||||
|---|---|---|---|---|---|---|
| S. aureus ATCC 6538 | S. epidermidis ATCC 12228 | E. coli ATCC 25922 | P. aeruginosa ATCC 15442 | P. hauseri ATCC 13315 | C. albicans ATCC 10231 | |
| 1 | 109 | 109 | 164 | 137 | 219 | >273 |
| 2 | 91 | 124 | 91 | 124 | 173 | >248 |
| 3 | 71 | 96 | 120 | 72 | 144 | >240 |
| 4 | 153 | 153 | 179 | 102 | 230 | >256 |
| 5 | 200 | 150 | 175 | 125 | 175 | >250 |
| AgSD | 252 | 224 | 84 | 56 | 84 | 56 |
The antimicrobial properties of complexes 1–5 towards Gram-negative bacteria and yeasts are insignificant. Their activity are about two times lower than commercially available AgSD.
The obtained results are in agreement with our previous studies,42 which indicated that silver(I) complexes of metronidazole displayed better antibacterial activity against Gram-positive bacteria than Gram-negative strains and yeasts.
Although silver(I) complexes with N-heterocyclic carbenes, described in the Introduction part,28–41 have completely different modes of coordination than complexes 1–5, both the groups show moderate to excellent antimicrobial activity. The MIC values of Ag–NHCs range from 0.25 to 6 μg mL−1 for the best ones,56 but usually from 25 to 200 μg mL1, depending on the tested strains. The activity of complexes 1–5 is similar and amounts to ca. 30–40, when expressed in μg mL−1. Overall, Ag–NHCs are effective against highly resistant bacteria strains, such as MRSA, P. aeruginosa, E. coli or B. subtilis.
Silver(I) complexes with imidazole or benzimidazole moieties are known to be effective antimicrobial drugs.57,58 Silver(I) complexes of 2-hydroxymethyl-N-alkylimidazoles displayed meaningful antimicrobial activity against E. coli and B. spizizenii depending on the alkyl chain length.24 Further study showed that the incorporation of the above mentioned ligands and their Ag(I) complexes into electrospun nylon 6 nanofibers produces attractive antimicrobial materials.25
Rowan et al.20 investigated the growth of inhibitory effects of silver(I) complexes containing imidazole derivatives against the pathogenic bacteria MRSA and E. coli and the fungal pathogen C. albicans. Amongst the five investigated Ag(I)-imidazole (imH) complexes, two of them, i.e. [Ag2(imH)4](salH)2 (salH2 = salicylic acid) and [Ag(MeNO2imH)2]ClO4, exhibited significant bacterial effects which were better or comparable of those of AgSD. All of them also showed excellent activity against C. albicans, especially the complex [Ag2(imH)4](salH)2, which was 47 times more active than ketoconazole.
Similarly, silver(I) complexes of 9-anthracenecarboxilic acid and N-substituted imidazoles demonstrated very high antifungal activity about 10 times better than the activity of ketoconazole, against C. albicans. These compounds also possess meaningful antibacterial activity toward E. coli and MRSA.21
Other Ag(I) complexes containing substituted imidazole ligands, i.e. bis(imidazole-2-yl)methane and its derivatives22 and (imidazol-1-yl)-2-phenylpropenenitrile,23 displayed only moderate activity against C. albicans.22
The antimicrobial activity of Ag(I) histidines (proteinogenic amino acids containing an imidazole functional group) against selected bacteria, yeasts and molds was compared of that of AgNO3, which was effective only against two Gram-negative bacteria. Most of the tested silver(I)-histidines showed a wider spectrum of activity than AgNO3.26,27 Significantly, all of the mentioned metal-free imidazole ligands were found to be inactive against the considered pathogens.
Conclusions
Silver(I) complexes containing the 4(5)-(hydroxymethyl)imidazole ligand (L) and various counter-ions were successfully synthesized and characterized by elemental, ESI-MS, NMR and IR analyses. X-ray crystal structures of complexes 2–4 were also determined. Complex 2 consists of a monomeric unit [Ag(L)2]ClO4 and a dimeric unit [Ag2(L)4](ClO4)2, while complexes 3 and 4 consist only of dimeric units [Ag2(L)4](CF3COO)2 (3) and [Ag2(L)4](BF4)2 (4), respectively. The crystal packing of 2 is dominated by a 3-D network of hydrogen bonds. The characteristic feature of molecular arrangements of 3 and 4 is 2-D sheets formed by hydrogen bonds and Ag⋯O interactions.The study on the antimicrobial activity of a series of novel silver(I) complexes of 4(5)-CH2OHimH and selected counter-ions, i.e. NO3−, ClO4−, CF3COO−, BF4− and CH3SO3−, was done. We evaluated the impact of particular counter-ions on the biological properties of the tested complexes. The results suggested that the variation of the counter-ions of the complexes affected their antimicrobial activity. The complex containing CF3COO− as a counter-ion had the highest activity against Gram-positive bacteria. It differs from our previous research42 in which we studied silver complexes containing metronidazole (MTZ) and various counter-ions. These data showed that the CH3SO3− counter-ion contributed to the highest antimicrobial activity against Gram-positive bacteria and yeasts. However, both series of the silver complexes, viz. containing 4(5)-CH2OHimH and MTZ ligands, displayed better activity against Gram-positive bacteria than Gram-negative strains and yeasts. It is probably associated with the differences in the structure of the cell wall and membrane of particular microorganisms.
Newly synthesized silver complexes exhibit significant antibacterial properties against Gram-positive bacteria, better than commercially available silver sulfadiazine. In future, they may represent attractive alternatives for existing drugs. In addition, all complexes are water soluble which is a desirable feature of potential antimicrobial drugs.
Acknowledgements
This work was supported by the Medical University of Łódź (Statute Fund No. 503/3-016-02/503-31-001) and by National Science Centre, Poland, No. UMO-2014/15/B/NZ7/00944.Notes and references
- The evolving threat of antimicrobial resistance- options for action, WHO Patient Safety Programme, World Health Organization, 2012.
- M. H. Miceli, J. A. Diaz and S. A. Lee, Lancet Infect. Dis., 2011, 11, 142–151 CrossRef PubMed.
- A. B. G. Lansdown, Silver in Healthcare: Its Antimicrobial Efficacy and Safety in Use, RCS Publishing, UK, 2010 Search PubMed.
- S. Silver, FEMS Microbiol. Rev., 2003, 27, 341–353 CrossRef CAS PubMed.
- I. Chopra, J. Antimicrob. Chemother., 2007, 59, 587–590 CrossRef CAS PubMed.
- A. B. G. Landsdown, Crit. Rev. Toxicol., 2007, 37, 237–250 CrossRef PubMed.
- G. G. de Gracia, Burns, 2001, 27, 67–74 CrossRef PubMed.
- R. J. White and R. Cooper, Wounds, UK, 2005, vol. 1, pp. 51–61 Search PubMed.
- F. W. Fuller, J. Burn Care Res., 2009, 30, 464–470 CrossRef PubMed.
- G. Sandri, M. C. Bonferoni, F. Ferrari, S. Rossi, C. Aguzzi, M. Mori, P. Grisoli, P. Cerezo, M. Tenci, C. Viseras and C. Caramella, Carbohydr. Polym., 2014, 102, 970–977 CrossRef CAS PubMed.
- M. L. Cooper, S. T. Boyce, J. F. Hansbrough, T. J. Foreman and D. H. Frank, J. Surg. Res., 1990, 48, 190–195 CrossRef CAS PubMed.
- M. L. Cooper, J. A. Laxer and J. F. Hansbrough, J. Trauma, 1991, 31, 775–782 CrossRef CAS PubMed.
- A.-M. Leuck, J. R. Johnson, M. A. Hunt, K. Dhody, K. Kazempour, P. Ferrieri and S. Kline, Am. J. Infect. Control, 2015, 43, 260–265 CrossRef PubMed.
- Y.-M. Chena, A.-P. Daib, Y. Shic, Z.-J. Liua, M.-F. Gongc and X.-B. Yin, Int. J. Infect. Dis., 2014, 29, 279–286 CrossRef PubMed.
- R. Thomas, A. P. Nair, K. R. Soumya, J. Mathew and E. K. Radhakrishnan, Appl. Biochem. Biotechnol., 2014, 173, 449–460 CrossRef CAS PubMed.
- J. H. Kim, A. R. Unnithan, H. J. Kim, A. P. Tiwari, C. H. Park and C. S. Kim, J. Ind. Eng. Chem., 2015, 30, 254–260 CrossRef CAS.
- B. Sadek, Pharma Chem., 2011, 3, 410–419 CAS.
- D. Sharma, B. Narasimhan, P. Kumar, V. Judge, R. Narang, E. De Clercq and J. Balzarini, Eur. J. Med. Chem., 2009, 44, 2347–2353 CrossRef CAS PubMed.
- B. F. Abdel-Wahab, G. E. A. Awad and F. A. Badria, Eur. J. Med. Chem., 2011, 46, 1505–1511 CrossRef CAS PubMed.
- R. Rowan, T. Tallon, A. M. Sheahan, R. Curran, M. McCann, K. Kavanagh, M. Devereux and V. McKee, Polyhedron, 2006, 25, 1771–1778 CrossRef CAS.
- M. McCann, R. Curran, M. Ben-Shoshan, V. McKee, A. A. Tahir, M. Devereux, K. Kavanagh, B. S. Creaven and A. Kellett, Dalton Trans., 2012, 41, 6516–6527 RSC.
- S. Abuskhuna, J. Briody, M. McCann, M. Devereux, K. Kavanagh, J. B. Fontecha and V. McKee, Polyhedron, 2004, 25, 1249–1255 CrossRef.
- M. McCann, B. Coyle, J. Briody, F. Bass, N. O'Gorman, M. Devereux, K. Kavanagh and V. McKee, Polyhedron, 2003, 22, 1595–1601 CrossRef CAS.
- P. Kleyi, R. S. Walmsley, M. A. Fernandes, N. Torto and Z. R. Tshentu, Polyhedron, 2012, 41, 25–29 CrossRef CAS.
- P. Kleyi, C. L. Frost, Z. R. Tshentu and N. Torto, J. Appl. Polym. Sci., 2014, 131, 39783–39793 CrossRef.
- N. C. Kasuga, Y. Takagi, S. Tsuruta, W. Kuwana, R. Yoshikawa and K. Nomiya, Inorg. Chim. Acta, 2011, 368, 44–48 CrossRef CAS.
- K. Nomiya, S. Takahashi, R. Noguchi, S. Nemoto, T. Takayama and M. Oda, Inorg. Chem., 2000, 39, 3301–3311 CrossRef CAS PubMed.
- W. J. Youngs, A. R. Knapp, P. O. Wagers and C. A. Tessier, Dalton Trans., 2012, 41, 327–336 RSC.
- F. Hackenberg and M. Tacke, Dalton Trans., 2014, 43, 8144–8153 RSC.
- M. Tacke, J. Organomet. Chem., 2015, 782, 17–21 CrossRef CAS.
- R. A. Haque, P. O. Asekunowo and S. Budagumpi, Inorg. Chem. Commun., 2014, 47, 56–59 CrossRef CAS.
- P. O. Asekunowo, R. A. Haque, M. R. Razali and S. Budagumpi, Appl. Organomet. Chem., 2015, 29, 126–137 CrossRef CAS.
- R. A. Haque, P. O. Asekunowo and M. R. Razali, Transition Met. Chem., 2014, 39, 281–290 CrossRef CAS.
- J. C. Garrison and W. J. Youngs, Chem. Rev., 2005, 3978–4008 CrossRef CAS PubMed.
- A. Kascatan-Nebioglu, A. Melaiye, K. Hindi, S. Durmus, M. J. Panzner, L. A. Hogue, R. J. Mallett, C. E. Hovis, M. Coughenour, S. D. Crosby, A. Milsted, D. L. Ely, C. A. Tessier, C. L. Cannon and W. J. Youngs, J. Med. Chem., 2006, 49, 6811–6818 CrossRef CAS PubMed.
- A. Kascatan-Nebioglu, M. J. Panzner, C. A. Tessier, C. L. Cannon and W. J. Youngs, Coord. Chem. Rev., 2007, 251, 884–895 CrossRef CAS.
- K. M. Hindi, T. J. Siciliano, S. Durmus, M. J. Panzner, D. A. Medvetz, D. V. Reddy, L. A. Hogue, C. E. Hovis, J. K. Hilliard, R. J. Mallet, C. A. Tessier, C. L. Cannon and W. J. Youngs, J. Med. Chem., 2008, 51, 1577 CrossRef CAS PubMed.
- M. J. Panzner, A. Deeraksa, A. Smith, B. D. Wright, K. M. Hindi, A. Kascatan-Nebioglu, A. G. Torres, B. M. Judy, C. E. Hovis, J. K. Hillard, R. J. Mallett, E. Cope, D. M. Estes, C. L. Cannon, J. G. Leid and W. J. Youngs, Eur. J. Inorg. Chem., 2009, 1739 CrossRef CAS PubMed.
- F. Hackenberg, G. Lally, H. Müller-Bunz, F. Paradisi, D. Quaglia, W. Streciwilk and M. Tacke, Inorg. Chim. Acta, 2013, 395, 135–144 CrossRef CAS.
- W. Streciwilk, J. Cassidy, F. Hackenberg, H. Müller-Bunz, F. Paradisi and M. Tacke, J. Organomet. Chem., 2014, 749, 88–99 CrossRef CAS.
- R. A. Haque, S. Y. Choo, S. Budagumpi, A. A.-A. Abdullah, M. B. K. Ahamed and A. M. S. A. Majid, Inorg. Chim. Acta, 2015, 433, 35–44 CrossRef CAS.
- U. Kalinowska-Lis, A. Felczak, L. Chęcińska, K. Zawadzka, E. Patyna, K. Lisowska and J. Ochocki, Dalton Trans., 2015, 44, 8178–8189 RSC.
- CrysAlis PRO, Agilent Technologies, Yarnton, Oxfordshire, England, 2013 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 CrossRef PubMed.
- B. Barszcz, Coord. Chem. Rev., 2005, 249, 2259–2276 CrossRef CAS.
- K. Kurdziel and T. Głowiak, J. Coord. Chem., 2007, 60, 877–889 CrossRef CAS.
- A. Dołga, K. Baranowska and Z. Jarząbek, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2008, 64, m1515 Search PubMed.
- A. Dołga, K. Baranowska, A. Pładzyk and K. Majcher, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2008, 64, m259–m263 Search PubMed.
- K. Kurdziel, T. Głowiak and J. Jezierska, Polyhedron, 2002, 21, 1857–1863 CrossRef CAS.
- B. Barszcz, T. Głowiak, J. Jezierska and K. Kurdziel, Inorg. Chem. Commun., 2002, 5, 1056–1058 CrossRef CAS.
- W. Liu, K. Bensdorf, A. Hagenbach, U. Abram, B. Niu, A. Mariappan and R. Gust, Eur. J. Med. Chem., 2011, 46, 5927–5934 CrossRef CAS PubMed.
- N. C. Kasuga, A. Sugie and K. Nomiya, Dalton Trans., 2004, 3732–3740 RSC.
- A. A. Massoud, Y. M. Gohar, V. Langer, P. Lincoln, F. R. Svensson, J. Jänis, S. T. Gårdebjer, M. Haukka, F. Jonsson, E. Aneheim, P. Löwenhielm, M. A. M. Abu-Youssef and L. R. Öhrström, New J. Chem., 2011, 35, 640 RSC.
- H. Wu, X.-W. Dong and J.-F. Ma, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2006, 62, m1227 CAS.
- P. Ovejero, E. Asensio, J. V. Heras, J. A. Campo, M. Cano, M. R. Torres, C. Núnez and C. Lodeiro, Dalton Trans., 2013, 42, 2107 RSC.
- B. D. Wright, P. N. Shah, L. J. McDonald, M. L. Shaeffer, P. O. Wagers, M. J. Panzner, J. Smolen, J. Tagaev, C. A. Tessier, C. L. Cannon and W. J. Youngs, Dalton Trans., 2012, 41, 6500–6506 RSC.
- U. Kalinowska-Lis, E. M. Szewczyk, L. Chęcińska, J. M. Wojciechowski, W. M. Wolf and J. Ochocki, ChemMedChem, 2014, 9, 169–176 CrossRef CAS PubMed.
- A. J. K. Atia, Molecules, 2009, 14, 2431–2446 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1411662 (2), 1411663 (3) and 1411664 (4). For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c5nj02514a |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2016 |